Abstract
Objective
Thousands of complex-disease single-nucleotide polymorphisms (SNPs) have been discovered in genome-wide association studies (GWAS). However, these intragenic SNPs have not been collectively mined to unveil the genetic architecture between complex clinical traits. The authors hypothesize that biological annotations of host genes of trait-associated SNPs may reveal the biomolecular modularity across complex-disease traits and offer insights for drug repositioning.
Methods
Trait-to-polymorphism (SNPs) associations confirmed in GWAS were used. A novel method to quantify trait–trait similarity anchored in Gene Ontology annotations of human proteins and information theory was developed. The results were then validated with the shortest paths of physical protein interactions between biologically similar traits.
Results
A network was constructed consisting of 280 significant intertrait similarities among 177 disease traits, which covered 1438 well-validated disease-associated SNPs. Thirty-nine percent of intertrait connections were confirmed by curators, and the following additional studies demonstrated the validity of a proportion of the remainder. On a phenotypic trait level, higher Gene Ontology similarity between proteins correlated with smaller ‘shortest distance’ in protein interaction networks of complexly inherited diseases (Spearman p<2.2×10−16). Further, ‘cancer traits’ were similar to one another, as were ‘metabolic syndrome traits’ (Fisher's exact test p=0.001 and 3.5×10−7, respectively).
Conclusion
An imputed disease network by information-anchored functional similarity from GWAS trait-associated SNPs is reported. It is also demonstrated that small shortest paths of protein interactions correlate with complex-disease function. Taken together, these findings provide the framework for investigating drug targets with unbiased functional biomolecular networks rather than worn-out single-gene and subjective canonical pathway approaches.
Keywords: Complex disease, SNP, gene ontology, protein-interaction networks, information theory, translational bioinformatics, complex disease, ontology, bioinformatcis, genetics, network, prostate cancer, protein networks, pathway analysis, network modeling, knowledge representations, uncertain reasoning and decision theory, languages and computational methods
Introduction
An essential objective of genome-scale sequencing and functional genomics is to improve on the paucity of associations between genetic variations and human diseases or other phenotypic traits (such as birth weight) and the impact of epigenetic modifications. From these upstream genetic causes, the ultimate goal is to achieve an improved form of personalized medicine based on individual patient's genetic variation.1 Genetic disorders are often categorized as single-gene diseases or as complex, multi-gene diseases such as cancers and diabetes. Typical examples of single-gene diseases are those of Mendelian inheritance, caused by mutations in an individual gene that result in an altered function or loss of its ability to properly interact with other genes.2 3 In contrast, complex diseases arise from the interplay of many different genes and single-nucleotide polymorphisms (SNPs). Although many of these diseases are common, their driving genetic mechanisms remain poorly understood on a molecular level.
To pinpoint the genes involved in complex diseases and elucidate their underlying genetic variations, hundreds of genome-wide association studies (GWAS) have been carried out and compare affected individuals with control cohorts. Despite the fact that many disease alleles have been discovered, most possess only a small effect size: OR<1.5.4 Therefore, it is unlikely that a few SNPs alone give rise to complex diseases, and it is more probable that an accumulation of large combinations of SNPs and other forms of genetic variations disrupt key biological mechanisms and consequently alter normal human physiology.5
Since the clinical functions of numerous intragenic trait-associated SNPs (SNPs located within gene regions) remain uncharacterized, the genetic architectures within and between these traits are thus also poorly understood. For instance, obesity is a disease that often fundamentally contributes to many other diseases such as diabetes and hypertension, and indeed obesity-associated genes have been prioritized in adult-onset diabetes GWAS.6 7 We therefore hypothesize that there should theoretically be some core shared SNPs, genes, or biological pathways that contribute to or cause common underlying traits. Such genetic architecture is evident in cancer, where gain-of-function mutations in oncogenes occur in the same genes across distinct cancers. Furthermore, these central genetic architectures can contribute to and link the diseases found within a particular metatrait, defined as a class of disorders clinically related in time (eg, one disease causally precedes another) or sharing common molecular functions and processes. For example, oncogenic processes leading to ‘cancer’ can together be considered a metatrait that comprises different types of specific cancers, as their somatic mutations often overlap genetically or functionally. Similarly, metabolic syndrome is considered a metatrait that includes insulin resistance, hypertension, obesity, hyperlipidemia, and hypercholesterolemia. Therefore, to elucidate this genetic underpinning, great emphasis is placed on shared characteristics, such as symptoms and drug responses, when developing disease networks8 9 and their causative genetic networks.
Numerous methods have been developed to construct human disease networks and can be categorized as either non-SNP-based or SNP-based methods. Information in electronic medical records, such as disease correlation or comorbidities, can be directly applied to construct disease networks.10 11 Furthermore, underlying biological disease data, such as mRNA expression profiles9 12 and protein–protein interactions (including protein complexes),11 13 14 can also be employed to infer disease networks. Additionally, metabolic data, such as adjacent or mutual biochemical reactions, have also been used in disease network development.15 Recently, with the dramatic increase in genetic variation data and GWAS results, shared intragenic SNPs and their host genes (the genes physically containing the variations)16 have been used to link distinct diseases, both single-gene inherited diseases17 18 and complex diseases.19 However, unlike single-gene diseases, these early complex-disease network studies that use simple SNP and gene overlaps have not obtained the expected modularization results (related diseases highly connected with each other) because of small dataset sizes. Specifically, many diseases were found to be isolated and totally disconnected from other diseases within the same disease class.19
Since previous disease network modeling methods have been mainly based only on analyzing gene overlap or clinical relatedness as found in the GWAS or the medical record rather than biological relatedness, only those diseases with obvious genetic or clinical connections have been highlighted. Furthermore, the majority of these networks used Mendelian inheritance facts from the Online Mendelian Inheritance in Man (OMIM) rather than complex inheritance patterns from GWAS. Many diseases are obviously clinically related, eg, the comorbidity between hypertension and obesity; however, no overlapping genes or SNPs have been discovered by GWAS to date. Therefore, more complex ways of relating two diseases and constructing disease networks must be designed in order to understand their common pathologies and relatedness, which can further our ability to treat diseases.
One application of such networks would be drug repositioning accomplished through the identification of shared biological mechanisms between one treatable disease and one for which no effective treatment exists. This can be conducted using network theoretic models in which biological mechanisms are used to relate diseases and their associated molecular structures. Novel methodologies that can mechanistically relate diseases that are observably clinically related but have little or no shared genetic or physiological underpinning may elucidate more complex mechanisms and point to therapies that can be repurposed between the two.
To address this issue, we propose a novel method that builds disease–disease networks that extend well beyond mere shared SNPs or host gene linkages. To this end, we exploit the semantic similarity among host genes of validated trait-associated SNPs in the National Human Genome Research Institute Catalog of Published Genome-Wide Association Studies (NHGRI GWAS Catalog)20 via existing annotations of host genes in Gene Ontology (GO)21 to build a similarity network of diseases with an information theory-based approach. Specifically, our method integrates genetic alteration (GWAS) data with standardized textual descriptions of gene functions and processes and their inter-relationships in order to characterize the mechanistic underpinnings of disease of complex inheritance. We hypothesize that similarity among clinically related diseases is reflected in the similarity between their constitutive deregulated processes and functions that can be investigated computationally through the biological annotations associated with their genes hosting intragenic GWAS SNPs (host genes). Thus, we use a novel application of GO to computationally examine and compare data derived from GWAS.
We further analyze our disease–disease network using protein interactions to create a disease–gene network (which also contains disease–disease, gene–gene, and disease–gene connections). Integration of protein interaction data allows the identification of functional similarity network relationships that can be explained straightforwardly at the protein level and those that are most likely due to higher scale biological processes (eg, cell proliferation associated with cancer disease). Further, this enhances the current paradigm of ‘targeted’ therapy repositioning, which implies a ‘protein target’ and is thus better understood at the protein level, with non-trivial and multi-scale biological mechanisms unveiled by similarity metrics (GWAS/SNP/GO). We have previously demonstrated that this gene information theoretic similarity (ITS) method can accurately predict protein functions in poorly characterized genes22 and, further, can exploit the shared genetic architecture of diseases by using their common interactions or interaction paths. Thus, we hypothesize that this sensitive similarity approach could allow the elucidation of non-trivial associations between trait-associated genes. Additionally, we constructed our network based on a much larger number of NHGRI intragenic SNPs than previous studies.
Methods
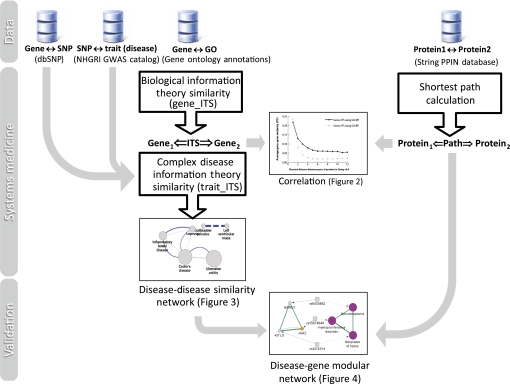
The workflow of our methodology is shown in figure 1, and each component is described as follows. Table 1 provides definitions of the major concepts and terms used in this paper.
Figure 1.
Workflow of our methodology. This study demonstrates the non-trivial modularity of biological mechanisms shared by some complex diseases. Specifically, complex diseases are connected using biological similarity computations between single-nucleotide polymorphisms (SNPs) associated by genome-wide association studies (GWAS) with complex traits. First, gene–gene similarity in Gene Ontology (GO) was calculated at a genome scale using an information theory similarity (ITS) measure validated previously. For a pair of genes in the STRING database, the shortest protein interaction distance is shown correlated with their biological similarity obtained by Gene_ITS using GO annotations (figure 2). Second, host genes were mapped to complex traits using the NHGRI GWAS Catalog and dbSNP database, and the trait–trait ITS of their associated traits was derived from results calculated in step 1. A disease similarity network was constructed by choosing the significant trait–trait similarity (figure 3). Metatrait modules were extended by their shortest paths between host genes with significant information similarities, due to the validated reverse correlation between them (figure 4).
Table 1.
Definitions and abbreviations
| Concept | Definition |
| Genome-wide association studies (GWAS) | Investigation of genes in the whole genome for a large number of individuals that tests the genetic variations differentially found between two contrasted groups (case vs control) with respect to a specific trait, such as a disease |
| Gene Ontology (GO) | Controlled vocabulary of annotations to gene and gene product attributes |
| Protein–protein interaction network (PPIN) | Graphic representation of protein–protein interactions on a large scale constructed in order to appreciate the network structure |
| Single-nucleotide polymorphism (SNP) | Single-nucleotide variation in the genomic sequence found to be different between individuals or between two chromosomes of the same individual |
| Intragenic SNP | A SNP located within a gene region |
| Intergenic SNP | A SNP located outside any gene region |
| Host gene of an intragenic SNP | The gene that physically contains the intragenic SNP in its genomic sequence |
| Trait | A characteristic phenotype or disease state of an individual, such as hair color or type II diabetes |
| Metatrait | A class of disorders clinically related in time or sharing common molecular mechanisms (eg, ‘metabolic syndrome’ is a metatrait for the traits ‘essential hypertension,’ ‘adult-onset diabetes mellitus’ and others) |
| Intertrait | Relationship found between two traits |
| Intra-metatrait | Connections between traits that belong to the same metatrait |
| Network modules | A subnetwork possessing some biological or medical implications whose nodes are densely connected inside the subnetwork but are sparsely connected with nodes outside of the subnetwork |
| Gene Ontology term | Standardized description of a biological concept, such as the molecular function, the biological processes or the subcellular localizations of a gene |
| Minimal ancestor of two GO terms | The most specific GO term that could summarize or contain the characteristics shared between a pair of GO terms |
| NHGRI GWAS Catalog | The National Human Genome Research Institute Catalog of Published Genome-Wide Association Studies (http://genome.gov/admin) |
| STRING | Search tool for the retrieval of interacting genes/proteins: the most comprehensive database of protein–protein interactions and associations |
| Fusion | A reliable protein–protein interaction prediction method based on the hypothesis that two proteins are more likely to interact with each other if they have been incorporated into a third protein as two domains during evolution |
| Shortest distance of two proteins | The minimum number of distinct edges found among all possible routes connecting two proteins in a protein–protein interaction network |
| Shortest path(s) of two proteins | All routes possessing the minimum number of distinct edges found among all possible routes connecting two proteins in a protein–protein interaction network |
Data and preprocessing
GWAS dataset file ‘gwascatalog.txt’ was downloaded on May 25, 2010 from the NHGRI GWAS Catalog (http://genome.gov/admin).20 It comprises well-validated associations between 350 complex traits and diseases and 2793 SNPs, of which 1355 are intergenic, 1137 are intronic, and 144 are exonic. Of the exonic SNPs, 113 are non-synonymous. Expert curation of the 271 distinct textual terms representing complex traits and diseases was performed in order to identify synonymous terms (eg, ‘hemoglobin’ and ‘hemoglobin level’) or highly related terms (eg, ‘glioma’ and ‘high grade glioma’), and aggregate their intragenic SNPs. We resolved these fairly redundant annotations through manual curation and thus increased the average number of SNPs associated with any single trait, which resulted in 177 conceptual entities (traits) from the 271 NHGRI catalog traits, during which 131 textual terms were merged with others as 37 conceptual entities, as described in online supplementary table 1. Hereafter, we refer to these 177 conceptual entities as ‘traits’ or ‘diseases’. A total of 1438 intragenic SNPs associated with these diseases were included in this study, while 1355 intergenic SNPs (see table 1 for definitions) were not.
Host genes of intragenic SNPs were assigned using the default definitions and parameters from the Single Nucleotide Polymorphism Database (dbSNP) annotations (file downloaded from ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606/ASN1_flat/ on October 19, 2009). In spite of the rare instances where a SNP could lead to two distinct genes (sense and anti-sense transcription), in dbSNP each SNP is uniquely mapped to a single host gene.16 To most accurately determine the inter- or intra-genic status of a SNP, dbSNP annotations were preferred, since the NHGRI GWAS Catalog does not delineate whether the SNP's associated gene is the nearest one or the actual host gene, making the status often indistinguishable. As a result, 1083 disease host genes were mapped from the 1438 distinct disease-associated intragenic SNPs.
GO hierarchies21 and Gene Ontology annotation files gene2go and gene_info were downloaded from http://www.geneontology.org/ontology/gene_ontology.obo and ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/ on May 19, 2009. GO terms classified as ‘GO molecular functions’ (GO:MF) and ‘GO biological processes’ (GO:BP) were identified and analyzed as two distinct semantic types using the similarity metrics described below. GO terms from the third semantic type, ‘GO Cellular Components’, were not included, as it was assumed that membership in a cellular component geneset is generally not sufficient to impute functional or mechanistic similarity between genes underpinning the same complex trait.
The interaction network of gene products (proteins) was downloaded from the search tool for the retrieval of interacting genes/proteins (STRING v8.0; http://string.embl.de, last update on November 17, 2008).23 The STRING database contains the most comprehensive dataset to date for protein–protein interactions and associations and was thus used as the database for physical protein interactions in humans. Protein interactions included in this study met the following criteria: (1) species: Homo sapiens; (2) physical interactions coded in STRING as ‘experimental’, ‘fusion’ (see details in table 1), and ‘other database based methods’ (excluding those exclusively derived from ‘text mining’ to avoid a bias introduced by utilizing protein interactions discovered via GWAS results); and (3) high confidence scores of at least ‘900’ (available range 0–999) to exclude lower quality ones. Consequently, 72 730 interactions between pairs found among 7677 proteins were obtained after 1 181 753 interactions had been filtered out. These data were used in figures 2 and 4.
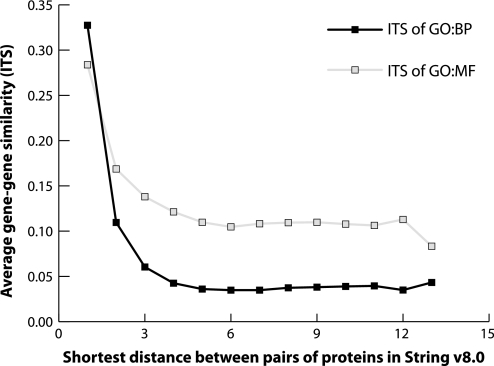
Figure 2.
Higher Gene Ontology (GO) similarity between proteins is associated with smaller shortest distance in protein interaction networks. Relationships are seen between average Gene_ITS values and the shortest distance between pairs of proteins in the protein–protein interaction network. An average information theory similarity value was calculated for groups of protein–protein pairs in STRING v8.0 with the same shortest distances (length value of shortest paths). As hypothesized, higher biological similarity in GO was associated with shorter distances in the protein interaction network (p<10−16, Spearman correlation, using the entire set of protein combinations with GO annotations and protein interactions, which were 5753 and 5955 for GO biological processes (GO:BP) and GO molecular functions (GO:MF), respectively). These results are reproducible in the subset of genes hosting the single-nucleotide polymorphisms associated with disease traits in the National Human Genome Research Institute Catalog of Published Genome-Wide Association Studies (data not shown).
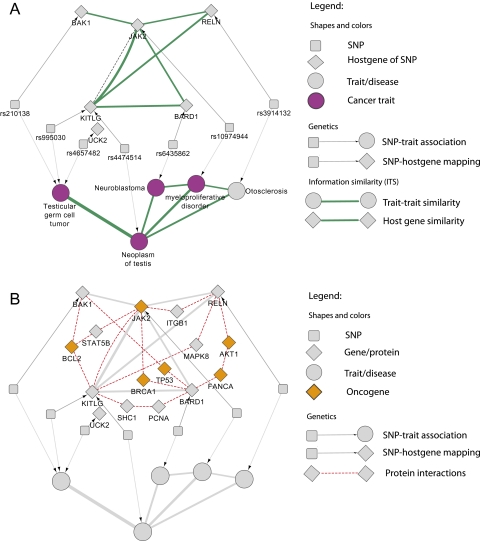
Figure 4.
Biological similarity between cancer traits annotated by shortest protein interaction path between host genes is enriched in oncogenes. The subset of figure 3 circled in orange corresponds to a biomodule of cancer traits. (A) provides the detailed view of their genome-wide association study-associated single-nucleotide polymorphisms (SNPs), their corresponding host genes, and their dense biological similarities (Gene_ITS ≥0.2; Trait_ITS ≥0.2; green lines; based on Gene Ontology biological processes). (B) provides an additional annotation of shortest protein interaction paths (red dotted lines) between host genes. Oncogenes (gold color) are statistically enriched in the shortest protein-interaction paths among pairs of SNP host genes associated with distinct traits that were paired by similarity measures (p=0.0001, Fisher's exact test). In addition, five out of six host genes associated with these cancer traits are either oncogenes or directly interact with oncogenes in the cancer-related modules in the disease network. Taken together, our metric produces multi-scale connections, as it contains protein interactions as well as biological similarities that both underlie the disease network connections and utilize different knowledge bases, thus validating one another.
Calculation of shortest path(s) and shortest distance between two proteins in a protein interaction network
In a protein interaction network consisting of nodes (proteins) and edges between these nodes (direct interactions between two proteins), the shortest paths between a pair of proteins are the routes with minimal number of edges among all possible routes connecting the two proteins.24 Note that different paths may meet the criteria of the shortest path, and thus more than one set of edges (paths) can result. The cardinality of the shortest path between two proteins is calculated as the count of its distinct edges and is termed the ‘shortest distance between two proteins’.8 A breath-first algorithm was implemented on the protein interaction network (undirected graph) in-house as classically described.24 The metric of shortest distance between two proteins was used as a gold standard in the validation provided in figures 2 and 4, since pairs of proteins with smaller ‘shortest distances’ have previously been established as more functionally related than those with longer ones.25 26 Furthermore, as described in the data subsection of the methods, the physical protein interaction network was designed to be independent of the discoveries stemming from GWAS and thus does not contain any interactions discovered by text mining of the literature after 2006 (initial period of GWAS publications).
GO-anchored information similarity between host genes of intragenic SNPs
Disease networks were built from trait–trait similarities that were calculated using the GO-anchored ITS between the traits' genes. Trait genes were the host genes that harbor the variant DNA sequence (SNPs) associated by GWAS with the trait. A brief explanation of the procedure used to measure gene–gene similarity will follow and is directly calculated using our published method (figures 2–4).22
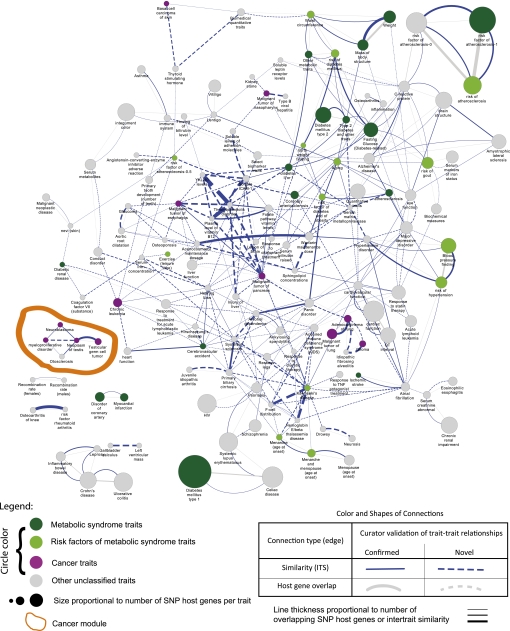
Figure 3.
Network of biological similarity between complex-disease traits calculated from genome-wide association study single-nucleotide polymorphisms (SNPs). The disease similarity network was calculated using Genome Ontology biological process similarity of the host genes of trait-associated intragenic SNPs with similarities ≥0.2 and an empirical p value <0.05. As shown, among 280 intertrait similarity connections (blue lines), 186 (66.4%) cannot be explained simply by shared host genes between the traits (gray lines) and are reported here for the first time. Therefore, this figure illustrates that our information theoretic similarity (ITS) method has found non-trivial relationships that would not have been found by conventional methods. As hypothesized, metabolic syndrome traits (green) and cancer traits (purple) are significantly enriched in connections with other traits in the same metatrait (p=3.5×10−7 and 0.001 respectively, Fisher's exact test). A significant module of cancer traits (circled in orange) is shown in greater detail in figure 4. Circles represent diseases or traits whose sizes are proportional to their number of associated intragenic SNP host genes. Green circles represent metabolic syndrome-related traits curated a priori (dark green for metabolic syndrome traits and light green for their risk factors), purple circles represent cancer traits, and gray circles represent other traits. Blue lines represent biological process similarities that are ≥0.2 and have a p value <0.05. Gray lines represent shared SNP host genes between diseases if their Trait_ITS is ≥0.2 (in other words overlapping connections between our information theoretic method and conventional gene overlapping method). Line thicknesses are proportional to Trait_ITS similarity values or number of shared genes. Solid lines have been validated as clinically meaningful by clinicians, while dotted lines have not.
The similarity of two genes is defined conceptually by the similarity of their GO annotations as measured by their shared information content. Specifically, the ITS of two terms is defined as the information content of their minimal ancestor in GO (common ancestor with maximal information content) divided by the average information content of the two terms, where the information content of a single term is the probability of the term and its sub-terms being selected randomly in GO.21 The formal definition of term–term similarity that we selected is Lin's metric,27 with straightforwardly interpretable scores that range from ‘0’ (no similarity) to ‘1’ (100% similar):22
| (1) |
| (2) |
where ic(a) is the information content of GO term a, ms(a,b) is the minimal ancestor of terms a and b, G(a) is the sub-graph of GO rooted at a, A is the root term of the GO, and the function ‘|G(a)|’ is the cardinality of G(a) measured as the count of distinct terms in this sub-graph.
The information content of a term is a non-negative value representing the specificity of the term. For terms hierarchically organized as an acyclic directed graph, the root term has zero information content, as no specific information is generated from this term because of its generality, while a leaf term with great depth (distance from the root) has the largest information content, since it is inherently more specific. Thus a term's information content is roughly related to its specificity. Term–term similarity is a value between 0 (for two terms having the root term as their only common ancestor) and 1 (for two terms sharing identical sub-graphs and identical sets of ancestors). More discussion on this GO-based similarity can be found in our previous publication22 and related review paper.28
The ITS of two genes (Gene_ITS), such as two SNP host genes, was measured by the average best-matching pair similarity between their annotated GO terms. For any GO term annotated to a gene, its best-matching term pair in another gene's GO annotation list is the one with the maximum term–term similarity as compared with all other terms from the other gene. Furthermore, only the most reliable subset of the best-matching term pairs across the two term sets is retained in the calculation, while all other term pairs are ignored because of the annotation noise in GO. Mathematically, the information similarity of two genes is defined as 22:
| (3) |
where α and β are two genes being annotated to two term sets, the included best-matching term pairs are represented as a relationship π with pairs ai and bi, and t is the similarity threshold for any term pair to be included in the calculation for further annotation noise reduction (set as 0.7 in our implementation22). The similarity of two genes is based on their number of shared GO terms and, if the terms were not identical, the term proximity in the GO graph. The information similarity of two genes is normalized to the range of 0 to 1, corresponding to genes with no similar annotations and genes with equivalent annotations, respectively. More details and examples can be found in our previous paper.22
Trait–trait information similarity and empirical distributions
Trait–trait ITS (Trait_ITS or intertrait similarity) was measured by the shared information between the host genes of the associated intragenic SNPs, specifically the average similarity of reciprocal best-matching host gene pairs from GWAS. The best-matching pair of a host gene (γ) with respect to another trait is the host gene (δ) of the other trait with maximum gene–gene similarity with the first host gene (γ). We required reciprocal maximal similarity between host genes from two traits for the most reliable relationship between traits, but did not use a threshold to control noise because host genes derived from intragenic SNPs are much more reliable than GO annotations and contain much less noise. We define trait–trait similarity formally as follows:
| (4) |
where U and V are two traits representing two sets of host genes identified by GWAS, and the reciprocal best-matching host gene pairs are represented as a relationship π' with pairs αi and βi. The information similarity of two traits ranges from 0 (for two traits with totally dissimilar host genes) to 1 (for two traits with identical or equivalently annotated host genes).
In order to prioritize Trait_ITS scores, we conducted a permutation resampling. We regarded the network directly derived from GWAS as a bipartite network, with one set of nodes being the diseases/traits (177) and the other set as the host genes (1083), and shuffled the edges in the network so that the number of degrees for any host gene or traits (nodes) remained the same. We created 10 000 such networks by permutation, and calculated an empirical p value for each specific trait–trait connection according to the rank of its observed Trait_ITS score among those of that specific relationship in the control networks.
Disease similarity networks
A disease–disease network was thus constructed from pairwise, intertrait similarities directly subjected to a certain joint similarity and statistical significance cut-off, where nodes in the network represent complex traits, and edges represent the significant biological similarity between two traits as calculated by ITS. The sizes of the nodes and edges were proportional to the number of host genes and the strength of the trait–trait similarities, respectively (figure 3).
Evaluation of metatrait biomodules
Clinically relevant modules in the disease–disease networks were identified on the basis of expert knowledge as well as their statistical significance (figure 4). Traits were categorized into metatraits by two clinicians blinded to the results (online supplementary table 1). An ITS metatrait module is defined as a group of traits known to be subsumed by a metatrait that are also predicted to be similar to one another by their Trait_ITS. We further conducted an enrichment study to identify if these ITS metatrait modules comprised more Trait_ITS connections than expected using Fisher's exact test, by considering two factors for all possible trait pairs: (1) whether the trait pair was completely within the metatrait of interest (intra-metatrait connection) or if at least one of the traits was found outside of the metatrait (non-intra-metatrait connection) and (2) whether the trait pair was within the network or not in the network. Calculations were made using a contingency table (table 2):
| (5) |
where M is a metatrait containing a set of related traits, T is the set of all traits in the disease network. CMM is all possible pairwise combinations of traits in M, and CTT is the total possible pairwise combinations between all traits in the network.
Table 2.
Fisher's exact test contingency table for enrichment of a specific type of intra-metatrait connections in disease networks
| Intra-metatrait connections | Non-intra-metatrait connections | Subtotal | |
| Observed in the network | NMM | NTT−NMM | NTT |
| Not observed in the network | CMM−NMM | CTT−CMM−NTT+NMM | CTT−NTT |
| Subtotal | CMM | CTT−CMM | CTT |
M, a metatrait containing a set of related traits; T, the set of all traits in the disease network; NMM, the number of observed connections between the two traits within the metatrait M; NTT, the number of connections in the disease network; CMM, all possible pairwise combinations of traits in M; CTT, the total possible pairwise combinations between all traits in the network.
Metatraits enriched in Trait_ITS connections are defined as enriched ITS metatrait modules (defined in table 2). Significant connections among ITS metatrait modules were examined in closer detail by adding the shortest protein interaction path between every pair of similar host genes associated with each pair of diseases. See the Results section for a detailed rationale on using the shortest protein interaction distance.
Overlaying a drug network
SNPs were associated with known drugs based on the Ingenuity Knowledge Base. Each SNP dataset of interest was uploaded into the application. Each identifier was mapped to its corresponding object in the Ingenuity Knowledge Base. SNPs with appropriately mapped genes were deemed to be ‘network eligible molecules’, and were overlaid on to a global molecular network developed from information contained in the Ingenuity Knowledge Base. Canonical drug information was then overlaid on to the SNP network (online supplementary figure 3).
Results
Biological significance of gene–gene similarity
Our initial study was designed to investigate the biological validity of the gene–gene ITS calculated from biological functions and processes annotated to genes. We hypothesized that biological similarity would increase with small shortest distance in protein interaction networks. A high-quality subset of the STRING database v8.0 (physical interactions) was used to generate a genome-wide protein–protein interaction network (PPIN), which includes 72 370 interactions for 7677 proteins23 (figure 2, table 3). In addition, we investigated this correlation between gene–gene ITS and protein–protein interactions in the subset of genes hosting SNPs associated with disease traits in the NHGRI GWAS Catalog16 which includes 1083 disease host genes and 1438 intragenic SNPs (online supplementary table 2).
Table 3.
Direct protein–protein interactions are enriched in gene–gene pairs with high biological similarity
| Biological similarity between pairs of genes (genome-wide) | ||||
| GO biological process | GO molecular function | |||
| ITS ≥0.7 | ITS <0.7 | ITS ≥0.7 | ITS <0.7 | |
| Direct protein interaction between pairs of genes | 7314 | 41 710 | 5413 | 47 112 |
| No direct protein interactions | 53 174 | 16 442 890 | 138 125 | 17 537 385 |
| OR | 53.7 | 14.6 | ||
| Fisher's exact test | p<10−16 | p<10−16 | ||
Table 3 shows the OR of biologically similar pairs of genes enriched in first-degree protein interactions on a genome scale, and online supplementary table 2 corroborates these results on the subset of genes associated with complex diseases through GWAS. Specifically, direct protein–protein interactions were found to be enriched in gene–gene pairs with similarities ≥0.7 (the empirical p values for the similarity ≥0.7 were 0.007 and 0.019 for GO:BP and GO:MF, respectively). For stratified Gene_ITS thresholds, we calculated their enrichment in first-degree interactions (OR ≥14.6, p<10−14 for Gene_ITS ≥0.7; Fisher's exact test) corresponding to (1) GO:BPs or (2) GO:MFs either (3) over a genome-wide scale (table 3) or (4) on the subset of genes associated with traits from the NHGRI GWAS Catalog (online supplementary table 2).
We then investigated the interacting protein pairs with small shortest distances and hypothesized that they are most likely to have corresponding gene pairs with large similarity values. Shortest protein interaction distances at either 1 or 2 (direct and indirect neighbors, respectively) were found to be enriched in gene pairs with a Gene_ITS ≥0.7 on a genome-wide scale (and at the NHGRI SNP level), and for GO:BP and GO:MF alike (OR >4.2 in the stringent PPIN; p<10−6, Fisher's exact test). These results are consistent with other reports that indirect neighbors may possess similar functions in PPINs.25
Additionally, the shortest protein interaction distance between two genes was found to inversely correlate with the ITS value (figure 2). In other words, smaller shortest protein interaction distances correlate with larger ITS scores between genes. The non-parametric Spearman correlations between information similarity and shortest distance in the stringent PPIN were −0.15 and −0.09 for similarity of GO:BPs and GO:MFs, respectively (p<2.2×10−16, using the entire set of protein combinations). Other versions of STRING led to similar results (data not shown), establishing the rationale for investigating shortest paths using gene similarity associated with complex traits in GWAS as seen in figure 4.
Disease–disease networks
Disease similarity networks were constructed from intertrait similarity at a certain threshold, as defined in Equation 4, which makes use of host gene–gene similarities as defined in Equation 3 (Methods). Two disease similarity networks were constructed using the information similarity of GO:BP and GO:MF terms separately, as shown in figure 3 and online supplementary table 3 at an ITS of two traits ≥0.2 and a p value <0.05 and in online supplementary figure 1 at an ITS of two traits ≥0.3 and a p value <0.05 (Trait_ITS thresholds chosen by the minimal value that guaranteed the statistical significance of most values above this cut-off). Additionally, the information connections created by GO:BP similarity show stronger associations with disease traits than those of GO:MFs. Thus, our subsequent analysis focused on the networks created using GO:BPs. Two hundred and eighty Trait_ITS relationships were selected as significant between traits (Trait_ITS ≥0.2; 138 of the 177 traits in the selected network, figure 3). The connectivity of traits with a large number of host genes is high, as expected. For example, ‘diabetes mellitus type 1’ is associated with 40 host genes in the NHGRI GWAS Catalog and is ranked fifth among 177 traits in terms of number of host genes before Trait_ITS analysis. Furthermore, after Trait_ITS analysis, the host genes of ‘diabetes mellitus type 1’ contributed significantly to the similarity network, which results in the greatest number of host genes in the network and is represented as the largest node in figure 3.
Traits in several metatraits were more likely to interact with other traits in the same metatrait in the disease network. For example, cancers (OR=4.0; Fisher's exact test p=0.001) and metabolic syndrome (OR=3.1; Fisher's exact test p=3.5×10−7) are shown to be enriched in significantly similar traits (figure 3; purple and green colors, respectively). By definition, metabolic syndrome includes the disorders hypertension, arteriosclerosis, adult-onset diabetes mellitus, hyperlipidemia, and obesity. Different metabolic syndrome component diseases are expected to share common genetic architecture, such as GO:BPs, and should thus be related to one another, as should distinct cancer types. A large metabolic syndrome module is shown in figure 3 (in green) along with two isolated cancer modules (in purple). The module located in the bottom left of figure 3 outlined in orange was examined in more detail in figure 4 because of its novelty and clinical relevance.
We also constructed a disease network with a conventional shared gene method using intragenic SNP host genes shared between diseases for comparison with our ITS method. The shared gene method has been previously reported in the literature, but with a much smaller set of GWAS data. The full disease network created using shared genes is shown in online supplementary figure 2. It contains 383 connections between 137 combined traits with 40 isolated traits. The ‘skeletal finding’ trait was associated with the greatest number of host genes (105, supplementary figure 2). Again, metabolic syndrome traits were found to be significantly more likely to interact with other metabolic syndrome traits in the whole network constructed using shared genes of trait-associated SNPs (OR=4.1, Fisher's exact test p=5.2×10−15), but cancer traits were not found to be significantly more likely to interact with other cancer traits (OR=1.7, Fisher's exact test p=0.22). This finding is mainly consistent with the disease similarity network we constructed, but is contrary to previous findings that most diseases in the same disease classes (eg, metabolic syndrome) are isolated and the whole network is only sparsely connected19—a conclusion that may have arisen from insufficient data. Gene overlap has been previously published by Barrenas et al,19 and interestingly our Trait_ITS method recapitulates these findings and indicates additional relationships for metabolic syndrome, such as the Trait_ITS relationship between ‘birth weight finding’ and ‘risk factor of diabetes and of obesity’. We later focused on the cancer module in figure 4 and provided details of the ITS-generated cancer biomodules showing underlying biological mechanisms that explain the high Trait_ITS scores. As a proof-of-concept, we also incorporated drug information into the aforementioned cancer-specific biomodule (online supplemental figure 3).
To better compare the disease networks created by our method and by the shared gene method, we plotted the shared gene connections in the disease similarity network in a gray color in figure 3. There were 114 traits in common between the two networks out of 138 total traits in the disease similarity network, indicating 82.6% trait coverage at a similarity threshold of 0.2. In particular, there were 101 out of 114 (88.6%) overlapping traits that shared at least one edge between these networks, with 94 total overlapping connections that shared at least one host gene and also possessed a GO:BP similarity ≥0.2. One hundred and eighty-six non-trivial trait–trait connections were identified by our ITS method because these pairs of traits did not share SNPs, nor host genes of their respective SNPs. To validate the accuracy of these newly discovered disease connections, we conducted a preliminary manual validation on the 280 similarity connections, and determined that 109 were clinically reasonable, according to two clinician reviewers (minimal precision of 38.9%). The genetic architecture of complex traits may explain the apparently clinically irrelevant relationships, as geneticists specializing in GWAS have observed, particularly in Expression Quantitative Trait Loci (eQTL) studies that show a subset of genes and SNPs that contribute to many clinically unrelated diseases. In other words, the Trait_ITS connections that do not corroborate obvious clinical relationships may, in part, reflect the common modularity of complex-disease gene inheritance as well. The comparative results suggested that our similarity network contains many (186) potential connections among complex traits that have not yet been discovered by GWAS, and thus demonstrate that our ITS method is able to capture non-trivial relationships that would not have been otherwise found by conventional methods.
Finally, we conducted a smaller study focusing on Trait_ITS scores derived exclusively from the 113 non-synonymous (exonic) SNPs from the NHGRI GWAS Catalog. Trait_ITS scores derived from this subset were significantly higher than those obtained from the whole collection and from intronic ones (p<2×10−16 and p<2×10−16, Mann–Whitney test; 113 non-synonymous SNPs versus (1) 2793 intragenic SNPs and (2) 1137 intronic SNPs; p values of Trait_ITS scores led to similar results). These results suggest that polymorphism is more likely to be associated with missense or stop codon mutations, and consequently mutated proteins are more likely to be functionally similar by Trait_ITS than intronic sequences. However, the number of intertraits evaluated thus far is relatively small, and future studies may require a larger collection of exonic non-synonymous SNPs.
Disease similarity biomodule: cancer module case study
We focused our attention on one of the more interesting cancer modules, and plotted the shortest paths among the disease-associated genes in the PPIN in which the similarity of biological processes was positive (as shown in figure 4) because of the correlation between gene–gene similarity and shortest protein interaction distance. This disease similarity biomodule contains four distinct disorders that suggest a common genetic architecture for a cancer ‘metatrait’ associated with germline mutations or those arising early in life (shown in figure 4A). Two of the diseases are cancers, specifically neuroblastoma and testicular cancer (neoplasm of testis and testicular germ cell tumor). Furthermore, another class of diseases in the network, myeloproliferative disorders, contains precancer and cancer, and includes polycythemia vera and chronic myelogenous leukemia, among others. The final constituent is otosclerosis, a disease causing hearing loss that has been found to be a T cell-mediated autoimmune disorder involving abnormal bone growth in the middle ear.29 Interestingly, both neuroblastoma and neoplasms of the testis are found in younger patients and most commonly involve germline mutations of specific genes that cause these phenotypes. Similarly, both otosclerosis and myeloproliferative disorders have specific childhood variations similarly arising from heritable mutations, causing manifestations in early life. Although seemingly disparate, these diseases share a strong underlying genetic and clinical component. Indeed, among the 16 proteins connecting the diseases in the shortest path network shown in figure 4B, six are oncogenes as curated by the Sanger Institute.30 This is statistically significant and corresponds to an OR of 10.8 (408 cancer genes in total) with a p value=0.0001 (Fisher's exact test). Moreover, except for the host gene, UCK2, which is not annotated by GO, all other host genes associated with the five diseases are either oncogenes or directly interact with oncogenes.
Furthermore, we recapitulate known oncogenic pathways in our model, such as KITLG and JAK2, between testicular cancer and myeloproliferative disease. We also highlighted other shared pathways. For example, the gene product of KITLG is a ligand of a tyrosine kinase receptor involved explicitly in neuronal and germ cell development as well as hematopoiesis, a process that continues through adulthood. KITLG and its associated SNP (rs995030) are directly connected to testicular cancer,31 but have also be shown to be involved in myeloproliferative disorders32 via their role in hematopoiesis, and are conceivably connected to neuroblastoma with its involvement in neuronal development and migration. RELN similarly functions to regulate the migration of neuroblasts and is directly connected to otosclerosis in our network as well as in the literature.33 Not surprisingly, RELN's importance in neuroblastoma has been previously documented,34 and, correspondingly, our disease similarity network created a putative linkage through protein associations with AKT1, FANCA, and BARD1, consecutively.
JAK2 is a tyrosine kinase associated with cytokine receptors involved in cell signaling pathways mediating gene transcription as well as other cellular functions. It is a necessary component of hematopoiesis, and thus specific gain-of-function mutations in JAK2 are the essential errors in polycythemia vera and are involved in all other myeloproliferative disorders.35 Similarly, other mutations in JAK2 can contribute to the formation of many different neoplasms, plausibly confirming the connections made in our diagram through JAK2 to both testicular cancer and neuroblastoma via several different pathways. Therefore, the disease similarity biomodule reveals commonalities on both the phenotypic and molecular levels such as their host genes' similarities and the intermediate genes connecting similar host genes.
Discussion and conclusion
In this study, we explored a method for understanding disease–disease similarities at both the phenotypic and molecular levels simultaneously. To do this, we took advantage of information theory metrics of trait similarity that are defined using host gene–gene similarities derived from GWAS intragenic SNP data. Thus, the similarity arising between these traits from GO annotations at the molecular level can be qualitatively validated by observing if a significant number of known related clinical diseases are also related by their genetic similarity. Accordingly, we demonstrated the potential of using intragenetic SNPs to explain disease associations in a cancer biomodule. Furthermore, to quantitatively validate GO functional similarity, we assumed that, among a variety of biological mechanisms relating genes, the shortest distance for each protein pair can be calculated independently. We indeed confirmed that the shortest distance in the entire disease–gene PPIN, calculated exclusively from protein interactions and independently from GO similarity, correlate with higher GO similarity between the genes of disease-associated intragenic SNPs.
To visualize these results in the context of disease, we generated a composite disease–gene network comprising similarity connections between traits and the shortest paths in the PPIN between disease genes. We found clear biomodules that have shared pathways and clinical traits and thus merit further exploration. By generating novel genetic connections between diseases, this technique can be used to focus the scope of investigation, and, by being computational in nature, has the potential to elucidate disease underpinnings with a reduced need for conventional wet-lab methods.
Moreover, we have demonstrated the ability to layer pharmacologic data on to our network for potential future hypothesis generation. In our analysis, the network-associated drugs may be useful in chemoprevention, as patients with the affected SNP were more likely to develop a disease. However, it is important to note that the very nature of GWAS indicates that the SNPs used in our study are associated with (and not causal of) disease susceptibility. Thus, while it is possible to speculate on the potential role of pharmacology in our network, it is critical that the underlying mechanisms in the SNP–gene–disease triad first be better understood. In other words, when causation is established, pharmacologic agents captured at the gene/protein level have a far higher likelihood of being effective agents. Nevertheless, despite this limitation of the SNP data, our method provides researchers with a crucial starting point for testing repurposed drugs in their model systems of disease in conjunction with previously validated methods. Previous informatics studies have shown the opportunity to reposition drugs using structural and functional similarity between proteins within the scope of ‘gene targeting’36 and through a new paradigm of genome-based drug repositioning.37–39
Another limitation of our study is that there are more intergenic SNPs (SNPs located outside of gene regions) than intragenic SNPs (found within genes) identified through the NHGRI GWAS Catalog, and our approach cannot, by design, find meaning for these intergenic SNPs. The ways in which intergenic SNPs influence disease are of particular interest to biologists and physicians. Thus, we are currently developing other measures for the similarity of those SNPs, using mechanisms such as microRNAs and other regulatory elements, which we believe strongly associate diseases with one another, since protein–protein interactions are not the only mechanism by which diseases are connected. Further, the biological similarity of two traits anchored on Information Theory is assigned according to the GO terms annotated to the genes hosting the GWAS-associated SNPs, and thus each pair of traits may have a different number of host genes and a different number of GO annotations to these genes. We completed comprehensive empirical distributions for each pair of traits to obtain a p value specific to the similarity score of each pair of traits. In future studies, intertrait similarity connections will be constructed by combining these empirical probabilities with much lower similarity scores to calculate thresholds rather than using a stringent threshold based mainly on the similarity score as we did in this study. We expect to cover more intertrait connections with shared genes at lower but significant ITS scores. Finally, there may be multiple shortest PPIN paths between similar host genes. In these cases, we selected one path, and we will explore alternates in the future.
In this study, we explored trait-associated intragenic SNPs discovered in GWAS and developed a novel measure for intertrait similarity based on the host genes of these SNPs. We found correlations between SNP-associated gene–gene ITS values and the shortest protein–protein interaction paths, which were then used to discover potential associations among diseases. The case study demonstrates the utility of such an approach and suggests a novel genetic architecture of complex diseases, beyond straightforward SNP or gene overlap, and canonical pathway analyses. Although identifying the mechanisms and genetic pathways that cause these diseases from GWAS SNPs has been vexing, our network provides a means of hypothesis generation by establishing mechanistic/functional groupings of traits by originating our study from these SNPs. Additionally, we achieved modularity of the mechanism/function-anchored similarity network by grouping these traits into possible metatraits that recapitulate, in part, known relationships such as ‘cancers’ and ‘metabolic syndrome’ and identify potential new connections. Thus, by delving into shared phenotypic and genetic similarity of biomodules, SNP-associated networks hold promise for understanding the genetic architecture of complex pathophysiological conditions. Further, the proposed network differs fundamentally from canonical ones that are manually curated by biologists, which by definition are limited to existing biologically validated knowledge. In contrast, unbiased ‘biological similarity’ and ‘protein interaction’ networks anchored on genetic polymorphisms allow exploration of non-canonical mechanisms underpinning diseases. One can use drugs developed for one disease for another with shared molecular mechanisms, which differs from current drug development paradigms reliant on single-gene targets and known canonical pathways. This approach also provides testable hypotheses for drug repositioning anchored on three scales of biology: biological function similarity, protein interactions, and genetic polymorphisms. We propose that unbiased ‘network targeting’ methods have the potential to invigorate the investigation of therapeutic targets. In summary, we report a biological similarity network that demonstrates the genetic architecture of complex diseases derived from intragenic SNPs.
Supplementary Material
Acknowledgments
We are grateful for valuable comments from Kelly Regan, Dr Xinan Yang, Gurunadh Parinandi, Colleen Kenost and Mike Burton, and suggestions from anonymous reviewers of AMIA 2011 Annual Symposium and from this journal. We are also grateful for the resources provided by the Computation Institute at the University of Chicago and Argonne National Laboratory, and specifically acknowledge the assistance of Lorenzo Pesce and Neil Bahroos.
Footnotes
Funding: This work was supported in part by NIH grants (UL1RR029879, 1S10RR029030-01 BEAGLE, and K22LM008308).
Competing interests: None.
Contributors: HL conducted the metrics design and computational analysis of the study, YL carried out the early development of the metrics, JLC and ER performed the case studies, JL made the visualization analysis, and YAL conceived and directed the project.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The data are provided as supplementary tables.
References
- 1.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol 2007;3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong Q, Simonis N, Li QR, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol 2009;5:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKusick VA. Mendelian inheritance in man and its Online version, OMIM. Am J Hum Genet 2007;80:580–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ioannidis JP, Castaldi P, Evangelou E. A compendium of genome-wide associations for cancer: critical synopsis and reappraisal. J Natl Cancer Inst 2010;102:846–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barabási AL. Network medicine—from obesity to the “diseasome”. New Engl J Med 2007;357:404–7 [DOI] [PubMed] [Google Scholar]
- 6.Meyre D, Delplanque J, Chevre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet 2009;41:157–9 [DOI] [PubMed] [Google Scholar]
- 7.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yildirim MA, Goh KI, Cusick ME, et al. Drug-target network. Nat Biotechnol 2007;25:1119–26 [DOI] [PubMed] [Google Scholar]
- 9.Suthram S, Dudley JT, Chiang AP, et al. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput Biol 2009;6:e1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidalgo CA, Blumm N, Barabasi AL, et al. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol 2009;5:e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J, Lee DS, Christakis NA, et al. The impact of cellular networks on disease comorbidity. Mol Syst Biol 2009;5:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol 2006;24:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sam L, Liu Y, Li J, et al. Discovery of Protein-interaction networks shared by diseases. Pacific Symposium on Biocomputing, Hawaii, USA: World Scientific; 2007:76–87 [PMC free article] [PubMed] [Google Scholar]
- 14.Lage K, Karlberg EO, Storling ZM, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol 2007;25:309–16 [DOI] [PubMed] [Google Scholar]
- 15.Lee DS, Park J, Kay KA, et al. The implications of human metabolic network topology for disease comorbidity. Proceedings of the National Academy of Sciences. 2008;105:9880–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sirota M, Schaub MA, Batzoglou S, et al. Autoimmune disease classification by Inverse association with SNP alleles. PLoS Genet 2009;5:e1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh KI, Cusick ME, Valle D, et al. The human disease network. Proc Natl Acad Sci 2007;104:8685–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrenas F, Chavali S, Holme P, et al. Network Properties of complex human disease genes identified through genome-wide association studies. PLoS One 2009;4:e8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 2009;106:9362–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao Y, Sam L, Li J, et al. Information theory applied to the sparse gene ontology annotation network to predict novel gene function. Bioinformatics 2007;23:i529–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen LJ, Kuhn M, Stark M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 2009;37(Suppl 1):D412–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas HC, Leiserson CE, Rivest RL, et al. All-Pairs Shortest Paths. Introduction to Algorithms. 2nd edn Cambridge, Massachusetts: MIT Press, 2001:620–42 [Google Scholar]
- 25.Chua HN, Sung WK, Wong L. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics 2006;22:1623–30 [DOI] [PubMed] [Google Scholar]
- 26.Hishigaki H, Nakai K, Ono T, et al. Assessment of prediction accuracy of protein function from protein–protein interaction data. Yeast 2001;18:523–31 [DOI] [PubMed] [Google Scholar]
- 27.Lin D. An Information-theoretic Definition of Similarity. 15th International Conference on Machine Learning. Madison, Wisonson, USA, Morgan Kaufmann; 1998:296–304 [Google Scholar]
- 28.Pesquita C, Faria D, Falcao A, et al. Semantic similarity in Biomedical Ontologies. PLoS Comput Biol 2009;5:e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrauwen I, Venken K, Vanderstraeten K, et al. Involvement of T-cell receptor-[beta] alterations in the development of otosclerosis linked to OTSC2. Genes Immun 2010;11:246–53 [DOI] [PubMed] [Google Scholar]
- 30.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer 2004;4:177–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanetsky PA, Mitra N, Vardhanabhuti S, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet 2009;41:811–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore S, McDiarmid LA. Stem cell factor and chronic myeloid leukemia CD34+ cells. Leuk Lymphoma 2000;38:211–20 [DOI] [PubMed] [Google Scholar]
- 33.Schrauwen I, Ealy M, Huentelman MJ, et al. A genome-wide analysis identifies genetic variants in the RELN gene associated with otosclerosis. Am J Hum Genet 2009;84:328–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evangelisti C, Florian MC, Massimi I, et al. MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J 2009;23:4276–87 [DOI] [PubMed] [Google Scholar]
- 35.James C, Ugo V, Le Couedic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005;434:1144–8 [DOI] [PubMed] [Google Scholar]
- 36.Hansen NT, Brunak S, Altman RB. Generating genome-scale candidate gene lists for pharmacogenomics. Clin Pharmacol Ther 2009;86:183–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirota M, Dudley JT, Kim J, et al. Discovery and preclinical validation of drug Indications using Compendia of Public gene expression data. Sci Transl Med 2011;3:96ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lussier YA, Chen JL. The emergence of genome-based drug repositioning. Sci Transl Med 2011;3:96ps35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudley JT, Sirota M, Shenoy M, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med 2011;3:96ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.