Abstract
Objective
The relationship between diseases and their causative genes can be complex, especially in the case of polygenic diseases. Further exacerbating the challenges in their study is that many genes may be causally related to multiple diseases. This study explored the relationship between diseases through the adaptation of an approach pioneered in the context of information retrieval: vector space models.
Materials and Methods
A vector space model approach was developed that bridges gene disease knowledge inferred across three knowledge bases: Online Mendelian Inheritance in Man, GenBank, and Medline. The approach was then used to identify potentially related diseases for two target diseases: Alzheimer disease and Prader-Willi Syndrome.
Results
In the case of both Alzheimer Disease and Prader-Willi Syndrome, a set of plausible diseases were identified that may warrant further exploration.
Discussion
This study furthers seminal work by Swanson, et al. that demonstrated the potential for mining literature for putative correlations. Using a vector space modeling approach, information from both biomedical literature and genomic resources (like GenBank) can be combined towards identification of putative correlations of interest. To this end, the relevance of the predicted diseases of interest in this study using the vector space modeling approach were validated based on supporting literature.
Conclusion
The results of this study suggest that a vector space model approach may be a useful means to identify potential relationships between complex diseases, and thereby enable the coordination of gene-based findings across multiple complex diseases.
Keywords: Translational bioinformatics, vector space modeling, information retrieval, complex diseases, knowledge representation, bioinformatics, phylogenetics
Introduction
The manifestation of genetic diseases is inherently linked to a causative network of genes. A given gene may be involved in the etiology of multiple, symptomatically related diseases. Thus, ascertaining the relationship between a network of genes and possibly related diseases can be a complex endeavor.1 Clinical interventions that are proven effective for a given disease may shed light on the treatment of related diseases. The field of medical genetics has long been challenged with developing approaches for determining how diseases relate to one another according to causative genes.2 Especially complicating the elucidation of relationships between causative genes and diseases is the fact that many diseases can be causally linked to a single gene, and multiple genes can be related to a single disease.3
Previous work has demonstrated the potential to link genes to disease phenotypes using similarity networks. For example, clustering techniques (similar to those used for gene expression analysis) have been shown to organize phenotypic information associated with genetic diseases.4 Additional studies have focused on studying common molecular pathways in complex diseases to identify potential genes of interest.5 More recently, relatedness between complex diseases has been examined using graph theoretic approaches.6 These methods have all depended on information that could be inferred from disease catalogs (eg, related disease entries in Online Mendelian Inheritance in Man (OMIM)) or archives of curated resources such as molecular pathway databases, such as the Kyoto Encyclopedia of Genes and the Genomes Pathway Database.
Vector space model approaches have been shown to be effective in the context of information retrieval systems.7 8 Most notably, the SMART system has been demonstrated to effectively enable reliable information retrieval compared with experts,9 including within the biomedical domain.10 Briefly, such approaches consider the degree of similarity according to specified weights, such as wqi and wij, respectively corresponding to vectors, such as q and dj, where q represents the ‘query’ and dj the resulting document set (j represents a given document). The similarity (sim) between vectors can be calculated using a similarity metric such as the cosine (θ):
Therefore, for t-dimensional vectors, the cosine similarity measure between a query q and given document dj is calculated as:
The vector space model approach is thus a clustering approach for identifying related vectors according to a specified weighting scheme (as reflected by wqi and wij above). Accordingly, the precision and recall of the returned results will vary according to the chosen weighting scheme.7 In the context of document retrieval, perhaps the most commonly known such weighting scheme is tf×idf (term frequency × inverse document frequency).11
The basic local alignment search tool (BLAST+)12 is a commonly used approach to determine the similarity between molecular sequences, and is often used to identify molecular sequences of interest from publicly accessible repositories (eg, GenBank13). Similarity in BLAST+ is reported as an ‘E-value,’ which reflects the expectation that a given result would be recovered by chance (ie, lower E-values indicate greater support for a given result, with 0.0 reflecting a result with high confidence).
GenBank is part of the International Nucleotide Sequence Data Consortium (along with EMBL and DDBJ) and provides access to the sum of the world's molecular sequence data (totaling over 140 million records). GenBank is part of the Entrez system, maintained at the National Library of Medicine's National Center for Biotechnology Information, and is thus linked to a wide array of resources, including Medline. Medline reflects the largest publicly available citation database of biomedical literature (∼20 million records). Previous work has demonstrated the feasibility and utility of linking GenBank to Medline records, mostly focusing on exploratory information retrieval systems.14 15
The present study develops a vector space model approach to identify potentially related diseases based on sequence similarity. GenBank–Medline linkages were used to infer diseases (as annotated according to medical subject heading (MeSH) descriptors) that may be associated with a given gene associated with a disease cataloged in OMIM records. The utility of the proposed vector space model approach was demonstrated for two etiologically different diseases with a genetic basis: Alzheimer's disease and Prader-Willi syndrome. The top results of this feasibility study were then manually evaluated for each disease according to published literature.
Methods
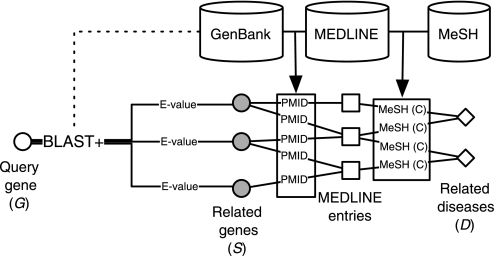
The goal of this study was to explore the potential of using a vector space model approach to identify potentially related genetic disorders. All scripts were written using the Ruby scripting language and made use of the BioRuby gem (an open source library of commonly used bioinformatics methods16) to leverage NCBI Entrez utilities. Local versions of the GenBank metadata and Medline databases, which were acquired through appropriate licensing agreements from the National Library of Medicine and parsed using a series of Ruby scripts and stored in a MySQL relational database, were accessed using the ‘mysql’ Ruby gem. The overall approach, shown in figure 1, was to retrieve related genes (S) for a given disease gene (G) using BLAST+. These retrieved related genes were then linked to potentially related diseases (D) on the basis of MeSH annotations of Medline records associated with the GenBank entries.
Figure 1.
Overall approach to identify related genes and associated diseases. For a given query gene (G), related genes (S) were retrieved using BLAST+. Related diseases (D) were then identified on the basis of MeSH annotations (C) for Medline citations associated with retrieved sequences.
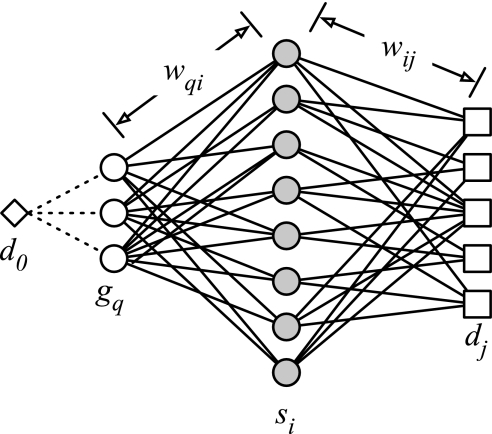
Within the scope of the present study, a vector space model approach was developed to identify related diseases (d; enumerated by j) based on sequence similarity to a set of related genes (s; enumerated by i) for a given set of genes (g; enumerated by q) known to be associated with a disease (d0). As shown in figure 2, the relationship between the vectors q and j is quantified using the respective weighting variables, wqi and wij.
Figure 2.
Overall approach to identify related diseases using a vector space model approach. The vector space model approach developed in this study aimed to link a query disease (d0) to a set of potentially related diseases (dj). The method uses two vectors: (1) a ‘gene vector’ (wqi), which is based on the number of related gene sequences (si) associated with genes (gq) that are directly linked with the query disease; and (2) a ‘disease vector’ (wij), which is a quantification of the relative relationships of diseases associated with the related gene sequences (si).
The OMIM resource was used to identify genes (via links to Entrez Gene records) known to be associated with a particular disease. For the purposes of this feasibility study, genes associated with two diseases were identified: (1) Alzheimer's disease (OMIM Record 104300): A2M, APOE, APP, PSEN1, and PSEN2; and (2) Prader-Willi syndrome (OMIM Record 176270): SNRPN and NDN. Nucleotide sequences were retrieved from GenBank using the following query: SYM[gene name] not ‘genome’ not ‘chromosome’, where ‘SYM’ was the gene symbol. Based on the nucleotide description (ie, as determined from the FASTA annotation), each retrieved sequence was manually verified to be the correct sequence associated for a given gene symbol. In sum, there were 14 A2M, 44 APOE, 275 APP, 22 PSEN1, and 17 PSEN2 sequences for Alzheimer's disease as well as 19 SNRPN and 11 NDN sequences for Prader-Willi syndrome retrieved and verified.
For each retrieved sequence, a Ruby script was used to mediate BLAST+ searches to identify similar sequences from GenBank. Based on the E-value similarity score (xqi), the individual weight (wqi) between a query sequence (gq) and GenBank sequence (si) was calculated as:
A ‘gene vector’ was then calculated for each retrieved gene relative to the query genes (t):
To determine the diseases associated with each related sequence, the local GenBank metadata database was queried for PubMed Identifiers (PMIDs). The PMIDs were then used to retrieve the associated MeSH descriptors, and filtered so that only MeSH descriptors in the ‘Disease [C]’ hierarchy were kept. The weight (wij) for each related sequence (si) and disease (dj) was then calculated as the ratio of the number of diseases associated with a given sequence (sd) and the total number of diseases retrieved (dj):
A ‘disease vector’ was then calculated for the retrieved genes that could be associated with at least one disease as inferred by MeSH annotations of Medline records:
Using a derivation of the previously described cosine similarity measure, the cosine similarity (simθ) between the set of query genes (G) and a given retrieved disease (dj) was then calculated using the gene vector and disease vector:
The overall similarity (sim[d0 →dj]) for a disease (dj) relative to a given disease (d0) was then calculated based on the number of genes (|G|) that linked the diseases to one another and the similarity score (simθ):
Medline searches were then performed to identify potentially supporting literature for each of the predicted related diseases for two etiologically different, yet with significant underlying genetic factors: Alzheimer's disease and Prader-Willi syndrome.
Results
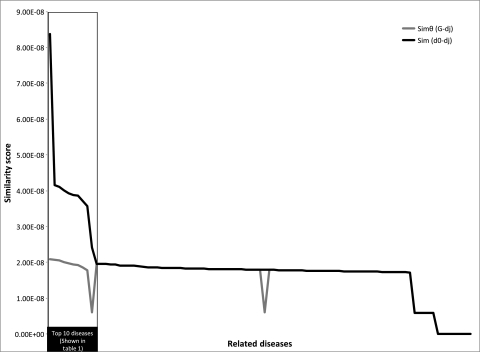
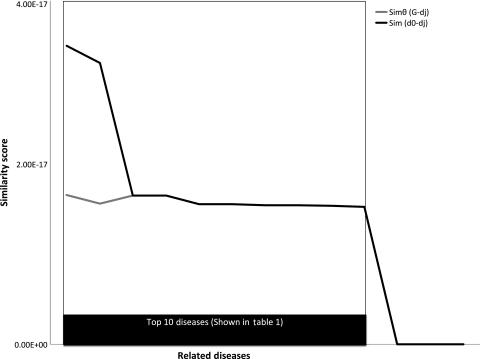
The vector space model approach was used to identify diseases related to the two chosen diseases for this study (Alzheimer's disease and Prader-Willi syndrome). The overall distribution of scores is respectively shown in figures 3 and 4 for Alzheimer's disease and Prader-Willi syndrome. The top 10 related diseases (according to the overall similarity score) identified along with the respective similarity scores for each disease are shown in table 1. Based on Medline literature searches, it was found that 90% of the returned results for Alzheimer's disease were potentially relevant; 80% of the returned results for Prader-Willi syndrome were potentially relevant. Based on Boolean searches for corroborating literature, there was direct support (ie, one or more articles with the MeSH descriptor for the query disease and the MeSH descriptor for the candidate disease) for all of the suggested related diseases except for ‘drug induced liver disease’ and ‘thyroid neoplasms’ for Prader-Willi syndrome.
Figure 3.
Similarity scores for Alzheimer's disease. The similarity score (y-axis) for each related disease (x-axis) is shown. Details for the top 10 diseases (highlighted in the box) are in table 1.
Figure 4.
Similarity scores for Prader-Willi syndrome. The similarity score (y-axis) for each related disease (x-axis) is shown. Details for the top 10 diseases (highlighted in the box) are in table 1.
Table 1.
Top 10 most related diseases, based on similarity score, to Alzheimer's disease and Prader-Willi syndrome based on the vector space model approach
| Related disease | Simθ (G →dj) | Sim (d0 →dj) | Corroborating Medline entries |
| Alzheimer's disease | |||
| Alzheimer's disease | 2.09E-08 | 8.38E-08 | 57 297 |
| Disease models, animal | 2.08E-08 | 4.16E-08 | 2718 |
| Polycystic kidney diseases | 2.06E-08 | 4.11E-08 | 2 |
| Down's syndrome | 2.01E-08 | 4.02E-08 | 1153 |
| Williams Syndrome | 1.97E-08 | 3.93E-08 | 2 |
| Neoplasms | 1.94E-08 | 3.87E-08 | 665 |
| Chromosome deletion | 1.93E-08 | 3.86E-08 | 10 |
| Polyploidy | 1.86E-08 | 3.72E-08 | 4 |
| Chromosome inversion | 1.79E-08 | 3.57E-08 | 2 |
| Melanoma | 6.03E-09 | 2.41E-08 | 15 |
| Prader-Willi syndrome | |||
| Genetic predisposition to disease | 1.74E-17 | 3.48E-17 | 2407 |
| Angelman syndrome | 1.64E-17 | 3.27E-17 | 478 |
| Drug-induced liver injury | 1.74E-17 | 1.74E-17 | 0 |
| Glioma | 1.73E-17 | 1.73E-17 | 4 |
| Neuroblastoma | 1.63E-17 | 1.63E-17 | 7 |
| Lupus erythematosus, systemic | 1.63E-17 | 1.63E-17 | 1 |
| Adrenal gland neoplasms | 1.62E-17 | 1.62E-17 | 1 |
| Thyroid neoplasms | 1.62E-17 | 1.62E-17 | 0 |
| Chromosome breakage | 1.61E-17 | 1.61E-17 | 26 |
| Melanoma | 1.60E-17 | 1.60E-17 | 7 |
Respective gene–disease and disease–disease similarity scores are shown, as well as the number of corroborating Medline entries (as determined by performing MeSH-based searches using a Boolean ‘AND’ to identify citations associated with both the related disease and the query disease). Literature searches were performed in August 2011.
For the scenario where related diseases were sought for Alzheimer's disease, five known associated genes were used as the query set. The most related disease was reported as Alzheimer's disease, which suggests that the algorithm is reliable in being able to recover the query disease itself. It also implies that the genes chosen are strongly affiliated with Alzheimer's disease and thus reflect genes of high relevance. The next high-ranking disease, polycystic kidney disease, shares the possibility of apoptotic processes having a role such as associated with Alzheimer's disease.17 Additionally, polymorphisms in the APOE gene are reported to have some association with both polycystic kidney diseases18 and Alzheimer's disease.19 The remaining related diseases return similar results when similar MeSH-based PubMed queries are carried out.
The second scenario, where the approach was used to search for related diseases to Prader-Willi syndrome, presented a slightly different set of results. The most relevant disease found using the vector space model approach developed here was Angelman syndrome, which is a known to involve the same chromosomal region as associated with Prader-Willi syndrome.20 While there is a genetic underpinning emerging for drug-induced liver injury,21 MeSH-based searches did not reveal any direct correlation with Prader-Willi syndrome. On the other hand, there is literature describing the role of SNRPN in the context of thyroid neoplasms22 (although not specific to the context of Prader-Willi syndrome).
Discussion
As there are many gene-to-disease and disease-to-gene relationships, the study of polygenetic diseases is indeed a complex endeavor. The exponential growth of molecular sequence data, made possible in large part by significant advances in sequencing technologies, provides a rich opportunity to explore the genomic space relative to complex diseases. The increase in availability of sequencing services also implies that there will be increased interest in studying an increasing range of diseases. Understanding the relationship between diseases may be of utility in identifying synergistic prophylaxes, diagnoses, or cures that could be more generally applied beyond single diseases. In the case of drug discovery, there has been increased interest in studying how drugs may be repurposed for use in the context of multiple diseases.23 24 On the other hand, although it is known that many genes can be associated with many diseases, there has been limited work in leveraging knowledge sources such as GenBank and Medline to recover some of these linkages.
Seminal work by Swanson demonstrated the potential to leverage secondary sources, such as biomedical literature, to identify possibly meaningful linkages within biomedicine.25 Initially shown to identify a previously undiscovered correlation (fish oil and Raynaud's syndrome26), subsequent development of the Arrowsmith system27 has shown the potential to validate epidemiological studies (eg, correlation between estrogen and Alzheimer's disease28). Literature mining approaches have also been shown to have potential for identifying potential gene candidates.29 30 The present study demonstrates an approach for enhancing literature-based information for putative relationship mining with an additional dimension of molecular sequence-based relationships. Future work would involve quantifying the impact of including molecular sequence information.
This study explored the potential to link potentially related complex diseases based on genetic relationships as determined by a vector space model approach. An important feature of the proposed vector space model is that, like many information retrieval paradigms (eg, tf×idf), it is specifically designed to account for signal-to-noise challenges. This is especially important in the light of the volume of potential data in exponentially growing databases such as GenBank. The possible relationships between diseases are based on the network of genes that relate to each other on the basis of sequence similarity (as determined using BLAST+). However, it is known that BLAST+ alone may not always recover the most related sequence,31 and sequence similarity may by itself not be a sufficient criterion of homology. Future work will thus incorporate more robust algorithmic techniques to identify potentially related sequences, such as those that leverage reciprocal sequence similarity (eg, COG,32 eggNOG,33 and InParanoid34) or phylogenetic relationships between potentially orthologous sequences (eg, OrthoDB,35 OrthologID,36 and Roundup37).
In this study, the relatedness between diseases, via this network of gene-based relationships, was determined using a cosine similarity metric. Of course, there are other metrics that might be worth exploring (eg, Dice38 or Jaccard39). Future work will include comparisons of the relative ranking of related diseases using such measures (eg, by comparing the relative precision and recall of retrieved results using different similarity metrics). Notably missing from the current feasibility study is the development of a statistical approach for quantifying the strength of the relationships; currently, the approach presents results as raw cosine scores. Akin to the E-value used by BLAST+ for assessing the confidence of results, a similar type of statistical metric will be needed for subsequent assessment of the results as the approach is used for additional diseases. In the absence of such a metric, it is difficult to compare the importance of suggested relationships between different diseases. Nonetheless, for the purposes of this feasibility study, potentially interesting relationships did emerge between suggested related diseases.
A gene's relationship to a particular disease is not necessarily exclusive (eg, APOE is indeed associated with many cases of Alzheimer's disease, but it is not universal). An advantage of the vector space model is that it accounts for not just one gene, but is specifically designed for finding related diseases based on multiple genes. In fact, the proposed vector space model would perform better at identifying potentially related diseases with more information about a broader list of genes that might be associated with a given disease. In this study, the genes of interest were manually selected on the basis of their descriptions and catalog information in OMIM; however, there may be additional resources to consider that would have additional genes of interest (eg, based on genome-wide association studies (GWAS), as cataloged at the NHGRI GWAS Catalog).
The two diseases used to explore the feasibility of the proposed methodology were chosen because of their dissimilarity, in terms of both their etiology and the number of cataloged genes referenced in OMIM. To this end, an important distinction between the disease searches was the number of query genes used: five for Alzheimer's disease versus two for Prader-Willi syndrome. As suggested by the results, more complex polygenic diseases may yield potentially more interesting results, with the complexity of the disease being a function of the number of genes potentially involved in the disease etiology. Further corroborating this notion is that there were 14 possibly related diseases returned for Prader-Willi syndrome, compared with 91 for Alzheimer's disease. Thus a limitation of the present study is that it only focused on the gene–disease vector space. There are plans to explore the search space of potentially related diseases as a measure of the number of genes involved in complex diseases, which will also include empirical measures of polymorphic rates within given genes (thus testing the hypothesis that complex disorders that are more difficult to study will have more potentially related genes).
The candidate diseases were based exclusively on MeSH descriptors associated with Medline-indexed articles. A significant challenge of this approach is that the MeSH descriptors used for Medline indexing may be too general to actually be meaningful to infer disease relationships. For this study, all MeSH descriptors in the ‘Disease [C]’ hierarchy were included. However, this resulted in a number of very generic disease concepts being associated with the query diseases. For example, in the case of Alzheimer's disease, ‘disease models, animal,’ ‘chromosome deletion,’ and ‘chromosome inversion’ were all returned as possibly related diseases. Although these may all be relevant concepts to consider in the light of Alzheimer's disease (and could be supported with Medline-indexed articles from a Boolean search), they may not necessarily be meaningful if one is seeking to identify related diseases. Similarly, restricting the possible valid MeSH descriptors to only those in the ‘Disease [C]’ hierarchy may also artificially restrict other potentially related concepts of interest (eg, concepts in other MeSH hierarchies, such as in ‘psychiatry and psychology [F]’). Future work will thus require a more controlled approach to determine which MeSH descriptors should be included in a query.
The number of corroborating Medline articles did not have a direct relationship with the relatedness between potentially related diseases. For example, in the case of polycystic kidney disease and Alzheimer's disease, very few (two) articles were found using MeSH-based Medline searches; however, using the vector space model approach described here, polycystic kidney disease ranked third most related. This type of finding may lead to future work that incorporates the number of MeSH-based results (or lack thereof) into an ‘interestingness’ score suggesting disease relationships that might warrant further investigation because of genetic evidence.
To date, much of the analysis carried out for gene-to-disease relationships has been based almost exclusively on data from curated resources, such as OMIM. Such studies depend on adequate levels of annotation and also on the correction of the putative relationships between diseases and the reported causative genes. This is undoubtedly a similar concern for the approach provided for the present study. However, where the present study does differ is leveraging the actual sequence information to explore the genetic relational space. Notably missing from this first vector space model approach are other factors that can have significant influence on genetic diseases (eg, epigenetic or environmental features). There has been significant work demonstrating the potential to study gene–environment features,40 which will form the basis for future work in adding a ‘gene–environment’ vector to the vector space model to complement the gene–disease and gene–gene vectors described in this study. The development of the gene–environment vector will be a significant endeavor, especially as currently available public datasets are generally not of the same depth as molecular sequence databases such as GenBank. Minimally, the gene–environment vector will need to incorporate relationships from resources such as the Genetic Association Database41 or within publicly available datasets in dbGAP42 that include environmental variables. Literature mining techniques may also be used to provide corroborating evidence to determine the strength of the relationships (eg, as cataloged by HuGEnet43). The resulting gene–environment vector could then be incorporated into the vector space model, thus enabling environmental variables to be incorporated to infer relationships between diseases (in addition to the gene-based relationships demonstrated in this feasibility study). A possible validation may be the ability to recover disease–disease relationships that have known common environmental relationships (eg, smoking and its influence on atherosclerosis and lung cancer).
Through the use of the vector space model approach, this study has demonstrated the potential to identify and rank relationships between diseases. Accordingly, it presents a mechanism for hybridizing information from predominantly genomic resources (eg, GenBank) and exploiting explicit linkages to literature-based knowledge (eg, as reported in Medline).
Conclusion
Vector space model approaches have been used predominantly in the context of information retrieval paradigms. Here, an adaptation of a vector space model approach is presented that enables the incorporation of sequence-based information. In the context of complex diseases, this study shows how the proposed approach could be used to identify potentially related diseases based on relationships to genes. The promising results of this feasibility study suggest a potentially powerful method for exploring the complex landscape of polygenetic diseases.
Footnotes
Funding: INS is supported in part by National Institutes of Health grant R01LM009725.
Competing interests: None.
Contributors: This study was conceived of, performed by, and written about by INS.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science 2002;298:2345–9 [DOI] [PubMed] [Google Scholar]
- 2.McKusick VA. On lumpers and splitters, or the nosology of genetic disease. Perspect Biol Med 1969;12:298–312 [DOI] [PubMed] [Google Scholar]
- 3.Biesecker LG. Lumping and splitting: molecular biology in the genetics clinic. Clin Genet 1998;53:3–7 [DOI] [PubMed] [Google Scholar]
- 4.Cantor MN, Lussier YA. Mining OMIM for insight into complex diseases. Stud Health Technol Inform 2004;107:753–7 [PMC free article] [PubMed] [Google Scholar]
- 5.Franke L, van Bakel H, Fokkens L, et al. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet 2006;78:1011–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barabási AL. Network medicine: from obesity to the “diseasome”. N Engl J Med 2007;357:404–7 [DOI] [PubMed] [Google Scholar]
- 7.Salton G, Buckley C. Term-weighting approaches in automatic text retrieval. Inform Process Manag 1988;24:513–23 [Google Scholar]
- 8.Salton G, Lesk M. Computer evaluation of indexing and text processing. J ACM 1968;15:8–36 [Google Scholar]
- 9.Salton G. The SMART Retrieval System; Experiments in Automatic Document Processing. Englewood Cliffs, NJ: Prentice-Hall, 1971 [Google Scholar]
- 10.Salton G. A new comparison between conventional indexing (MEDLARS) and automatic text processing (SMART). J Am Soc Inf Sci 1972;23:75–84 [Google Scholar]
- 11.Salton G, Yang C. On the specification of term values in automatic indexing. J Doc 1973;28:11–21 [Google Scholar]
- 12.Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson DA, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res 2011;39(Database issue):D32–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller H, Norton CN, Sarkar IN. GenBank and PubMed: how connected are they? BMC Res Notes 2009;2:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar IN, Schenk R, Miller H, et al. LigerCat: using “MeSH Clouds” from journal, article, or gene citations to facilitate the identification of relevant biomedical literature. AMIA Annu Symp Proc 2009;2009:563–7 [PMC free article] [PubMed] [Google Scholar]
- 16.BioRuby: Open Source Bioinformatics Library for Ruby. http://bioruby.open-bio.org/ (accessed Nov 2011).
- 17.Woo D. Apoptosis and loss of renal tissue in polycystic kidney diseases. N Engl J Med 1995;333:18–25 [DOI] [PubMed] [Google Scholar]
- 18.Bennet AM, Di Angelantonio E, Ye Z, et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA 2007;298:1300–11 [DOI] [PubMed] [Google Scholar]
- 19.Brouwers N, Sleegers K, Van Broeckhoven C. Molecular genetics of Alzheimer's disease: an update. Ann Med 2008;40:562–83 [DOI] [PubMed] [Google Scholar]
- 20.Cassidy SB, Schwartz S. Prader-Willi and Angelman syndromes. Disorders of genomic imprinting. Medicine (Baltimore) 1998;77:140–51 [DOI] [PubMed] [Google Scholar]
- 21.Andrade RJ, Robles M, Ulzurrun E, et al. Drug-induced liver injury: insights from genetic studies. Pharmacogenomics 2009;10:1467–87 [DOI] [PubMed] [Google Scholar]
- 22.Ohosone Y, Mimori T, Griffith A, et al. Molecular cloning of cDNA encoding Sm autoantigen: derivation of a cDNA for a B polypeptide of the U series of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci U S A 1989;86:4249–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu K, Cote TR. Database identifies FDA-approved drugs with potential to be repurposed for treatment of orphan diseases. Brief Bioinform 2011;12:341–5 [DOI] [PubMed] [Google Scholar]
- 24.Ekins S, Williams AJ, Krasowski MD, et al. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov Today 2011;16:298–310 [DOI] [PubMed] [Google Scholar]
- 25.Swanson DR. Medical literature as a potential source of new knowledge. Bull Med Libr Assoc 1990;78:29–37 [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson DR. Fish oil, Raynaud's syndrome, and undiscovered public knowledge. Perspect Biol Med 1986;30:7–18 [DOI] [PubMed] [Google Scholar]
- 27.Smalheiser NR, Swanson DR. Using ARROWSMITH: a computer-assisted approach to formulating and assessing scientific hypotheses. Comput Methods Programs Biomed 1998;57:149–53 [DOI] [PubMed] [Google Scholar]
- 28.Smalheiser NR, Swanson DR. Linking estrogen to Alzheimer's disease: an informatics approach. Neurology 1996;47:809–10 [DOI] [PubMed] [Google Scholar]
- 29.Hettne KM, Weeber M, Laine ML, et al. Automatic mining of the literature to generate new hypotheses for the possible link between periodontitis and atherosclerosis: lipopolysaccharide as a case study. J Clin Periodontol 2007;34:1016–24 [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez G, Uribe JC, Tari L, et al. Mining gene-disease relationships from biomedical literature: weighting protein-protein interactions and connectivity measures. Pac Symp Biocomput 2007;39:28–39 [PubMed] [Google Scholar]
- 31.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol 2001;52:540–2 [DOI] [PubMed] [Google Scholar]
- 32.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science 1997;278:631–7 [DOI] [PubMed] [Google Scholar]
- 33.Muller J, Szklarczyk D, Julien P, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res 2010;38(Database issue):D190–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostlund G, Schmitt T, Forslund K, et al. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res 2010;38(Database issue):D196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterhouse RM, Zdobnov EM, Tegenfeldt F, et al. OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res 2011;39(Database issue):D283–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu JC, Lee EK, Egan MG, et al. OrthologID: automation of genome-scale ortholog identification within a parsimony framework. Bioinformatics 2006;22:699–707 [DOI] [PubMed] [Google Scholar]
- 37.Deluca TF, Wu IH, Pu J, et al. Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics 2006;22:2044–6 [DOI] [PubMed] [Google Scholar]
- 38.Dice L. Measures of the amount of ecologic association between species. Ecology 1945;26:297–302 [Google Scholar]
- 39.Jaccard P. Étude comparative de la distribution florale dans une portion des Alpes et des. Bulletin de la Société Vaudoise des Sciences Naturelles 1901;37:547–79 [Google Scholar]
- 40.Liu YI, Wise PH, Butte AJ. The “etiome”: identification and clustering of human disease etiological factors. BMC Bioinformatics 2009;10(Suppl 2):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker KG, Barnes KC, Bright TJ, et al. The genetic association database. Nat Genet 2004;36:431–2 [DOI] [PubMed] [Google Scholar]
- 42.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 2007;39:1181–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu W, Gwinn M, Clyne M, et al. A navigator for human genome epidemiology. Nat Genet 2008;40:124–5 [DOI] [PubMed] [Google Scholar]