Abstract
The National Center for Biomedical Ontology is now in its seventh year. The goals of this National Center for Biomedical Computing are to: create and maintain a repository of biomedical ontologies and terminologies; build tools and web services to enable the use of ontologies and terminologies in clinical and translational research; educate their trainees and the scientific community broadly about biomedical ontology and ontology-based technology and best practices; and collaborate with a variety of groups who develop and use ontologies and terminologies in biomedicine. The centerpiece of the National Center for Biomedical Ontology is a web-based resource known as BioPortal. BioPortal makes available for research in computationally useful forms more than 270 of the world's biomedical ontologies and terminologies, and supports a wide range of web services that enable investigators to use the ontologies to annotate and retrieve data, to generate value sets and special-purpose lexicons, and to perform advanced analytics on a wide range of biomedical data.
Keywords: Collaborative technologies, knowledge representations, knowledge acquisition and knowledge management, controlled terminologies and vocabularies, ontologies, knowledge bases, applications that link biomedical knowledge from diverse primary sources (includes automated indexing), statistical analysis of large datasets, methods for integration of information from disparate sources, discovery, and text and data mining methods, automated learning, information retrieval, HIT data standards, representing, identifying, and modeling biological structures, developing and refining ehr data standards (including image standards)
Mission
Advances in computing power and new computational techniques have changed the way researchers approach biology, medicine, and indeed all of science. In biomedicine, one of the most fruitful approaches has been to use software tools and knowledge resources known as ‘ontologies’—machine-processable descriptions of scientific domains—that can promote the integration of disparate data sources. We have shown that such resources can enable data aggregation, improve search, and allow the detection of new associations that were previously not detectable. It is now possible to demonstrate computationally correlations among genes, diseases, treatments, and outcomes, to use these correlations to efficiently direct research into potentially fruitful areas, and to translate the insights from this research to the practice of medicine. Achieving these integrative analyses requires software systems that take advantage of the semantics of these areas and that can intelligently negotiate domains and knowledge sources, identifying commonality across systems that use different and conflicting vocabularies, while understanding apparent differences that may be concealed by the use of superficially similar terms.1 An appropriate ontology provides the cornerstone of software for bridging systems, domains, and resources.2 Ontologies are the foundation of all semantic technologies in e-science, and are a critical component of multi-disciplinary and translational research in biomedicine.3
The National Center for Biomedical Ontology (NCBO) has become a leading scientific organization for bringing semantic technology to biomedicine. With core performance sites at Stanford University, the Mayo Clinic, the University of Victoria, and the University at Buffalo, our team works to create and disseminate national infrastructure that supports the use of computer-stored knowledge in the form of ontologies. Our overall mission comprises four main objectives:
to create and maintain a repository of biomedical ontologies and terminologies;
to build tools and web services to enable the use of ontologies and terminologies;
to educate our trainees and the scientific community broadly about biomedical ontology and about NCBO technology;
to collaborate with a variety of groups who develop and use ontologies and terminologies in biomedicine.
Outputs of the NCBO
The outputs of our Center can be best described in terms of the overall objectives of our work.
Repository of biomedical ontologies
The NCBO's BioPortal provides access to more than 270 biomedical ontologies and controlled terminologies.4 5 Users come to the BioPortal website to browse biomedical ontologies and to search for specific ontologies that have terms that are relevant for their work. A cancer biologist may learn from BioPortal that the Gene Ontology offers the best coverage for annotating her experimental data with terms related to cell division, or that she can access more precise terms in the National Cancer Institute (NCI) Thesaurus. She may discover that the Mouse Adult Gross Anatomy Ontology can be used to describe the body parts from which her experimental specimens were obtained, or that the National Drug File–Reference Terminology provides valuable information about the properties of the drugs used in her experiments.
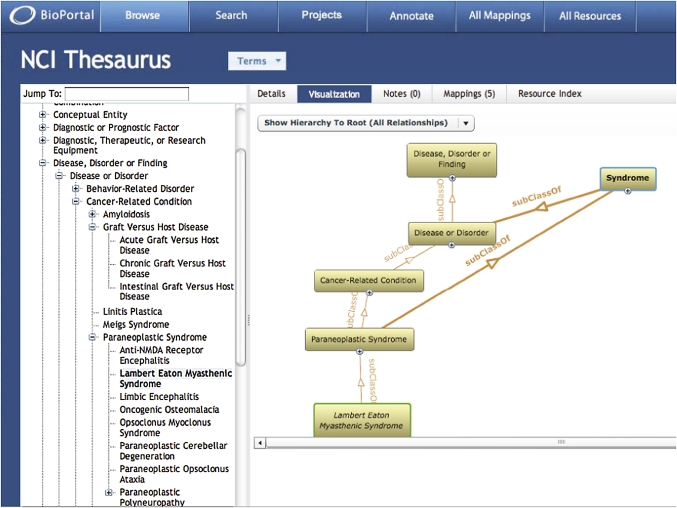
BioPortal enables users to navigate ontologies using a standard tree browser. Users can also visualize resources in BioPortal using special tools that offer cognitive support for understanding the complexities of large ontologies (figure 1).
Figure 1.
The BioPortal ontology repository. In the figure, the user is browsing the National Cancer Institute Thesaurus. A tree browser along the left-hand side of the screen allows the user to navigate the taxonomic hierarchy of the ontology. The visualization window on the right facilitates exploration of complex relationships—here, the pathway between the selected term (Lambert–Eaton myasthenic syndrome) and its superclasses in the hierarchy. The menu bar above the visualization window allows the user to change the view to examine the details of the selected term, end-user notes regarding the term or its descendants, mappings between the term and related terms in other ontologies, or links between the selected term and the data sources referenced in the National Center for Biomedical Ontology Resource Index.
When users need to understand the relationships between terms in two different ontologies, BioPortal provides mappings between the ontologies to enable direct comparisons. The mappings can inform the user that the term ‘lung’ in the Mouse Adult Gross Anatomy Ontology is related to the term ‘lung’ in the Foundational Model of (human) Anatomy or that the term ‘limb’ in the NCI Thesaurus is related to the term ‘extremity’ in the Mouse Adult Gross Anatomy Ontology.
The mappings between ontologies in BioPortal not only allow users to compare the use of related terms in different ontologies, but also allow analysis of how whole ontologies compare with one another. They allow us to identify ontologies that cluster together6 and to identify the degrees of overlap among ontologies.7 Like the UMLS metathesaurus,8 the mappings in BioPortal facilitate automated translation of terms among ontologies, but entail much more content. The mappings in BioPortal form the basis for what we refer to as the NCBO ‘mega’-thesaurus.
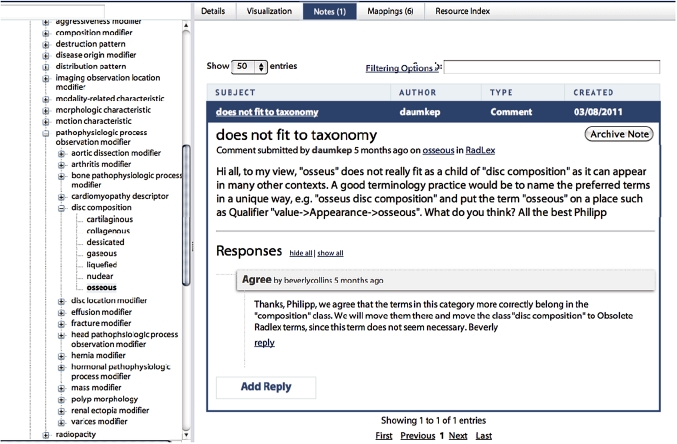
BioPortal is much more than an ontology repository, however. We have created the system as the nexus of an online community of ontology developers and ontology users who use BioPortal to view, comment on, and discuss the content of biomedical ontologies (figure 2). Registered users of BioPortal can not only upload new ontology content, but also mark up their content (or that of any other user) with highly granular comments about any ontology.9 Users can indicate where they believe ontologies may reflect inappropriate modeling decisions, and other users can respond to those comments in threaded discussions that the entire BioPortal community can monitor. These threaded conversations allow BioPortal to behave very much like a wiki for making annotations to ontology content, and they enable new users to locate regions of BioPortal's ontologies where modeling decisions have been particularly controversial and ontology developers to identify elements of their work that may benefit from refactoring in future versions of their ontologies. They also allow users to identify those groups of resources, such as are maintained by the Open Biological and Biomedical Ontologies (OBO) Foundry initiative,10 that have been subjected to a process of external review designed to ensure compliance with an evolving set of best practice principles.
Figure 2.
Notes in BioPortal. Registered users of BioPortal can comment on any of the ontologies in the repository. They can point out what they believe to be errors or can make suggestions for changes. Other users can respond to these comments and begin a threaded discussion. In the figure, a user has left a note in the RadLex Ontology suggesting that the term ‘osseous’ may be misclassified. Another user has left a note agreeing that the term needs to be relocated in a future version of RadLex.
BioPortal allows users themselves to post overarching reviews of the system's ontologies—and to post online very specific proposals for changes that ontology developers might want to consider in future revisions. BioPortal thus adopts Web 2.0 conventions to allow its users to communicate with one another about the NCBO's hosted ontologies in a highly interactive manner. The outcome of these capabilities is that BioPortal offers the equivalent of online, open, community-based peer review for the BioPortal ontology content.9
We are developing BioPortal so that computer-based ontology-development tools can access all its content programmatically—including the mappings between ontology terms and the notes about the ontology content contributed by members of the user community. Thus, users of the web-based version of the Protégé ontology editor11 can view BioPortal content directly from within the Protégé browser window, copy terms and other content from existing ontologies into new ontologies, review the notes and comments about previous versions of ontologies uploaded to BioPortal by their users, and act on those notes as they develop new versions. This integration of ontology authoring with community-based access to ontologies through BioPortal has been particularly important to groups developing large ontologies in an open, distributed fashion. For example, the World Health Organization is now using NCBO technology routinely in its global effort to develop the next edition of the International Classification of Diseases (ICD-11).12
Tools and web services
In addition to providing a comprehensive library of biomedical ontologies and terminologies, the NCBO develops tools and services that use those ontologies to aid biomedical investigators in their work. Although these tools are all available through a web-browser interface, most users access our software programmatically via web services.
NCBO Annotator
Perhaps the most widely used tool created by the NCBO is one that maps arbitrary keywords and natural-language text to standardized ontological terms. The NCBO Annotator thus takes as input some specified text and generates as output a set of terms derived from BioPortal-stored ontologies, such that the terms refer to concepts that the NCBO Annotator identifies in the text.13 It provides a mechanism to determine what the text is ‘about’ in terms of standardized, ontological entities. The structure of the ontologies in BioPortal permits the NCBO Annotator to associate the text not only with particular terms (eg, ‘adenocarcinoma of the lung’ from the NCI Thesaurus), but also with more general terms (eg, ‘neoplasm’). As a result, users are offered an extremely rich set of descriptors for the corresponding text, at different levels of granularity and generality.
NCBO Resource Index
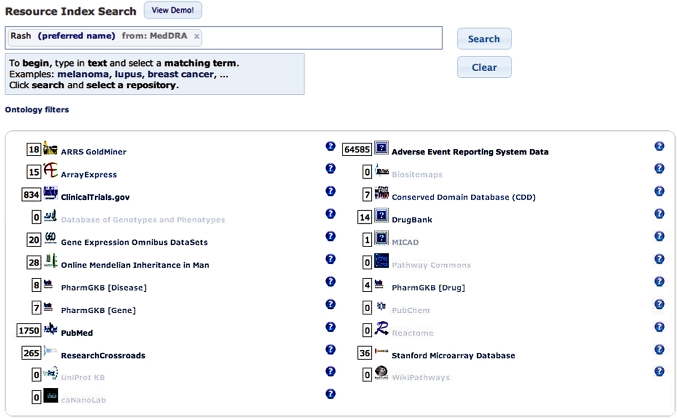
A common use of the NCBO Annotator is to ascribe ontological terms to the textual metadata that are associated with experimental datasets. The NCBO automatically runs the Annotator on a large collection of online datasets, linking the textual metadata associated with those data to all relevant ontological terms in BioPortal. The result is an enormous database of all the terms (and abstractions of those terms) that relate to the textual metadata (or text descriptions) found in a growing set of online data resources (such as the microarray datasets in the Gene Expression Omnibus or the individual protein descriptions in UniProt). We refer to this database as the NCBO Resource Index.14 A web-based interface allows investigators to search the Index, using terms in BioPortal-stored ontologies to locate relevant datasets from online repositories (figure 3). The Index offers the biomedical community a common interface for information retrieval, linking the dozens of ontologies in BioPortal to dozens of biomedical data resources. Thus, if an investigator is interested in learning what experimental data may have been archived online in public repositories that might be relevant to a particular term or set of terms, they can use the Index to search for the relevant data. The NCBO development team is linking new online data resources to the Resource Index on an ongoing basis.
Figure 3.
The user interface for the National Center for Biomedical Ontology's Resource Index. The Resource Index is a database that links each term of each ontology in BioPortal to online data and knowledge resources that may reference that term. In the figure, the user has entered a particularly vague term—‘rash’, as used in MedDRA. The system uses the Resource Index and the underlying ontological structure in which the term appears to allow the user to locate some 18 images in the American Roentgen Ray Society's GoldMiner repository of radiographs, 20 microarray datasets in the Gene Expression Omnibus, 28 records in Online Mendelian Inheritance in Man, and so on. Each of the associated datasets refers to patients with some kind of rash. Clicking on a particular resource description in the user interface allows the user to navigate to the actual data records that have been indexed.
NCBO Ontology Recommender Service
Investigators are often unsure about which of the dozens of ontologies in BioPortal provide the best coverage for capturing the entities in a particular application area. The NCBO Ontology Recommender Service15 takes as input representative textual data relevant to a domain of interest and returns as output an ordered list of ontologies available in BioPortal, the terms of which would be most appropriate for annotating the corresponding text.
NCBO Lexicon Builder
Users often turn to the terms of biomedical ontologies to create the ‘value sets’ that constitute the basis of ‘pick lists’ that allow users to make selections from menus when filling in computer-based forms. The NCBO Lexicon Builder16 also allows users to obtain more manageable subsets of large ontologies that are amenable to particular analyses, and to combine portions of different ontologies to create specialized collections of terms. The latter functionality is of particular interest to members of the natural-language processing community, who often need hand-crafted lexicons to drive named-entity recognition in particular domains.
Web widgets
Many NCBO services are called automatically through small collections of HTML program code that our Center makes available to web developers who wish to take advantage of our offerings. Developers can embed these ‘widgets’ in their code so that their web pages can immediately access BioPortal ontologies, value sets, mappings, and other resources.
Detailed information for developers who wish to access and employ NCBO tools, services, and widgets is available on a wiki maintained by the Center.17
Education and outreach
A full-time NCBO outreach coordinator has multifaceted responsibilities that include serving as a liaison to collaborating projects, shepherding new collaborations, and presenting NCBO technology to the scientific community. Our outreach coordinator hosts a very well attended, biweekly ‘Webinar’ series, in which members of the NCBO and the larger biomedical ontology community discuss their research; video recordings of past Webinars are archived on the NCBO website.18
The NCBO has an active dissemination program of custom-tailored workshops and tutorials. The Center is also a major sponsor of the International Conference on Biomedical Ontology.
Collaborative projects
The NCBO's tools and services are designed for use in support of the informatics activities of biomedical researchers. As a result, our collaborators tend to be well versed in biomedical informatics and understand the power that ontologies can offer their work in data annotation and indexing, natural-language processing, data mining, and decision support.
The NCBO has supported a series of Driving Biological Projects that have provided important use cases for the NCBO Annotator (eg, in the annotation of rat genome data19) and for the NCBO Resource Index (eg, to enable retrieval of information about therapeutic nanoparticles20). The use of ontology-driven analytics has allowed collaborators to interpret high-throughput data in novel ways,21 22 making both methodological and biological contributions. Other collaborators have used the rich content availability in BioPortal as a starting point for quality assurance of ontologies23 and for further enrichment of biomedical ontologies by processing text from electronic health records.24 Finally, our collaborations have led to the development of a burgeoning number of new ontologies for use by the biomedical community.10 12 25 26
The vast majority of biomedical investigators who take advantage of NCBO technology are not explicit collaborators, however. Most of the Center's users simply browse the BioPortal website or invoke NCBO web services as a routine element of their investigative work. Currently, some 16 000 users browse ontologies via BioPortal each month. During the same period of time, NCBO servers respond to more than 3 million programmatic web-service requests. It is extremely gratifying to the members of the Center that the NCBO apparently has become an indispensible technology resource for such a large community of biomedical scientists, and that the vast majority of these users have come to take the Center's services for granted.
Future goals
A major initiative of the NCBO in the coming years will concentrate on ensuring the scalability of our technology. As BioPortal acquires increasing numbers of ontologies (with increasing numbers of inter-ontology mappings and end-user notes), as more and more biomedical data resources are linked to the NCBO Resource Index, and as the number of users who access our technology via web browsers or via web services continues to grow, the NCBO must be able to accommodate the corresponding demand. Much of the Center's activity concerns ensuring a robust infrastructure for its technology, accommodating more content, more users, and more demands in as seamless a manner as possible.
The scientific work of the Center will focus on support for the management of the complete ontology life cycle, allowing users of ontology-development systems (such as Protégé,11 OBO-Edit,27 and LexWiki28) to integrate with BioPortal. The authors of ontologies will be able to publish their work directly in BioPortal and take advantage of end-user notes when revising their ontologies in subsequent versions.29 We will merge the processes of ontology authoring and ontology dissemination, and investigate whether the open peer-review process offered by BioPortal can lead to improvements in biomedical ontologies and enhanced adoption of the resultant ontologies by the community.
Other work will concentrate on new uses of the ontologies in BioPortal in the interpretation of high-throughput experimental data.30 We are also optimistic that the ontology-oriented techniques that we are developing will enable investigators to analyze data from electronic patient records in novel ways.31
More information about the NCBO is available from the Center's website.32
Footnotes
Funding: The National Center for Biomedical Ontology is supported by the NIH Common Fund, the National Human Genome Research Institute, and the National Heart, Lung, and Blood Institute through grant U54 HG004028.
Competing interests: None.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1.Shah NH, Musen MA. Ontologies for formal representation of biological systems. In: Studer S, Staab S, eds. Handbook on Ontologies. New York: Springer-Verlag, 2009:445–62 [Google Scholar]
- 2.Uschold M, Gruninger M. Ontologies: principles, methods, and applications. Knowl Eng Rev 1996;11:93–136 [Google Scholar]
- 3.Lussier YA, Bodenreider O. Clinical ontologied for discovery applications. In: Baker CJO, Cheung K-H, eds. Semantic Web: Revolutionizing Knowledge Discovery in the Life Sciences. New York: Springer-Verlag, 2006:101–19 [Google Scholar]
- 4.Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res 2009;37:W170–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whetzel PL, Noy NF, Shah NH, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res 2011;39:W541–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghazvinian A, Noy NF, Jonquet C, et al. What four million mappings can tell you about two hundred ontologies. Proceedings of the Eighth International Semantic Web Conference (ISWC 2009). Washington, DC, September, 2009. New York: Springer-Verlag, 2009 [Google Scholar]
- 7.Ghazvinian A, Noy NF, Musen MA. How orthogonal are the OBO Foundry ontologies? J Biomed Semantics 2011;2(Suppl 2):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32:D267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noy NF, Dorf MV, Griffith N, et al. Harnessing the power of the community in a library of biomedical ontologies. Proceedings of the Workshop on Semantic Web Applications in Scientific Discourse. Eighth International Semantic Web Conference (ISWC-2009). Chantilly, VA, 2009 [Google Scholar]
- 10.Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 2007;25:1251–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tudorache T, Vendetti J, Noy NF. WebProtégé: A lightweight OWL ontology editor for the Web. Proceedings of the Fifth OWLED Workshop on OWL: Experiences and Directions. Seventh International Semantic Web Conference (ISWC-2008). Karlsruhe, Germany, 2008 [Google Scholar]
- 12.Tudorache T, Falconer SM, Nyulas CI, et al. Supporting the Collaborative Authoring of ICD-11 with WebProtégé. AMIA Annu Symp Proc 2010:802–6 [PMC free article] [PubMed] [Google Scholar]
- 13.Shah NH, Nipun B, Jonquet C, et al. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinformatics 2009;10(Suppl 9):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah NH, Jonquet C, Chiang AP, et al. Ontology-driven indexing of public datasets for translational bioinformatics. BMC Bioinformatics 2009;10(Suppl 2):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonquet C, Musen MA, Shah NH. Building a biomedical ontology recommender web service. J Biomed Semantics 2010;1(Suppl 1):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parai GK, Jonquet CM, Xu R, et al. The Lexicon builder Web service: Building custom lexicons from two hundred bomedical ontologies. AMIA Annu Symp Proc 2010:587–91 [PMC free article] [PubMed] [Google Scholar]
- 17.http://www.bioontology.org/wiki/index.php/Main_Page (accessed 30 Jul 2011).
- 18.http://www.bioontology.org/webinar-series (accessed 30 Jul 2011).
- 19.http://qtlhighlighter.hmgc.mcw.edu/ (accessed 30 Jul 2011).
- 20.Thomas DG, Pappu RV, Baker NA. NanoParticle Ontology for cancer nanotechnology research. J Biomed Inform 2011;44:59–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mort M, Evani US, Krishnan VG, et al. In silico functional profiling of human disease-associated and polymorphic amino acid substitutions. Hum Mutat 2010;31:335–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar IN. Leveraging biomedical ontologies and annotation services to organize microbiome data from Mammalian hosts. AMIA Annu Symp Proc 2010:717–21 [PMC free article] [PubMed] [Google Scholar]
- 23.Verspoor K, Dvorkin D, Cohen KB, et al. Ontology quality assurance through analysis of term transformatons. Bioinformatics 2009;25:i77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K, Hogan WR, Crowley RS. Natural language processing methods and systems for biomedical ontology learning. J Biomed Inform 2011;44:163–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winslow RL, Saltz J, Foster I, et al. The CardioVascular Research Grid (CVRG) project. AMIA Annu Symp Proc 2010:77–81 [Google Scholar]
- 26.Tenenbaum J, Whetzel PL, Anderson K, et al. The Biomedical Resource Ontology (BRO) to enable resource discovery in clinical and translational research. J Biomed Inform 2011;44:137–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Day-Richter J, Harris MA, Haendel M, et al. OBO-Edit—an ontology editor for biologists. Bioinformatics 2007;23:2198–200 [DOI] [PubMed] [Google Scholar]
- 28.Jiang G, Solbrig H. LexWiki framework and use cases. The First Meeting of Semantic MediaWiki Users. Boston, MA, 2008 [Google Scholar]
- 29.Noy NF, Tudorache T, Nyulas CI, et al. The ontology life cycle: Integrated tools for editing, publishing, peer review, and evolution of ontologies. AMIA Annu Symp Proc 2010:552–6 [PMC free article] [PubMed] [Google Scholar]
- 30.Tirrell R, Evani U, Berman AE, et al. An ontology-neutral framework for enrichment analysis. AMIA Annu Symp Proc 2010:797–801 [PMC free article] [PubMed] [Google Scholar]
- 31.LePendu P, Racunas S, Iyer S, et al. Annotation analysis for testing drug safety signals. The Fourteenth Annual Bio-Ontologies Meeting. Conference on Intelligent Systems for Molecular Biology (ISMB-2011). Vienna, AT, July 2011. http://www.bio-ontologies.org.uk/programme (accessed 30 Jul 2011).
- 32.http://bioontology.org (accessed 30 Jul 2011).