Abstract
iDASH (integrating data for analysis, anonymization, and sharing) is the newest National Center for Biomedical Computing funded by the NIH. It focuses on algorithms and tools for sharing data in a privacy-preserving manner. Foundational privacy technology research performed within iDASH is coupled with innovative engineering for collaborative tool development and data-sharing capabilities in a private Health Insurance Portability and Accountability Act (HIPAA)-certified cloud. Driving Biological Projects, which span different biological levels (from molecules to individuals to populations) and focus on various health conditions, help guide research and development within this Center. Furthermore, training and dissemination efforts connect the Center with its stakeholders and educate data owners and data consumers on how to share and use clinical and biological data. Through these various mechanisms, iDASH implements its goal of providing biomedical and behavioral researchers with access to data, software, and a high-performance computing environment, thus enabling them to generate and test new hypotheses.
Keywords: Privacy technology, cloud computing, electronic health records, genomics, natural language processing, informatics, machine learning, predictive modeling, statistical learning, knowledge bases, translational research—application of biological knowledge to clinical care, developing/using clinical decision support (other than diagnostic) and guideline systems, knowledge acquisition and knowledge management, privacy technology, cloud computing, data mining, detecting disease outbreaks and biological threats, statistical analysis of large datasets, discovery, and text and data mining methods, automated learning, measuring/improving patient safety and reducing medical errors, monitoring the health of populations
Mission
Although it has been 10 years since the publication of the first complete draft of the human genome,1–3 relatively few examples of scientific team effort have followed, in part due to inadequate computational environments that enable collaborative projects. iDASH (integrating data for analysis, anonymization, and sharing) was conceived as a computational collaborative environment that could fill gaps in the methods for accessing biomedical data, software, and sophisticated computational infrastructure (figure 1). As a result, iDASH was designed to allow as many researchers as possible to leverage other researchers' work and accelerate discoveries. Because a critical component of responsible data sharing is to preserve the privacy of individuals whose data are being shared,4–7 we have undertaken the challenge of developing a secure environment in which access is granted on the basis of privacy technology and policies that are enforced according to constraints imposed by laws and regulations, institutional policies, and data contributors.
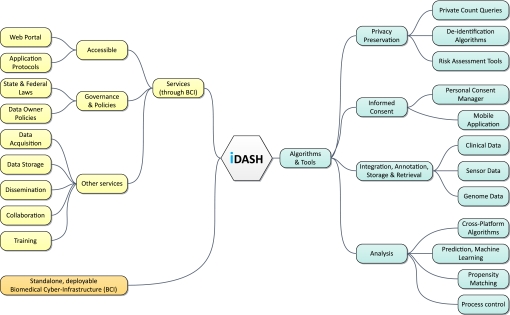
Figure 1.
Integrating data for analysis, anonymization, and sharing. iDASH, the newest National Center for Biomedical Computing, is a scientific team effort that brings together a multi-institutional team of quantitative scientists (information and computer scientists, biostatisticians, mathematicians, and software engineers) to develop algorithms and tools, services, and a biomedical cyber-infrastructure for use by biomedical and behavioral researchers. An illustration of the major components within each area is depicted in this figure. BCI, biomedical cyber-infrastructure.
Needs of biomedical and behavioral researchers that drive iDASH research and development
The biomedical or behavioral research projects (Driving Biological Projects or DBPs) that currently drive the development of our tools represent the multiplicity of data-rich projects funded by the National Institutes of Health.
In Molecular Phenotyping of Kawasaki Disease (KD), our collaborators are studying the molecular mechanisms of vasculitis and aneurysm formation8 to identify new therapeutic targets and tailor treatment. In iDASH's secure cloud, the research is enabled by a translational bioinformatics9 infrastructure that integrates data from genotyping (single-nucleotide polymorphism array data and whole-genome sequence for selected patients), gene expression (microRNA and RNA arrays and sequencing), and proteomic measurements with demographics, laboratory values, images, therapeutic interventions, and clinical phenotypes. Because KD is relatively rare, aggregating data from as many sources as possible is very important. iDASH will make the data available to the research community through proper distribution policies and extensive annotation to allow real and meaningful data reanalysis. We have developed compression tools10 to allow whole-genome sequence KD data to be included in electronic health records. The inclusion of genetic data requires extra attention to privacy preservation. We are thus developing algorithms to protect the privacy of disclosed data.11
In Individualized Intervention to Enhance Physical Activity, our collaborators are investigating how personal patterns of motion throughout the day associate with morbidity in order to create an interventional device that performs real-time behavior pattern recognition using machine learning algorithms.12 This team is iteratively designing the intervention system with the goal of having an interactive system that easily integrates into an individual's daily living. However, there is considerable risk of privacy breach when remote sensors are used without the proper infrastructure. Analogous to the needs of the KD project, continuous sensor data generated from this project need to be annotated, anonymized, and shared through the iDASH cyber-infrastructure in a privacy-preserving manner.
In Multi-institutional Surveillance of Medications, our collaborators are monitoring medication safety in a distributed environment. Post-market safety surveillance of newly approved medications is a complex task compounded by rapid diffusion of new medications into previously unstudied patient populations. Early detection of event rate elevations is promoted by an increased rate of patient accrual in this type of environment. In addition, because some of these events are rare, large numbers of patients and statistical process controls that are guided by event rate estimates obtained from calibrated predictive models are needed to reliably detect them.13 This multi-institutional team is utilizing iDASH's tools for observational cohort outcome surveillance. The researchers are leveraging iDASH's natural language processing (NLP) tools to extract findings from narrative text14 and will leverage tools from a related project (SCAlable National Network for Effectiveness Research or SCANNER, http://scanner.ucsd.edu). These tools will automate policies to enable access to data from institutions operating under different state laws and local policies for data sharing (one private, one federal-owned, and one state-owned institution).
Algorithms and tools for integrating data for analysis
SlimGene and genome query language
We have developed a tool for compressing high-throughput sequencing datasets driven by the needs of the KD project. On datasets with read information only (no quality values), the tool can achieve 400× compression, making the preservation of genomic information for an individual a trivial task.10 However, current sequencing technologies often have high error rates, and return quality values (log odds of probability of error) for each nucleotide. Compression of quality values is very challenging, and the developed tool still achieves 4–5× compression based on a Markov encoding of quality value dependencies. A theoretical framework is also provided to argue that a few bits are sufficient to encode quality values without a significant loss of performance. While compression makes it possible to archive large datasets, it must also allow querying for genetic variation. We are working at two levels to address this. First, we are developing an abstraction of software layers that allows genome analysis tools to interact with distributed and compressed datasets. Second, we are specifying and developing a genome query language that will allow researchers to query the genomic data for variations in a seamless and efficient manner.
AnyExpress
Another tool we have developed is AnyExpress,15 a toolkit that combines and filters cross-platform gene expression data. The cross-platform analysis of KD gene expression data, specifically motivated by the KD project, requires multiple, intricate processing at different layers on various platforms. To study the pathogenesis of KD, microRNA and RNA expression levels are determined through next-generation sequencing experiments in addition to existing expression data from heterogeneous microarray platforms that were generated through our collaborators' work with the International KD consortium. In contrast with other tools, which could provide incorrect results because of their tight coupling with specific versions of reference databases, AnyExpress handles multiple platforms with flexible software that supports custom changes, such as new statistical methods for preprocessing, updated versions of the reference genome, and new platform releases. AnyExpress also allows users to select reference sources according to their preferences.
Observational cohort event analysis and notification system (OCEANS)
We are developing a suite of flexible open-source statistical applications for the identification of event rate outliers related to a particular exposure within observational cohorts. An important challenge in monitoring safety of medications and medical devices is to determine appropriate expectations for rates of adverse events in the population receiving the new medication or device.16 Our statistical applications automate the risk-adjusted sequential surveillance of a structured summary of the electronic health record. This suite includes statistical methods such as risk-adjusted statistical process control, risk-adjusted sequential probability ratio testing, and Bayesian hierarchical logistic regression.17–19 These algorithms incorporate continuous risk adjustment using risk prediction models and automated propensity matching, as well as adjustment for repeated measurement α error inflation through multiple mechanisms.20 The toolkit also includes functions to study the diffusion of new medications into particular subpopulations of patients, to allow exploration of safety signals within these at-risk sub-populations, and to generate safety alerts in monitoring structured clinical pharmacologic datasets. These analysis applications will be used within the Multi-institutional Surveillance of Medications DBP to evaluate the real-time accumulation of medication safety rates from three participating healthcare systems in different states, which collectively provide care to millions of patients.
Information models
We are developing information modeling and meta-data definitions for different modalities of existing data standards and ontologies—for example, utilizing the process of developing an information model documented by the Biomedical Research Integrated Domain Group21 and the Observational Medical Outcomes Partnership.22 We will use best practices and tools developed by the National Center for Biomedical Ontology for encoding data with standardized terminologies. For example, the Ontology Recommender Service23 24 and the National Center for Biomedical Ontology Annotator25 26 can guide us to identify the most suitable terminology systems for encoding the data in the iDASH domain and to automate concept mapping to those terminologies.
Content-based image retrieval system
In January 2011, iDASH sponsored its first workshop in imaging informatics, in which researchers and other stakeholders discussed how to create a collaborative environment for sharing biomedical images. Feedback from biomedical researchers determined that the repository must provide functionality for deidentifying, annotating, and retrieving similar images. Query and retrieval of medical images can be based on either the meta-data associated with the image or on the content of the image itself. In medical images, the Digital Imaging and Communications in Medicine (DICOM) standard defines a large set of meta-data (both free text and coded data) to describe of whom an image was obtained, how an image was obtained (physical processes and mathematical manipulations), and how the image is stored. Reports and annotations generated by physicians interpreting the images can also be viewed as meta-data. All these data need to be protected by privacy-preserving algorithms. Additionally, the richest expression of a disease remains in the actual pixels of the image, and consequently research is also devoted to developing techniques to identify and retrieve ‘similar’ images based on the image content itself while ensuring that the identity of the individuals whose images are being shared is protected. Since KD is the most image-oriented of our DBPs, our initial focus is on developing cardiovascular disease models for retrieval.
NLP Ecosystem
In May 2011, the biomedical NLP community gathered in an iDASH Workshop to discuss the development of an NLP Ecosystem to provide resources to aid in both development and application of NLP to clinical text. Integrating data from electronic health records is required in our DBPs, but data often consist of narrative text. Progress in this area can be accelerated by collaborative efforts.27 Our NLP Ecosystem (figure 2) will comprise a repository of clinical text, a repository of manual and automated annotations on the texts, downloadable resources and tools, and web services. Developers will be able to access our Ecosystem for shared collaboration on code development and can evaluate and benchmark their code on datasets housed in iDASH. Researchers will be able to access community standards and conventions, search for and download existing NLP tools, or interactively call web services running those tools. In addition, domain experts can collaboratively create knowledge bases for a particular extraction, run NLP tools using that knowledge base, and view the output.
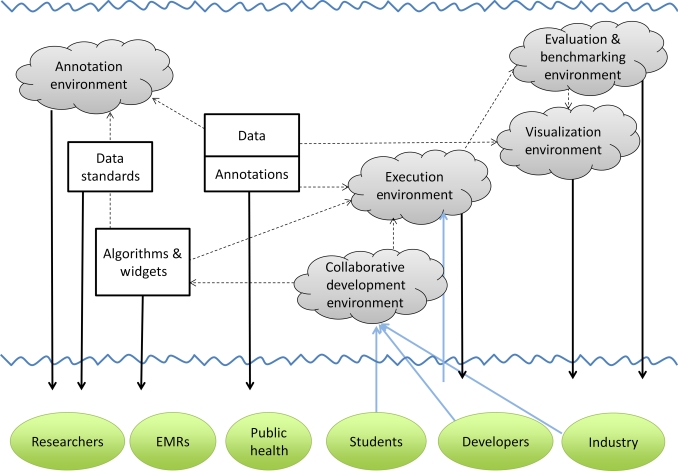
Figure 2.
iDASH natural-language processing (NLP) Ecosystem. Among the multiple resources that iDASH is developing is an NLP Ecosystem to allow collaborative development and testing of algorithms and tools, which structure data from clinical text. This Ecosystem will enable a diverse group of users (eg, researchers, students, and developers) to access public and private datasets, annotations on the data, downloadable tools (eg, algorithms and widgets), and web services. The cyber-infrastructure will provide a computing environment for evaluating code against existing datasets (execution, evaluation, and benchmarking environments), visualizing system output (visualization environment), and performing manual annotations on the textual data (annotation environment). EMR, electronic medical record.
Algorithms and tools for privacy-preserving data sharing
Informed consent management system
We are developing an extensible scheme for codifying consented uses of data for research. The coding scheme is multidimensional to reflect the many aspects of a subject's concerns,28 representing, for example, confidentiality of the subject's identity, the sensitivity of research, concerns about which researchers or institutions should be able to use the data, and concerns about frequency of future contacts. The major benefit of this codification scheme is to provide a common language to capture consented uses of data and specimens, enabling sharing of these across repository boundaries. The coding scheme is being developed as an ontology in OWL.29 We have implemented a prototype web-based informed consent management system, which is accessed via a touch-screen tablet computer. The objectives of the application are to improve the presentation of the information about the study, to offer the subject with tiered options as described by others30 for the uses of their data and how they participate in a study, and to record their consent in a codified manner in an electronic consent registry. We are designing a consent broker that will help an investigator determine if a proposed use of data in a study complies with consent, or as a corollary, select those data for the investigator's study that are compliant with the subjects' consented uses and the investigator's permissions.
Privacy technology and policy
Even in queries that only return aggregate results (eg, counts of patients with a particular diagnosis, gender, and age), there is a risk of individual reidentification of individuals.31 Previous work in privacy technology32–41 has described the difficulties in quantifying the reidentification risk in disclosed datasets. The last decade has seen renewed efforts to provably quantify cumulative risks to individuals from the application of information extraction methods and to develop algorithms to prevent reidentification of individuals.42–47 We are developing access methods that provably bound the risks to individuals over time,11 as well as tools that minimize the risk of reidentification.48 We are also working to support an infrastructure that can keep track of data donors' sharing policies, continuously track privacy risks incurred for individuals, and optimize sharing of information. In addition, we have been working toward developing versions of linear dimension reduction methods and machine learning methods that preserve privacy of individuals and institutions.
Cyber-infrastructure
We are building upon work on Rich Services,49 an architectural blueprint that promotes encapsulation, separation of concerns, reusability, and service orientation, while enabling direct and easy deployment mapping to runtime systems such as Enterprise Service Buses and Web Services. This work allows us to build the iDASH architecture in a hierarchical fashion, simply by following the same blueprint when decomposing the constituent application or infrastructure services. The iDASH architecture is implemented in a HIPPA-compliant hosting environment housed within the San Diego Supercomputer Center's secure data center, a high-performance computing facility protected by video monitoring, controlled access, and biometric controls. The iDASH system leverages physical and virtualized computational resources that allow segregated access to multiple projects within iDASH. These resources are protected by both physical and virtual firewalls and are managed by enterprise-grade applications through an isolated management network (figure 3).
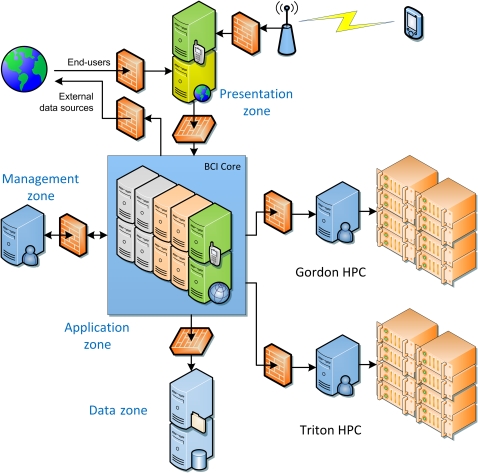
Figure 3.
iDASH cyber-infrastructure. iDASH is architecting a Health Insurance Portability and Accountability Act (HIPAA)-compliant biomedical cyber-infrastructure to enable data sharing and analysis with high-performance computing (Gordon and Triton HPC). Security is maintained through separation of layers (presentation, application, data, and management zones) and firewalls that secure communication between the zones. The presentation zone, which separates the user from the application, data, and management zones, provides the primary gateway to the overall system and contains the main components of the biomedical cyber-infrastructure framework, such as load balancers and web servers. The application zone provides the business applications and logic to the presentation zone and contains various application servers. The data zone contains all sensitive data repositories, which are accessed by the application zone. Finally, the management zone, which is only accessible to authorized administrators from approved workstations, provides system management and monitoring, security infrastructure, backup tools, and administration tools. BCI, biomedical cyber-infrastructure.
Education and outreach
iDASH directs education and outreach toward five areas: (1) hosting monthly webinars (http://idash.ucsd.edu/); (2) creating bibliographies that can be shared and supplemented by community members; (3) organizing workshops; (4) sponsoring internships; (5) supporting graduate education programs. The inaugural iDASH summer internship program hosted 21 high school, undergraduate, and graduate interns with diverse backgrounds such as biomedical informatics, computer science, electrical engineering, and linguistics. Examples of research topics included algorithms for face deidentification from medical images, anonymization of institutions in a shared data environment, and text mining.
Current and future goals
iDASH is about to complete its first year of establishment. It leverages tools developed by other National Centers for Biomedical Computing, as well as its own, to enable multi-institutional collaborative research. iDASH algorithms and open-source tools for structuring, analyzing, anonymizing, and sharing data are being developed to serve the biomedical and behavioral research community. In addition, iDASH provides an environment for computer scientists and engineers to benchmark and test new algorithms and tools. The Health Insurance Portability and Accountability Act (HIPAA)-compliant iDASH cyber-infrastructure supports high-performance computing in a private biomedical data cloud and is designed to be used at iDASH facilities, or exported to other centers. Besides serving our current DBPs, we are actively seeking collaborators to help test our tools and collaborative environment.
The scientific community has never produced as much data as it does today. However, access to these data and to facilities that can efficiently process them is often limited to a small group of researchers. iDASH reduces these barriers by providing a much larger community of researchers with a level playing field in which to begin the race for cures and discoveries and equally participate in ‘big science.’
Acknowledgments
We thank iDASH team members and advisors Winston Armstrong, Natasha Balac, Jane Burns, James Chen, Rex Chisholm, Richard Cope, Sanjoy Dasgupta, Cynthia Dwork, Robert El-Kareh, Fern Fitzhenry, Anthony Gamst, Amilcare Gentili, Peter Good, Amarnath Gupta, Mayuko Inoue, Ronald Joyce, Ingolf Krueger, Grace Kuo, Jennie Larkin, Karen Messer, Lalit Nookala, Greg Norman, Keith Norris, Kiltesh Patel, Paulina Paul, Pavel Pevzner, Kevin Patrick, Sergei Pond, Jialan Que, Susan Rathbun, Susan Robbins, Anand Sarwate, Chisato Shimizu, Heidi Sofia, Peter Tarczy-Hornoch, Dallas Thornton, Florin Vaida, Faramarz Valafar, George Varghese, Nicole Wolter, Cindy Wong, Mona Wong, and Alex Zambon.
Footnotes
Funding: iDASH is supported by the National Institutes of Health through the NIH Roadmap for Medical Research Grant U54 HL108460.
Competing interests: None.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1.Kanehisa M, Bork P. Bioinformatics in the post-sequence era. Nat Genet 2003;(33 Suppl):305–10 [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921 [DOI] [PubMed] [Google Scholar]
- 3.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science 2001;291:1304–51 [DOI] [PubMed] [Google Scholar]
- 4.Malin B, Benitez K, Masys D. Never too old for anonymity: a statistical standard for demographic data sharing via the HIPAA Privacy Rule. J Am Med Inform Assoc 2011;18:3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loukides G, Denny JC, Malin B. The disclosure of diagnosis codes can breach research participants' privacy. J Am Med Inform Assoc 2010;17:322–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malin B, Karp D, Scheuermann RH. Technical and policy approaches to balancing patient privacy and data sharing in clinical and translational research. J Investig Med 2010;58:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karp DR, Carlin S, Cook-Deegan R, et al. Ethical and practical issues associated with aggregating databases. PLoS Med 2008;5:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimizu C, Jain S, Davila S, et al. Transforming growth factor-beta signaling pathway in patients with Kawasaki disease. Circ Cardiovasc Genet 2011;4:16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar IN, Butte AJ, Lussier YA, et al. Translational bioinformatics: linking knowledge across biological and clinical realms. J Am Med Inform Assoc 2011;18:354–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozanitis C, Saunders C, Kruglyak S, et al. Compressing genomic sequence fragments using SlimGene. J Comput Biol 2011;18:401–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhuri K, Hsu D. Sample complexity bounds for differentially private learning. Proceedings of the 24th Annual Conference on Learning Theory (COLT 2011), 2011 [PMC free article] [PubMed] [Google Scholar]
- 12.Dasgupta SD. Two faces of active learning. Theor Comput Sci 2011;412:1767–81 [Google Scholar]
- 13.Jiang X, Osl M, Kim J, et al. Smooth isotonic regression: a new method to calibrate predictive models. AMIA Summits on Translational Science. San Francisco, CA, USA: American Medical Informatics Association, 2011 [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman BE, Lee S, Kang HP, et al. Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm. J Biomed Inform. Published Online First: 1 April 2011. doi:10.1016/j.jbi.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Patel K, Jung H, et al. AnyExpress: integrated toolkit for analysis of cross-platform gene expression data using a fast interval matching algorithm. BMC Bioinformatics 2011;12:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidi VD, Matheny ME, Donnelly S, et al. An evaluation of a distributed medical device safety surveillance system: the DELTA network study. Contemp Clin Trials 2011;32:309–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matheny ME, Morrow DA, Ohno-Machado L, et al. Validation of an automated safety surveillance system with prospective, randomized trial data. Med Decis Making 2009;29:247–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matheny ME, Arora N, Ohno-Machado L, et al. Rare adverse event monitoring of medical devices with the use of an automated surveillance tool. AMIA Annu Symp Proc 2007;2007:518–22 [PMC free article] [PubMed] [Google Scholar]
- 19.Matheny ME, Ohno-Machado L, Resnic FS. Risk-adjusted sequential probability ratio test control chart methods for monitoring operator and institutional mortality rates in interventional cardiology. Am Heart J 2008;155:114–20 [DOI] [PubMed] [Google Scholar]
- 20.Resnic FS, Gross TP, Marinac-Dabic D, et al. Automated surveillance to detect postprocedure safety signals of approved cardiovascular devices. JAMA 2010;304:2019–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biomedical Research Integrated Domain Group http://www.bridgmodel.org/ (accessed 29 Jul 2011).
- 22.Observational Medical Outcomes Partnership, 2011. http://omop.fnih.org/ (accessed 15 May 2011). [Google Scholar]
- 23.Jonquet C, Musen MA, Shah NH. Building a biomedical ontology recommender web service. J Biomed Semantics 2010;1(Suppl 1):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ontology Recommender Web Service http://www.bioontology.org/wiki/index.php/Ontology_Recommender_Web_service (accessed 3 Aug 2011).
- 25.Roeder C, Jonquet C, Shah NH, et al. A UIMA wrapper for the NCBO annotator. Bioinformatics 2010;26:1800–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NCBO Annotator http://bioportal.bioontology.org/annotator (accessed 1 Apr 2011).
- 27.Chapman W, Nadkarni P, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Amer Med Inf Assoc 2011;18:540–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bathe OF, McGuire AL. The ethical use of existing samples for genome research. Genet Med 2009;11:712–15 [DOI] [PubMed] [Google Scholar]
- 29.Smith M, Welty C, McGuiness D. OWL Web Ontology Language Guide, 2004. http://www.w3.org/TR/owl-guide/ [Google Scholar]
- 30.Eiseman E. Case Studies of Existing Human Tissue Repositories "Best Practices" for a Biospecimen Resource for the Genomic and Proteomic Era. Santa Monica, CA: Rand Corporation, 2003 [Google Scholar]
- 31.Dinur I, Nissim K. Revealing information while preserving privacy. PODS '03: Proceedings of the Twenty-Second ACM SIGMOD-SIGACT-SIGART Symposium on Principles of Database Systems. New York, NY: Association for Computing Machinery, 2003:202–10 [Google Scholar]
- 32.Ohno-Machado L, Silveira PS, Vinterbo S. Protecting patient privacy by quantifiable control of disclosures in disseminated databases. Int J Med Inform 2004;73:599–606 [DOI] [PubMed] [Google Scholar]
- 33.Ohrn A, Ohno-Machado L. Using Boolean reasoning to anonymize databases. Artif Intell Med 1999;15:235–54 [DOI] [PubMed] [Google Scholar]
- 34.Vinterbo SA. Privacy: a machine learning view. Ieee T Knowl Data En 2004;16:939–48 [Google Scholar]
- 35.Sweeney L. Privacy-preserving surveillance using databases from daily life. Ieee Intell Syst 2005;20:83–4 [Google Scholar]
- 36.Sweeney L. k-anonymity: a model for protecting privacy. Int J Uncertain Fuzz 2002;10:557–70 [Google Scholar]
- 37.Malin B, Sweeney L. Re-identification of DNA through an automated linkage process. Proc AMIA Symp 2001:423–7 [PMC free article] [PubMed] [Google Scholar]
- 38.Malin B, Sweeney L. Determining the identifiability of DNA database entries. Proc AMIA Symp 2000:537–41 [PMC free article] [PubMed] [Google Scholar]
- 39.Sweeney L. Guaranteeing anonymity when sharing medical data, the Datafly System. Proc AMIA Symp 1997:51–5 [PMC free article] [PubMed] [Google Scholar]
- 40.Vaidya J, Yu HJ, Jiang XQ. Privacy-preserving SVM classification. Knowl Inf Syst 2008;14:161–78 [Google Scholar]
- 41.Yu H, Vaidya J, Jiang XQ. Privacy-preserving SVM classification on vertically partitioned data. Lect Notes Artif Int 2006;3918:647–56 [Google Scholar]
- 42.Chawla S, Dwork C, McSherry F, et al. Toward Privacy in Public Databases. Theory of Cryptography Conference. Cambridge, MA, USA: Springer Verlag, 2005:363–85 [Google Scholar]
- 43.Barak B, Chaudhuri K, Dwork C, et al. Privacy, accuracy and consistency too: a holistic solution to contingency table release. Proceedings of Principles of Database Systems, Association for Computing Machinery, New York, NY, USA, 2007 [Google Scholar]
- 44.Dwork C. Differential privacy. ICALP 2006:1–12 [Google Scholar]
- 45.Chaudhuri K, Monteleoni C, Sarwate AD. Differentially private empirical risk minimization. JMLR 2011;12:1069–109 [PMC free article] [PubMed] [Google Scholar]
- 46.Blum A, Liqett K, Roth A. A learning theory approach to non-interactive database privacy. Conf Rec Annu ACM Symp Theor Comput 2008;2008:609–17 [Google Scholar]
- 47.Lasko TA, Vinterbo SA. Spectral anonymization of data. IEEE T Knowl Data En 2010;22:437–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang X, Cheng S, Ohno-Machado L. Quantifying record-wise data privacy and data representativeness. 17th ACM Conference on Knowledge Discovery and Data Mining (SIGKDD), workshop on Data Mining for Medicine and Healthcare, Association for Computing Machinery, San Diego, CA, USA, 2011 [Google Scholar]
- 49.Arrott M, Demchak B, Ermagan V, et al. Rich Services: The Integration Piece of the SOA Puzzle. In: Proceedings of the IEEE International Conference on Web Services (ICWS), Salt Lake City, Utah, USA: IEEE, Jul 2007, pp 176–183. [Google Scholar]