Abstract
Informatics for integrating biology and the bedside (i2b2) seeks to provide the instrumentation for using the informational by-products of health care and the biological materials accumulated through the delivery of health care to conduct discovery research and to study the healthcare system in vivo. This complements existing efforts such as prospective cohort studies or trials outside the delivery of routine health care. i2b2 has been used to generate genome-wide studies at less than one tenth the cost and one tenth the time of conventionally performed studies as well as to identify important risk from commonly used medications. i2b2 has been adopted by over 60 academic health centers internationally.
Keywords: Analysis, automated learning, bioinformatics, clinical informatics, data models, discovery, display, genomics, image representation, information storage and retrieval (text and images), knowledge representations, linking the genotype and phenotype, medical records, natural-language processing, processing, text and data mining methods, visualization of data and knowledge
Health care has grown so large that it encompasses multiple national agendas. Such a large presence requires instrumentation of the healthcare system to understand what is happening to us, the recipients of health care, and to be able to efficiently conduct research to improve healthcare delivery and to improve the state of biomedicine by advancing its science. Informatics for integrating biology and the bedside (i2b2) seeks to provide this instrumentation for using the informational byproducts of health care and the biological materials accumulated through the delivery of health care. This complements existing efforts to create prospective cohort studies or trials outside the delivery of routine health care. In the words of then director of the Congressional Budget Office, Peter Orzag,1 ‘…Clinical trials could be more persuasive but also more time consuming, and there is probably a limit to how many comparative trials could be undertaken effectively at any given time… if the issues of access and privacy could be addressed… [electronic medical records] could provide more comprehensive information both about the health histories of different patients and about their health outcomes. That additional information would make controlling for differences among patients receiving different treatments easier and would allow studies to address a broader set of outcomes than mortality.’ His report then makes it clear that using such data is not straightforward. Demonstrating how and when to use this hard-won clinical data is our primary mission and challenge.
Outputs
When we first defined the mission in 2002, the proposition was generally received as somewhat far-fetched and ill-fitting between the alternatives of prospectively organized clinical studies or cohort studies and the ongoing use of claims data to inform health services researchers. We were fortunate at the time we wrote the original proposal, as we explicitly recognized in the proposal itself, that we were in a position to leverage a large pre-existing investment in health information technology. Nonetheless, at the time we did not have any broadly shared tools nor a mechanism to allow the conduct of research over millions of patients including measurements of their biosamples. Six years later, i2b2 software has been adopted in over 60 academic health centers in the USA, including over half of the Clinical Translational Science Award (CTSA) awardees and 10 international medical centers. We now support an academic users' group of over 250 members from over 65 independent institutions that meets biannually for code workshops, discussion of application issues, preview of coming software, and networking (http://www.i2b2.org/work/aug.html).
The i2b2 workbench communicates with the i2b2 hive through XML-based web services. The i2b2 hive is an extensible architecture that allows new cells to be added and their functionality revealed through plug-ins in the i2b2 workbench and the i2b2 web client. The same web services are also used for all communication between cells within the i2b2 hive itself.
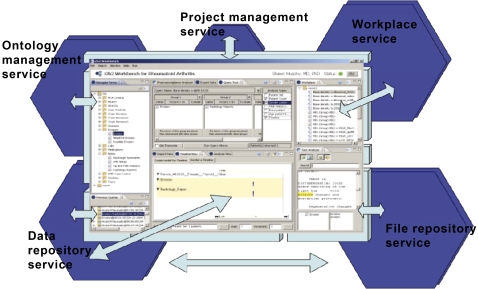
The i2b2 toolkit includes a set of software components (‘i2b2 cells’) organized into collections (‘i2b2 hives’, see figure 1) some of which are core (eg, authentication, database services, ontology services) and others of which are optional (eg, natural language processing; NLP). i2b2's adoption of a star schema architecture that communicates by XML messaging has enabled many i2b2 adopters to easily develop related cells that are in turn shared with the broader community.2 Moreover, the user-facing analytical functionality (the i2b2 workbench) comes in two flavors: a Java application and a Javascript web client, both of which allow the addition of user-developed additional functionality (‘plug-ins’).
Figure 1.
Informatics for integrating biology and the bedside (i2b2) workbench: the user-facing component of the i2b2 software system.
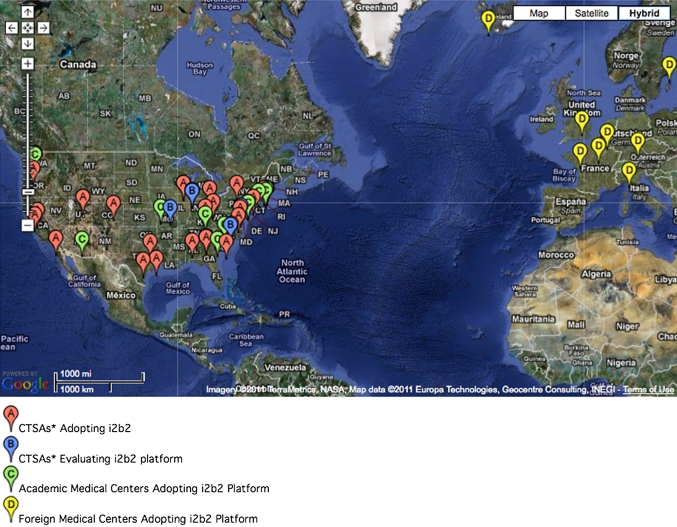
Stimulated by the need at our local clinical translational science center (Harvard catalyst) to link its own heterogeneous and competing hospitals and challenged by concerns of competitiveness and patient privacy inherent in a ‘one mega-database’ solution, we chose to build on top of previous efforts in distributed queries (W3EMRS,3 shared pathology informatics network)4–6 to create an interface that to the end-user would appear identical to a standard i2b2 query interface but would instead be a dynamic query to multiple i2b2 database instances (shared health research informatics network, SHRINE).7 SHRINE is a general purpose clinical querying protocol that can be adapted to many other types of data repositories. Following implementation of the individual i2b2 instances at the four main Harvard hospitals, the catalyst SHRINE entered α testing in 2008 and went into production in January 2011 for the sharing of aggregate counts of patients with defined inclusion/exclusion criteria for laboratories, diagnoses, demographics, and medications. We have also successfully supported i2b2 adopters at the University of Washington, Seattle (UWash), University of California, San Francisco (UCSF), and University of California, Davis (UCD) CTSA to install and demo a SHRINE instance that linked these CTSA to Harvard catalyst. The west coast application (http://www.i2b2cictr.org)8 is now fully active for selected queries. More recently, a pediatric rheumatology consortium (CARRAnet), a registry of patients across 60 institutions, and a pediatric inflammatory bowel consortium of at least 40 institutions, each represented by its own i2b2 repository, established their own SHRINE systems. Other members of the i2b2 network (see figure 2) are now considering creating or joining regional or national SHRINE networks.
Figure 2.
Geographical distribution of over 60 academic health centers (50 in the USA). Some locations (eg, San Francisco and Boston) have more sites than can be shown at the map's resolution. The map does not include the participants in the CARRAnet or pediatric inflammatory bowel registries.
Natural language processing
From the outset, we recognized that sufficiently accurate and detailed phenotyping for our proposed genomic studies would require that we engage in a significant investment in the methodologies of NLP. It was also clear that there were many research groups that could contribute to moving forward the state of the art. To encourage the development of NLP technologies, in addition to our own, for analysis of medical records, in 2006 we organized the first NLP shared task on clinical records. This shared task, held in conjunction with the annual AMIA meeting, focused on technologies for automatic de-identification and for automatic evaluation of the smoking status of patients based on the information contained in narrative patient records. We prepared this shared task by putting together a collection of actual medical discharge records, which were scrubbed for Protected Health Information (PHI), first automatically and then manually. We generated the ground truth for automatic de-identification and for automatic evaluation of the clinical status of the patients. We made part of the generated ground truth available to research communities and invited the development of systems for a new challenge task. The number of teams competing, drawn from a diverse international group with both academic and commercial participants, has grown every year, currently exceeding 30 teams representing 38 organizations. These efforts have resulted in over two dozen publications.9–17
Collaborations
During the past 5 years new efforts directly related to i2b2 have resulted in at least 27 new collaborative grants. As envisioned by the original Request For Applications (RFA) these represent extensions of work begun with National Center for Biomedical Computing (NCBC) funding, development of analytical tools for integration with the i2b2 hive, and support to CTSA and academic health centers to install, enhance, and/or apply their own i2b2 instances for clinical research studies.
Educating the next generation
We elected from the outset to contribute to the next generation of computational scientists, integrative genomicists, and bioinformaticians by enticing undergraduate students with strong analytical backgrounds into graduate studies in these fields. We did so by establishing de novo a summer program (Summer Institute in Bioinformatics and Integrative Genomics, http://www.i2b2.org/training/index.html) in partnership with the Harvard–MIT Division of Health Sciences and Technology that offers qualified students a 9-week intensive immersion through didactic lectures and case studies with top researchers, a communications tutorial and a mentored research project. Since our first class in year 2 (2005), we have graduated 82 students, including 38 women and 30 underrepresented minorities. Of the 60 who have graduated from college, at least 33 are now in graduate programs, including four in our health sciences and technology PhD program of the same name. Funding for this i2b2 program was procured from multiple sources.
Applications
When we first proposed to conduct genome-scale studies on populations using the informational and biological by-products of healthcare delivery, the idea was greeted with skepticism. Nine years later we just completed a review18 of the field of electronic health record-driven genomic research (EDGR), a recognition that this domain of genomic research had come into its own. This review, not coincidentally, was issued on the heels of a flurry of publications from i2b2 and others, particularly the eMERGE network describing successful applications of EDGR.19–29 Among the highlights of our work and that of the eMERGE network (the leaders of which sit on our external advisory committee) is the convincing validation of findings made in other studies in a broad array of phenotypes ranging from rheumatoid arthritis, major depressive disorders, and several other diseases. In all these studies, not only was the directionality of the OR of the Single Nucleotide Polymorphism (SNP) reproduced, but within 95% confidence limits the magnitude of the effects were comparable, all at least at an order of magnitude lower time and financial costs.21 Moreover, unlike the previous conventional cohort studies we were able to leverage the source of data of EDGR to identify and measure the effect size of these same SNP in populations other than those originally studied. In particular, we were able to see a partial overlap of the effect sizes in African-American and Hispanic populations in, for example, anti-citrullinated protein antibody-positive individuals with rheumatoid arthritis. Unlike conventional cohort studies in which it is challenging to reach sufficient representation of underrepresented minorities, by virtue of the overrepresentation of the same minorities in the patient population pool of large academic health centers, we were able to achieve very substantial study sizes in these otherwise neglected populations, consonant with the mandate of Collins et al30 in 2003. Furthermore, of very important significance for the scalability of this approach, the reproducibility of the phenotypes from codified data and natural language processed terms was very high and over 90% across Harvard and Vanderbilt Medical Center hospitals (R. Plenge, personal communication). That these phenotypes tuned for performance in one institution worked well in another without extensive re-tuning bodes extremely well for the future of EDGR.
Over the past 5 years i2b2 investigators have succeeded in achieving several notable demonstrations of the use of electronic health records system data for pharmacovigilance. First, an important proof of concept was the identification of a very large spike in cardiovascular mortality that after the fact we were able to convincingly associate with cyclooxygenase 2 inhibitor treatment, notably Vioxx.31 We were then able, in the middle of the controversy around the use of the oral hypoglycemic agent Avandia (rosiglitazone), to identify high RR for myocardial infarction with this drug even compared with others in the same class and prescribed for exactly the same indications, namely Actos (pioglitazone).32 Our study was cited among the handful of studies by the US Food and Drug Administration (FDA) in selecting to ‘black box’ Avandia. This use of electronic health records data rather than claims data, in which we leverage our access to the deeper clinical characterization of the health record, was a breakthrough demonstration that has stimulated many other such efforts.
It was in this context that our colleague, Russ Altman at the Stanford Symbios NCBC, chose to explore a finding from the FDA voluntary reporting database that suggested that it might be an interaction between one statin and one particular selective serotonin re-uptake inhibitor, pravastatin and paroxetine,33 that was associated with hyperglycemia. To validate this finding he contacted both the i2b2 team in Boston and the Vanderbilt team to see if we could validate the findings in the FDA database. Remarkably, in less than 1 month and with only a small number of phone calls and emails, we were able to validate these findings using the i2b2 methodological armamentarium. These results bode well for future efforts to detect population-level pharmacological effects.
Current and future goals
We have set for ourselves as the next methodological challenge the development of virtual cohort studies (VCS) encompassing the population of a healthcare system as study subjects and asking questions of efficacy, unforeseen adverse events and the identification of clinically relevant subpopulations. This is a substantial step forward from the pharmacovigilance studies carried out for drugs for which there were already previous suspicions and EDGR studies for association studies with clear case definitions. We will determine what findings that are obtained in the conventional cohorts are reproducible in the VCS and which findings in VCS provide insights not available to those studies. If we are successful, then the use of electronic health records for comparative effectiveness studies and as an alternative to conventional study design can be envisaged.
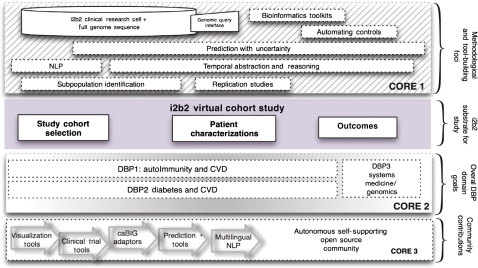
Developing VCS informs the direction of methodological developments. We are planning several efforts to improve the performance of i2b2 NLP techniques to capture ever more finely grained phenotypes such as disease activity and in a semi-supervised manner that requires far less technical oversight than previously. To give better insights into the temporal sequence of events across populations we will develop temporal abstraction and reasoning tools on top of the rudimentary time-stamped representation of most clinical databases. To address foursquare the methodological challenges of running VCS, we have assembled a joint team of leading biostatisticians, computer scientists and biomedical informaticians to devise, and find the limitations of, prediction methods, automated subpopulation identification, to assess the incremental value of conventional clinical and genomic/epigenomic markers, and to estimate effect sizes in the i2b2 convenience populations. Previously in i2b2 projects, clinical data were analyzed and patient populations were selected through the i2b2 workbench but the associated SNP genotype data were analyzed in separate third-party bioinformatics applications. We are now adopting open source bioinformatics analytical and workflow frameworks to integrate genotypic and full sequence data into the i2b2 workbench. To find unanticipated events across large multivariate databases, we will be investigating scalable solutions for hypothesis-free probabilistic modeling and evaluating their robustness in VCS as well as in the conventionally constructed cohort studies. As in the past, our driving biology projects (DBP) continue to serve both as a vital laboratory for the methodological work conducted by our interdisciplinary team, but also as essential proof of principle exemplars. By serving as β testers of our evolving software, our clinical researchers serve both as evaluators of existing tools and definers of additional needed functionality as they endeavor to create the crisp virtual cohorts required for successful deep dives from phenotype to genotype.22 23 As diagrammed below, we have extended our disease-specific foci with a series of DBP designed to investigate shared systemic themes of inflammation as expressed by cardiovascular disease. We will attempt to conduct integrative analyses across the clinical, genetic and epigenetic components of these themes across disparate diseases classes (in the third DBP) to advance our understanding of the underlying pathobiology.
Broadly, we have sought to leverage the creativity of the hundreds of members of our academic users group to take the additions that they have built or are about to build for added functionality for i2b2 (a sampling of which is listed in table 1). By establishing shared open-source governance mechanisms and the resources to incorporate these multiple highly useful and sought-after modules, we plan to generate a stable and enduring i2b2 ecosystem (see figure 3).
Table 1.
Sampling of tool/cell development from the i2b2 community
| Institution | i2b2 Tool/cell development |
| UCSF and UW CTSA | Health ontology mapper |
| Brigham and Women's Hospital | Sample acquisition and management cell |
| Cincinnati Children's Hospital Medical Center CTSA | i2b2-based patient registry and toolkit |
| Brigham and Women's Hospital | Genomics analysis results library integrated cell |
| NCBO/Stanford University | Ontology cell |
| University of Washington CTSA | Clinical trials cohort selection cell |
| Universities of Erlangen and Goettingen | Universal setup script |
| Boston University CTSA | Temporal modeling and health outcome monitoring and evaluation cell |
| University of Pavia, Italy | ONCO-i2b2: a bioinformatics tool to integrate biobank information and clinical data in oncology |
CTSA, Clinical and Translational Science Award; i2b2, informatics for integrating biology and the bedside; NCBO, National Center for Biomedical Ontology; UCSF, University of California, San Francisco; UW, University of Washington, Seattle.
Figure 3.
The informatics for integrating biology and the bedside (i2b2) roadmap forward. The top segment illustrates the tasks of core 1 in i2b2 in developing methodologies to support virtual cohort studies. The segment below outlines the components of virtual cohort studies and below that the three driving biology projects (DBP) described in the next. The bottom segment describes the various components of the open source community contributions to i2b2 that will be supported and/or integrated by core 3. CVD, Cardiovascular disease; NLP, natural language processing.
Footnotes
Funding: The authors were supported in part by NIH funding for National Centers for Biomedical Computing, U54 LM008748.
Competing interests: None.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1.Orzag P. Research on the comparative effectiveness of medical treatments: issue and options for an expanded federal role. A CBO Paper. Washington, DC: Congressional Budget Office, 2007:1–48 [Google Scholar]
- 2.Murphy SN, Mendis M, Hackett K, et al. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc 2007:548–52 [PMC free article] [PubMed] [Google Scholar]
- 3.Kohane IS, Greenspun P, Fackler J, et al. Building national electronic medical record systems via the World Wide Web. J Am Med Inform Assoc 1996;3:191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Namini AH, Berkowicz DA, Kohane IS, et al. A submission model for use in the indexing, searching, and retrieval of distributed pathology case and tissue specimens. Stud Health Technol Inform 2004;107:1264–7 [PubMed] [Google Scholar]
- 5.Holzbach AM, Chueh H, Porter AJ, et al. A query engine for distributed medical databases. Medinfo 2004;2004:1519 [Google Scholar]
- 6.Drake TA, Braun J, Marchevsky A, et al. A system for sharing routine surgical pathology specimens across institutions: the Shared Pathology Informatics Network. Hum Pathol 2007;38:1212–25 [DOI] [PubMed] [Google Scholar]
- 7.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:624–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson N, Chilana P, Anderson K, et al. Implementing cross-institutional clinical discovery for population based translational research. AMIA Spring Congress. Orlando, FL, 2009 [Google Scholar]
- 9.Uzuner Ö, Luo T, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007;14:550–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savova G, Ogren P, Duffy P, et al. Patient smoking status identification within Mayo Clinic Life Sciences System. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uzuner Ö, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uzuner Ö, Sibanda TC, Luo Y, et al. A de-identifier for medical discharge summaries. Artif Intell Med 2008;42:13–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uzuner Ö. Recognizing obesity and co-morbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uzuner Ö, Mailoa J, Sibanda T. Semantic Relations for Problem-Oriented Medical Records. Fall Symposium of the American Medical Informatics Association (AMIA 2009). San Francisco, CA, 2009:661 [Google Scholar]
- 15.Uzuner Ö, Zhang X, Sibanda T. Machine learning and rule-based approaches to assertion classification. J Am Med Inform Assoc 2009;16:109–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein I, Uzuner Ö. Specializing for predicting obesity and its co-morbidities. J Biomed Inform 2009;42:873–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savova G, Clark C, Zheng J, et al. The Mayo/MITRE system for discovery of obesity and its comorbidities. AMIA 2nd i2b2 challenge workshop. Washington, DC, 2008 [Google Scholar]
- 18.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet 2011;12:417–28 [DOI] [PubMed] [Google Scholar]
- 19.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Himes BE, Klanderman B, Kohane IS, et al. Assessing the reproducibility of asthma genome-wide association studies in a general clinical population. J Allergy Clin Immunol 2011;127:1067–9 [DOI] [PubMed] [Google Scholar]
- 21.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res 2009;19:1675–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurreeman F, Liao K, Chibnik L, et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet 2011;88:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010;26:1205–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation 2010;122:2016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumitrescu L, Ritchie MD, Brown-Gentry K, et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet Med 2010;12:648–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulley J, Clayton E, Bernard GR, et al. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci 2010;3:42–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010;86:560–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins FS, Green ED, Guttmacher AE, et al. US National Human Genome Research Institute A vision for the future of genomics research. Nature 2003;422:835–47 [DOI] [PubMed] [Google Scholar]
- 31.Brownstein JS, Sordo M, Kohane IS, et al. The tell-tale heart: population-based surveillance reveals an association of rofecoxib and celecoxib with myocardial infarction. PLoS One 2007;2:e840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brownstein JS, Murphy SN, Goldfine AB, et al. Rapid identification of myocardial infarction risk associated with diabetic medications using electronic medical records. Diabetes Care 2010;33:526–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011;90:133–42 [DOI] [PMC free article] [PubMed] [Google Scholar]