Abstract
Background and objective
With recent breakthroughs in high-throughput sequencing, identifying deleterious mutations is one of the key challenges for personalized medicine. At the gene and protein level, it has proven difficult to determine the impact of previously unknown variants. A statistical method has been developed to assess the significance of disease mutation clusters on protein domains by incorporating domain functional annotations to assist in the functional characterization of novel variants.
Methods
Disease mutations aggregated from multiple databases were mapped to domains, and were classified as either cancer- or non-cancer-related. The statistical method for identifying significantly disease-associated domain positions was applied to both sets of mutations and to randomly generated mutation sets for comparison. To leverage the known function of protein domain regions, the method optionally distributes significant scores to associated functional feature positions.
Results
Most disease mutations are localized within protein domains and display a tendency to cluster at individual domain positions. The method identified significant disease mutation hotspots in both the cancer and non-cancer datasets. The domain significance scores (DS-scores) for cancer form a bimodal distribution with hotspots in oncogenes forming a second peak at higher DS-scores than non-cancer, and hotspots in tumor suppressors have scores more similar to non-cancers. In addition, on an independent mutation benchmarking set, the DS-score method identified mutations known to alter protein function with very high precision.
Conclusion
By aggregating mutations with known disease association at the domain level, the method was able to discover domain positions enriched with multiple occurrences of deleterious mutations while incorporating relevant functional annotations. The method can be incorporated into translational bioinformatics tools to characterize rare and novel variants within large-scale sequencing studies.
Keywords: Translational bioinformatics, protein networks, text-mining
Introduction
As next-generation sequencing technologies continue to increase in throughput and decrease in cost, the next challenge for enacting whole-genome-based personalized medicine is to fully explain the functional contributions of genetic variations to human disease at the molecular level. The first clinical assessment of a personal genome recently demonstrated the potential of personalized medicine.1 The study evaluated the possible impact of variants both known and novel, rare and common, and with likely pharmacogenomic effects in combination with the patient's clinical and family history in order to suggest individualized treatment strategies. However, lacking knowledge of the functional mechanism by which a variant might contribute to disease, the researchers took a gene-based approach to prioritizing rare or novel non-synonymous variants, focusing on variants that occur in genes previously associated with diseases or with drug response and with predicted deleterious effects from variant effect prediction tools.
A recent study demonstrated the limitation of the gene-based approach to variant prioritization, noting that proteins function through interaction networks, and that mutations that cause a complete loss of a protein (node removal) are often phenotypically distinct from those that disrupt specific interactions without loss of the protein (edgetic perturbations).2 The same study also showed several examples where non-synonymous mutations or small, in-frame insertions or deletions in different domains in the same protein produce distinct disease phenotypes by disrupting different protein functions and interactions. Thus, inferring the effect of a mutation based only on its presence in the same gene as a previously disease-associated mutation is likely to be misleading. These results emphasize the need to functionally characterize individual variants in order to accurately predict their associations to disease, and demonstrate the potential for protein domains to provide the necessary functional information for variant characterization.
Protein domains are the structural and functional subunits of proteins. Different domains confer proteins with different functions, and unique combinations of domains confer proteins with the wide variety of protein function seen today. In addition, protein domains mediate most (approximately 75%) protein interactions.3 Individual domains also contain distinct functional features like binding sites, active sites, and post-translational modification sites. By mapping mutations to their relative positions within protein domains, the disruption of specific functional features or protein interactions can be revealed, providing a detailed explanation for the molecular contribution of the mutation to disease. In addition, aggregating mutations from all proteins at the domain level can also reveal individual positions within the domain that are highly susceptible to disease-causing mutations, providing a significant aid to variant prioritization. To visualize the aggregation patterns of disease mutations at the protein and domain levels, we recently developed the Domain Mapping of Disease Mutations database (DMDM), freely available at http://bioinf.umbc.edu/dmdm/.4
In this work, we study the patterns of aggregated cancer and non-cancer disease mutations at the protein domain level. A recent study showed that there are differences in the tendencies of Mendelian disease-related mutations and cancer somatic mutations to occur at solvent accessible positions within proteins,5 while another study showed different tendencies for mutations in oncogenes and tumor suppressors to cluster at functional sites on the protein and within the three-dimensional protein structure.6 Using domain visualizations provided by the DMDM database, we were able to confirm the clustering of disease mutations at individual domain positions and functional feature sites for both cancers and Mendelian diseases, and for known oncogenes and tumor suppressors. These observations motivated our decision to separate cancer and non-cancer mutations, and further cancer mutations in oncogenes and tumor suppressors, to determine if distinct patterns of mutation aggregation exist at the domain level. We first developed a methodology to identify significantly mutated positions within individual protein domains. In order to calculate the domain significance score (the DS-score) for each position within each domain, we mapped all known, disease-associated mutations to their relative positions within the domains. The DS-score is based on the probability for the current position within the domain to contain the number of disease mutations found, given the domain length and the total number of disease mutations mapping to the domain. A significant DS-score for a position implies that a mutation at the position is highly likely to contribute to disease in any protein containing the domain. In addition to this position-based DS-score, we also developed a feature-based DS-score. Using the feature-based DS-score methodology, the position-based DS-score for a significant position annotated as part of a specific functional feature (eg, binding site or active site) is distributed to all other positions annotated as part of the same functional feature in the domain under the assumption that the entire functional feature is critical to the normal function of the protein.
We find that disease mutations form significant cluster hotspots for both cancers and non-cancers. Furthermore, we find distinct differences between the DS-score distributions for both cancers and non-cancers. While cancers and non-cancers display a similar distribution at lower DS-scores, the DS-scores for cancers form a bimodal distribution, with hotspots in oncogenes forming a second peak at higher DS-scores, and hotspots in tumor suppressors have scores more similar to non-cancers. We also find that mutation hotspots in cancers tend to occur significantly more often at functional feature positions than non-cancers, while mutations in both sets show similar overlap with highly conserved positions in protein domains.
Finally, the DS-scores for individual protein domain positions can be used as predictors of the effect of uncharacterized, non-synonymous single nucleotide variants (nsSNVs) from sequencing studies. Numerous methods for predicting the effect of nsSNVs have been developed over the last 10 years that use a variety of features, including the evolutionary history of the mutated position, the physicochemical properties of the resulting amino acid substitution, and the predicted effect of the mutation on protein structure, as well as other features and combinations thereof.7–25 These have been recently reviewed.26–29 The DS-score method is novel in that it uses the domain positions of known disease mutations to predict the effects of unclassified variants. Variants can be mapped to their domain positions, and the domain positions checked for the presence of significant clusters of known disease mutations. The occurrence of a variant at a domain position with a significant cluster of disease mutations implies that the variant is likely to be deleterious, even if it occurs in a protein formerly unassociated with disease.
Domain sequences are highly conserved through evolution; therefore we compared the DS-scores for significant mutation hotspots to a simple measure of conservation for those positions to ensure that the DS-score did not simply identify highly conserved positions. We also compared the performance of the DS-score method as a predictor of the effect of nsSNVs on an independent mutation set to that of SIFT,7 a widely used method that bases its predictions on the conservation of the mutated positions, and to that of another domain-based approach, the LogR.E-value method.25 The LogR.E-value method bases its predictions on the change in alignment scores for the wild-type and mutated protein sequences to a hidden Markov model of the domain sequence. Due to the currently limited number of disease mutations listed in public databases, the DS-score has very low sensitivity compared to the SIFT and LogR.E-value methods because they do not restrict their predictions of damaging mutations to domain positions of known disease mutations. However, the DS-score has a significantly higher precision, outperforming SIFT and the LogR.E-value on mutations predicted to alter protein function. In addition, when we combine the DS-score prediction with that of SIFT or the LogR.E-value, the precision increases even more, to over 95% for the position-based DS-score, a characteristic important for the potential application of the method in a clinical setting. To facilitate the use of the DS-scores as an aid to variant classification by the scientific community, pre-computed DS-scores for all domains and domain positions are freely available on our FTP site (http://bioinf.umbc.edu/ds-score/ftp/), as well as a Perl script for mapping mutations from protein to domain positions.
Materials and methods
Databases
A human protein database containing 54 372 proteins was created with 33 963 proteins from RefSeq30 and 20 409 proteins from Swiss-Prot31 downloaded via NCBI's E-utilities. Since the RefSeq and Swiss-Prot databases contain many redundant protein entries, we selected only one representative protein for each unique Entrez gene ID, either the longest Swiss-Prot protein, or the longest RefSeq protein if no Swiss-Prot protein was listed for the gene ID. A protein domain set was obtained from the Conserved Domain Database (CDD)32 (version 2.25), which includes domains from CDD and the SMART,33 COG,34 and Pfam35 databases, with a total of 23 632 protein domains, 10 925 of which map to at least one human protein. Functional feature information was collected for CDD domains from the ‘cddannot.dat’ file located in the CDD FTP directory (ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd), totaling 1727 unique functional features. The non-overlapping set of human, non-synonymous disease mutations was created from the OMIM36 and Swiss-Prot variant databases obtained from E-utilities and UniProt's FTP directory (http://www.uniprot.org/docs/humsavar), respectively. Mutations were classified as ‘cancer’ or ‘non-cancer’ using a controlled vocabulary and manual curation. The resulting non-cancer dataset consists mostly of mutations related to diseases with Mendelian patterns of inheritance, but also contains a small number of complex disease mutations. Randomized datasets were created for both the cancer and non-cancer mutation sets by randomizing the domain position for each mutation using a uniform probability distribution.
Mapping mutations to protein domains
Hidden Markov models for protein domains from SMART, COG, CDD, and Pfam were built using multiple sequence alignments from CDD with the hmmerbuild tool (HMMer version 2.3.2).37 HMMer's hmmpfam tool was then used with the global option to search for complete domains in human proteins from the RefSeq and Swiss-Prot databases. Protein mutations were distributed to protein domain positions using HMMer's alignment output and assigning mutations that fall on gap regions of the domain model to the last position before the gap. Each mutation was mapped only to the representative protein for each unique gene in the dataset. The methods for mapping domains to human proteins and disease mutations to their domain positions were previously described for our DMDM tool.4 After mapping the mutations to domain positions, 39.2% of human protein domains contained at least one disease mutation from either the cancer or non-cancer sets.
Estimating conservation of domain positions
The program AL2CO38 was used to estimate the entropies for each column j in a protein domain alignment.
| (1) |
where p(ai,j) is the frequency of amino acid ai at position j. A threshold for identifying highly conserved positions was estimated by averaging the AL2CO scores for all domain positions and adding one SD.
Estimating domain significance scores (DS-scores)
We developed a method to estimate a position-based DS-score for each domain position. Let X be the number of mutations and X(k) be the kth order of number of mutations. P{X(k)=x} is from n independent observations and only depends on the probability of three events, a=P{X<x}, b=P{X=x}, c=P{X>x}, and a+b+c=1. From the multinomial distribution, the probability of X(k)=x if and only if there are no more than k−1 observations less than x and no more than n−k observations greater than x is.
| (2) |
The position-based DS-score is the probability of observing a cluster of a particular size given the number of available positions in a domain and the total number of mutations observed,
| (3) |
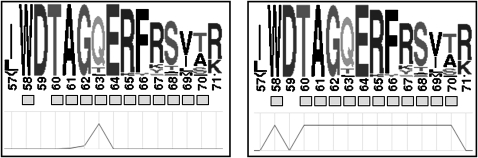
where L is number of positions in the domain, and m is number of mutations which are tied at maximum. We used a binomial probability of observing a cluster with size less than k in a domain with n mutations. Domain disease hotspots, or disease hotspots, were defined for those positions with DS-scores ≥1.3 (significant with a p value ≤0.05). The feature-based DS-score was created by distributing the highest position-based DS-score for each functional feature to all other positions annotated with the same functional feature in the domain. Figure 1 illustrates how the DS-score is disseminated to all functional feature positions. Perl and R were used to calculate and assign the DS-scores.
Figure 1.
Visual representation of the distinction between position-based and feature-based DS-scores. A position-based DS-score hotspot (left) is represented as a peak of the DS-score (line graph at bottom) at domain position 63. The DS-score for the hotspot at position 63 is distributed to all other functional feature positions (boxes below the sequence logos) to create the feature-based DS-scores (right). This figure is produced in colour in the online journal–please visit the website (www.jamia.org) to view the colour figure.
Estimating the background distribution of significant DS-scores
We created two additional sets of mutations by randomizing the domain position of each mutation in the cancer and non-cancer sets. This process was repeated 1000 times for each set of mutations. We then estimated the average and SD of the number of significant domain positions, that is, those with DS-scores ≥1.3, found in each randomized set of mutations.
Comparison of DS-score performance to SIFT and the LogR.E-value
To compare the performance of the DS-score method to other methods for predicting the effect nsSNVs, we used the set of all single amino acid substitutions in human proteins extracted from the Protein Mutant Database (PMD)39 used in Bromberg et al.20 PMD contains information from the literature on the effects of naturally occurring and experimentally induced mutations on protein activity and association with disease when applicable. Mutations noted to increase or decrease protein activity were classified as ‘function-altering’, whereas mutations noted to cause no change in function were classified as ‘neutral’. Neutral mutations were added to the PMD set in order to balance the number of function-altering and neutral mutations. To do so, highly sequence similar (>40% identity) enzymes with experimentally annotated functions in Swiss-Prot were aligned by pairwise BLAST,40 and positions containing different amino acids were assumed unlikely to affect protein function, and were thus labeled as neutrals.
The set of all PMD and added neutral mutations was used to benchmark the performance of the DS-score methods and to compare it with the SIFT7 and LogR.E-value25 predictors. SIFT predictions of ‘tolerated’ or ‘damaging’ were considered as neutral or function-altering, respectively. A LogR.E-value threshold of 1.0 (neutral < LogR.E-value 1.0 ≥ function-altering) was used to classify mutations, which was the threshold suggested by the authors of the LogR.E-value method to maximize precision in identifying deleterious mutations. Of the 15 182 PMD mutations including added neutrals, 9049 (59.6%) occurred within identified protein domain regions and produced prediction results with both the SIFT and LogR.E-value methods. We calculated the sensitivity, specificity, and precision of the SIFT and LogR.E-value predictions in reference to the PMD-derived classifications, and compared to the performance of the position-based and feature-based DS-scores for the mutation positions using a DS-score threshold of 1.3 (neutral < DS-score 1.3 ≥ function-altering). The DS-scores from the cancer and non-cancer sets were combined and used to classify the mutations. We also compared the performance of a simple method using a threshold of two known disease mutations at the domain position (neutral < 2 mutations ≥ function-altering) to classify the PMD and added neutral mutations. Additionally, because the DS-score method only considers the domain position of the mutation, and not the actual amino acid change, we calculated the precision for combining the DS-score predictions with the SIFT or LogR.E-value predictions, which do consider the amino acid change. To do so, mutations were classified as function-altering when both the DS-score prediction and the SIFT or LogR.E-value prediction classified the mutation as function-altering, otherwise the mutation was classified as neutral.
Results
Disease mutations on protein domains
After mapping all available disease mutations in human proteins to their corresponding domain positions, we found that 97% of disease mutations, including both cancer and non-cancer mutations, are located within a protein domain. In addition, both mutation sets display a significantly higher tendency to cluster at individual protein domain positions with respect to the random sets, with 54.4% and 58.8% (p value for both sets ≈0.0) of the cancer and non-cancer mutations, respectively, located in protein domain positions that contain two or more mutations, as shown in table 1.
Table 1.
Mutation, position-based, and feature-based hotspot counts; results for the random sets show the average numbers and their standard deviations over 1000 randomizations
| Cancer | Non-cancer | Randomized cancer | Randomized non-cancer | |
| Total mutations | 33 688 | 205 174 | 33 688 | 205 174 |
| Total position-based hotspots | 986 | 2004 | 7 (±3) | 10 (±6) |
| Total feature-based hotspots | 11 031 | 8556 | 18 (±20) | 18 (±20) |
| Mutations at position-based hotspots | 13.7% | 6.4% | 0.06 (±0.03)% | 0.05 (±0.04)% |
| Mutations at feature-based hotspots | 29.2% | 10.5% | 0.07 (±0.04)% | 0.06 (±0.04)% |
| Mutations at domain positions with ≥2 mutations | 54.4% | 58.8% | 33.2 (±2.2)% | 30.8 (±3.1)% |
Domain disease hotspots
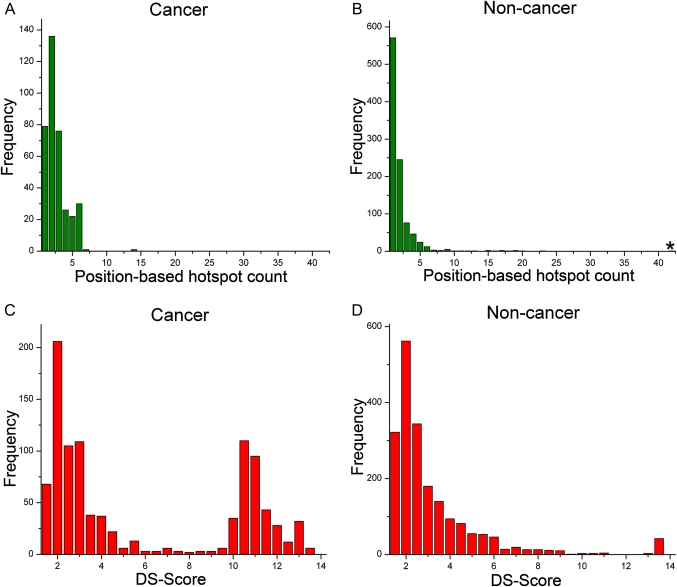
We developed a method for identifying significant mutation hotspots at individual protein domain positions or at annotated functional feature positions within the domain. The position-based DS-score was used to identify 986 and 2004 domain hotspots for the cancer and non-cancer mutation sets, respectively. As shown in table 1, these results are notably higher (p value ≈ 0.0 for both sets) with respect to what is expected by chance. The average number of position-based hotspots in the randomized sets were only eight (cancer) and 10 (non-cancer). Alternatively, the feature-based DS-score identifies significant clusters of mutations at annotated functional feature positions within the domain (see figure 1). Using the feature-based DS-score, we identified 11 031 feature-based hotspots in the cancer mutation set and 8556 in the non-cancer set. The corresponding randomized mutation sets yielded significantly lower counts, with only 18 feature-based hotspots in each of the random sets. The higher number of hotspots found using the feature-based score than using the position-based score was expected, as significant position-based scores are distributed throughout functional features to assign the feature-based scores. We also counted the number of position-based hotspots per domain in each mutation set (figure 2A,B). The non-cancer set yielded a higher overall number of position-based hotspots, likely due to the higher number of mutations in the non-cancer set. The distributions of the number of hotspots per domain for the cancer and non-cancer datasets were similar, except for an extreme outlier with 42 hotspots in the non-cancer set. Protein domains with the highest number of position-based hotspots for the cancer and non-cancer sets are shown in tables S1-A and S1-B, respectively. As expected, kinase and RAS domains are significantly represented in the cancer set.
Figure 2.
Number of position-based hotspots per protein domain and the distribution of DS-scores for the cancer and non-cancer datasets. The number of position-based hotspots was counted for each domain, and the distribution is shown in (A) and (B) for the cancer and non-cancer datasets, respectively. The asterisk (*) in (B) denotes one domain with 42 hotspots. The distribution of DS-scores for significant domain positions is shown for the cancer and non-cancer mutation sets in (C) and (D), respectively.
The vast majority of the mutations in the non-cancer set are from diseases with likely Mendelian patterns of inheritance. While both the cancer and non-cancer (mainly Mendelian) mutations in our study show significant patterns of aggregation at the protein domain level, figure 2C,D shows that there are significant differences in the distributions of the position-based DS-score for these datasets. The cancer mutations contain a second peak, indicating that cancer mutations have a significantly higher tendency to cluster at specific protein domain positions. The specific domains where these highly significant clusters of cancer and non-cancer mutations occurred are listed in tables S2-A and S2-B, respectively. These results confirm the significance of kinase domain mutations in cancer, but also point to the significance of other domains, such as EGF and collagen domains to the non-cancer diseases. The results in figure 2C,D were obtained using the DS-scores from all domains included in the CDD, Pfam, SMART, and COG databases in order to include all domains, including those exclusive to each domain set. We also computed the DS-scores and plotted the distributions using each domain database individually, and obtained similar DS-score distribution patterns for the cancer and non-cancer mutations. Figures S1-A and S1-B show the DS-score distributions for cancer and non-cancer mutations using only domains from the CDD domain set.
Hotspots at conserved and functionally annotated positions
As shown in table 2, more than 50% of the position-based and feature-based hotspots occur at highly conserved domain positions. For example, 58.1% (cancer) and 51.2% (non-cancer) of the position-based hotspots are in highly conserved positions. A slightly higher percentage, 67.6% (cancer) and 61.7% (non-cancer), of the feature-based hotspots are highly conserved. All p values estimated by the Fisher test for the comparison of these results against the background, randomized sets were significant (p values <0.05). To measure the correspondence of the DS-scores and entropy-based conservation scores, we calculated the Pearson correlation coefficient for all position-based hotspots in each set, resulting in a correlation coefficient of 0.19 for cancer and 0.10 for non-cancer. Additionally, to assess the functional significance of the disease hotspots, we calculated the percentage of position-based hotspots occurring at annotated functional feature positions. Also shown in table 2, 69.8% of the cancer hotspots occur at known, functional feature positions, while the majority of hotspots in the non-cancer set have no functional annotation. Only 35.9% of the non-cancer hotspots occur at a functional feature site. As expected, the vast majority of feature-based hotspots (97.6% for cancer and 90.3% for non-cancer) occur at functionally annotated features within the domain due to the nature of the feature-based DS-score assignment. Tables S3-A and S3-B list the functional features with the highest number of hotspots for the cancer and non-cancer datasets, respectively.
Table 2.
Percentage of hotspots occurring at highly conserved and annotated functional feature positions
| Cancer | Non-cancer | |
| Position-based hotspots at conserved positions | 58.1% | 51.2% |
| Feature-based hotspots at conserved positions | 67.6% | 61.7% |
| Position-based hotspots at functional features | 69.8% | 35.9% |
| Feature-based hotspots at functional features | 97.6% | 90.3% |
DS-score prediction performance
We measured the performance of the position-based DS-scores and feature-based DS-scores as predictors of the effect of nsSNVs on protein function and compared to the performance of two well known predictors: SIFT and the LogR.E-value. As expected, due to the low number of mutations in public databases with known disease phenotype, the sensitivities of the DS-score based methods (3.3% for the position-based DS-score, 5.6% for the feature-based DS-score) are extremely low compared with those of SIFT (64.1%) and the LogR.E-value (56.0%) which do not depend on the existing number of known disease mutations. However, the sensitivity increased by relaxing the parameters to predict deleterious domain positions. Using a simple model that classified any mutation in a domain position with two or more known disease mutations as likely to alter protein function, the sensitivity increased significantly to 20.5%. The specificities for the DS-score methods were very high in comparison to SIFT and the LogR.E-value methods: 99.5% for the position-based DS-score, 98.6% for the feature-based DS-score, 94.2% for the simple model, 76.2% for SIFT, and 78.2% for the LogR.E-value.
A comparison of the precision of the methods is shown in table 3. The highest precision, 91.6%, was obtained by the position-based DS-score, and there were only small decreases in precision when the DS-score hotspots were extended to aggregate mutations within the same functional features (87.2% for the feature-based DS-score), or when the simple model was used (85.6%). All methods had precisions higher than either SIFT (82.0%) or the LogR.E-value (81.3%). We also combined the DS-score method predictions with the SIFT and LogR.E-value predictions to determine the impact on precision. The SIFT and the LogR.E-value methods both consider the actual amino acid change of the mutation, while the DS-score methods only consider the domain position of the mutation. By requiring that both the DS-score and the SIFT or LogR.E-value method under consideration agreed that a mutation was predicted to alter function, all methods increased in precision with only small decreases in sensitivity. For example, combining the DS-score with the LogR.E-value predictions resulted in increases in precision of 4.1% and 3.8% for the position-based and feature-based DS-scores, respectively, with accompanying decreases in sensitivity of 0.9% and 1.7%. The precision for the simple model also increased (up 6.3%), but had a larger decrease in sensitivity (down 8.4%).
Table 3.
Benchmarking the precision of domain significance score (DS-score) based methods for classifying human non-synonymous single nucleotide variants of known functional effect from the Protein Mutant Database
| Method | Precision (%) | Precision of method combined with SIFT (%) | Precision of method combined with LogR.E-value (%) |
| SIFT | 82.0 | N/A | N/A |
| LogR.E-value | 81.3 | N/A | N/A |
| Position-based DS-score | 91.6 | 95.1 | 95.7 |
| Feature-based DS-score | 87.2 | 89.4 | 91.0 |
| Domain positions with ≥2 mutations | 85.6 | 91.2 | 91.9 |
Precision was calculated as the percentage of mutations predicted to alter function that were correctly predicted.
Discussion
As the field of personalized medicine develops, there is a growing need for new methods to identify deleterious mutations from the millions of variants present in each individual's genome. Personalized treatment for individuals found to harbor harmful mutations depends on the accurate assessment of mutation-specific disease risk.41 In our approach to identify functionally deleterious mutations, previously disease-associated mutations from all human proteins were aggregated at the protein domain level in order to identify and analyze disease hotspots. We recently developed a tool to visualize the aggregation patterns of disease mutations at the protein and domain levels, the DMDM database. We also developed a statistical measure, the position-based DS-score, to identify significantly mutated positions within each domain, based on the probability of observing the specific number of mutations at each position, given the length of the domain and the total number of mutations mapping to the domain. A similar method, mCluster,42 was recently developed to identify disease hotspots at the domain level, with a focus on distinguishing driver mutations from passenger mutations in cancer tumor sequencing data. However, in order to distinguish passenger and driver mutations, mCluster inherently includes somatic mutations from COSMIC43 and additional cancer sequencing studies with unknown disease relevance. The DS-score method utilizes only mutations of known disease relevance, consisting primarily of germline mutations in addition to a much smaller number of validated, cancer-associated somatic mutations from OMIM and Swiss-Prot. By using only validated, disease-associated mutations, the DS-score method is likely to be highly specific in identifying true domain disease hotspots, as evidenced by the highly significant enrichment (p value ≈0.0) of hotspots in the cancer and non-cancer sets in comparison to the random mutation sets (table 1).
Many current tools for predicting the impact of missense mutations use sequence conservation as a feature to help classify mutations as either neutral or deleterious including SIFT,7 PolyPhen,8 SNAP,20 and others. Using an entropy-based conservation score, we calculated the percentage of times that significant DS-score hotspots overlapped with highly conserved positions inside protein domains. As shown in table 2, we did not find a perfect correspondence between highly conserved positions and disease hotspots in either the position-based scores for the cancer (58.1%) and non-cancer (51.2%) sets or in the feature-based scores for the cancer (67.6%) and non-cancer (61.7%) sets. We also found very low correlation coefficients between the DS-scores and conservation scores for each set (0.19 for cancer and 0.10 for non-cancer), demonstrating that the DS-score method for identifying mutation hotspots goes beyond simply identifying mutations at highly conserved domain positions. Our DS-score methodology therefore incorporates additional information in its calculation of disease hotspots that could be used to aid in the characterization of rare or novel variants.
In addition to identifying significant disease mutation hotspots in protein domains, the DS-score provides an inherent functional context for explaining how mutations at the hotspot contribute to disease. Domains confer proteins with specific functional capabilities, and knowing that a specific capability is potentially disrupted by a mutation is critical to the design and implementation of future treatment strategies. Tools that predict the impact of non-synonymous mutations, like SIFT, do not currently provide this additional functional context provided by the DS-score methodology. To further leverage the functional context provided by the domain position of a mutation, we also created the feature-based DS-score. NCBI's Conserved Domains Database provides manually curated annotations for individual domain positions that contribute to specific functional features like active sites, binding sites, and phosphorylation sites. The feature-based DS-score is assigned by distributing the highest position-based DS-score for each functional feature to all other positions in the same functional feature in the domain. Thus, the feature-based DS-score expands the ability of the position-based DS-score to identify significant mutation hotspots under the assumption that any mutation in a functional feature already known to be disrupted in a disease (via a significant position-based DS-score) is highly likely to also contribute to disease.
The creation of the DS-score methodology allowed us to study the patterns of disease mutation clustering for both the cancer and non-cancer sets. We did not see significant differences in the number of hotspots per domain between the two sets (figure 2A,B). Surprisingly, we found a considerable difference in the position-based DS-score distributions for the cancer and non-cancer sets (figure 2C,D). Both sets show a similar distribution at lower DS-scores with a peak around 2.5, then rapidly dropping off as the score increases. The cancer set, however, shows a second peak of DS-scores higher than 9.5. When we compared the sets of genes containing mutations at the hotspots scoring below 9.5 to the set scoring above 9.5 (table S4), we found that only putative oncogenes were present in the set scoring above 9.5, while both putative oncogenes and tumor suppressor genes were present in the set scoring below 9.5. Of the genes we could classify as either putative oncogenes or tumor suppressors, the majority of genes containing mutations at significant DS-score hotspots (17 out of 22) are known to be oncogenes. These results suggest that mutations in oncogenes tend to cluster more significantly than mutations in tumor suppressors, and that mutations in tumor suppressors are more similar to mutations in non-cancer genes, typically associated with Mendelian disorders, than to mutations in oncogenes.
A recent study of the clustering patterns of somatic mutations in cancer that allowed for variable length clusters found much longer length clusters in tumor suppressors than in oncogenes,44 providing additional evidence consistent with our results. Another recent study by Stehr et al of the structural impact of somatic mutations in oncogenes and tumor suppressors6 found significantly higher enrichment of clustering for mutations in the three-dimensional structures of domains in oncogenes than in tumor suppressors, also supporting our results. The same study found that mutations in oncogenes were significantly more likely to occur at solvent accessible sites and at functional feature sites than were mutations in tumor suppressors. In addition, mutations predicted to destabilize the protein were highly enriched in tumor suppressors, but were highly depleted in oncogenes. Stehr et al proposed an explanation for these results, suggesting that activating mutations in oncogenes tend to occur at specific functional features on the protein surface, while mutations in tumor suppressors are more likely to be destabilizing mutations spread throughout the protein core. A study by Talavera et al provides support for this hypothesis, showing an enrichment of oncogenic driver mutations at functional sites on the surface of the protein.45 Our results also show that a higher percentage of hotspots occur at functional features in cancer (69.8%) than in non-cancer (35.9%) (table 2). Taken together, our findings of significant differences between the clustering profiles for mutations in cancers and non-cancers, and between oncogenes and tumor suppressors, are in close agreement with these studies. As expected due to their well-known relevance to cancer, domains found primarily in the RAS family of GTPases and in protein kinases dominate the list of domains with the highest number of significant hotspots in the cancer set (table S1-A). Additionally, different members of the family of catalytic domains of protein kinases have the 10 highest scoring hotspots in the cancer set (table S2-A). A recent study by Dixit et al of mutations in kinases supports our finding of significant clustering of cancer mutations at specific positions in the kinase catalytic core domain.46 The study also found that these hotspots were enriched for oncogenic, driver mutations, also in agreement with our findings.
The domain in the non-cancer set containing both the highest DS-score position (table S2-B), and the highest number of significant positions (table S1-B) was the calcium-binding EGF-like domain (EGF_CA, smart00179). The EGF_CA domain has 42 positions with a DS-score of 13.45. Both the score and the number of hotspots are significant outliers in the non-cancer distributions (figure 2B,D). The EGF_CA domain is relatively short in length, containing only 84 amino acids, and is commonly found in varying numbers of tandem repeats in membrane-bound and extracellular proteins.47 Fibrillin-1, for example, contains 43 EGF_CA domains, and NOTCH3 contains 27 EGF_CA domains. The short length of the domain and the tandem repeat configuration likely contributes to the high number of hotspots in the EGF_CA domain as disease mutations spread throughout individual proteins hit multiple copies of the domain, aggregating to the limited number of positions in the domain model. This configuration of a very large number of domain repeats in a single protein is not typical, however, as the average protein has three or fewer domains.48
Interestingly, our domain-based approach also enables us to detect common patterns of mutation for proteins involved in different biological processes sharing a common domain. Mutations in proteins containing the EGF_CA domain have been linked to a number of diseases including hemophilia B, Marfan syndrome, retinitis pigmentosa, and hypercholesterolemia.47 EGF_CA domains have a calcium binding site at the N-terminal end of the domain in addition to six highly conserved core cysteine residues which form three disulfide bridges.49 Disease mutations in the domain tend to cluster around the core cysteine residues, likely causing a disruption in the domain structure and loss of calcium binding which has been shown to be critical for maintaining the biological activity of the protein.47 Loss of calcium binding has been shown to disrupt protein interactions in the coagulation factor IX protein (associated with hemophilia) and structural rigidity in fibrillin-1 (associated with Marfan syndrome).47 While the extremely large number of hotspots in the EGF_CA domain is not typical, this example clearly demonstrates how knowledge of the domain position of a mutation can aid in its functional characterization and in the prediction of its potential impact on protein function regardless of the protein the mutation actually occurs in.
Finally, our results collectively show that the DS-score methodology could be used as a highly precise and specific predictor of the effect of uncharacterized, rare or novel coding variants from large-scale sequencing studies. Variants of interest could be mapped to their domain positions, and the positions checked for the presence of significant clusters of known, disease-associated mutations identified by significant DS-score hotspots. The occurrence of a variant at a domain position with a significant cluster of validated, disease-associated mutations implies that the variant is likely to be deleterious, even if it occurs in a protein formerly unassociated with disease. Unfortunately, due to the low number of disease-associated mutations currently available from public databases, this also contributes to a relatively low predicted sensitivity for the position-based DS-score, as only 13.7% and 6.4% of the cancer and non-cancer mutations fall in significant hotspots, respectively (table 1). By distributing the position-based DS-scores to all other positions in the same functional features, the feature-based DS-score can help to boost the sensitivity of the method, increasing the percentage of mutations covered by hotspots in the cancer and non-cancer sets to 29.2% and 10.5%.
To evaluate the performance of the DS-score methods as predictors, we benchmarked the methods on the independent PMD mutation dataset. While the sensitivities of the DS-score methods were confirmed to be low on the PMD dataset, we also confirmed that both the position-based and feature-based DS-score methods had extremely high specificities, over 98% for both methods. In addition, we found that the methods had very high precision, over 91% and 87% for the position-based and feature-based DS-score methods, respectively, outperforming both SIFT (82.0%) and the LogR.E-value method (81.3%) as shown in table 3. The precision of the methods was further increased when we combined the DS-score prediction with that of SIFT or the LogR.E-value method to account for the actual amino acid change resulting from the mutation. These increases in precision resulted from correctly reclassifying several mutations from likely to alter protein function to likely to be neutral due to relatively conservative amino acid substitutions. Since the PMD dataset contains mutations known to alter protein function, both with and without association to disease, the high precision of DS-score methods also demonstrates that the methods are highly promising, not only for predicting which variants are likely to cause disease, but also more generally for predicting which variants will disrupt normal protein function.
The number of annotated disease mutations in public databases continues to grow50 as disease association studies move to large-scale, whole genome-based designs. In the near future, a large number of novel disease-associated mutations will likely be identified, in addition to an increasing number of rare disease-associated mutations as methods to discover these low frequency mutations become more sophisticated.51 Therefore, we expect the sensitivity of the DS-score method to improve substantially as the number of known disease mutations increases and new domain disease hotspots are found. In the meantime, the application of text mining methods for automatically extracting previously identified mutations with disease association from the literature52 can be applied to supplement the mutation datasets from manually curated databases like OMIM and Swiss-Prot. To evaluate the potential for the DS-score method to increase in sensitivity as the number of known disease mutations increases, we calculated the percentage of mutations at domain positions with at least two disease mutations: 54.4% for cancer and 58.8% for non-cancer, which was substantially higher than the percentage of mutations at position-based or feature-based DS-score hotspots (table 1). We also confirmed a substantial increase in sensitivity for the simple predictor model that classified any mutation at a position with two or more known disease mutations as likely to alter protein function, from less than 6% for the position and feature-based DS-score methods to over 20% for the simple model. Therefore, the DS-score method is very likely to increase in sensitivity while maintaining a high specificity as new disease-associated mutations are identified in the future.
Conclusions
Through the development of our novel DS-score methodology for identifying specific protein domain positions with significant clustering of disease mutations, we performed a systematic analysis of mutations related to both cancer and non-cancer diseases. We show that cancer and non-cancer mutations both form significant mutation hotspots. In addition, cancer mutations, and more specifically mutations in known oncogenes, show a higher tendency to cluster at individual domain positions, while non-cancer mutations and mutations in tumor suppressor genes show lower tendencies to form significant mutation hotspots. We also demonstrate the application of the DS-score method as a highly specific and precise predictor of the effect of non-synonymous mutations in domain regions, particularly when used in combination with either the SIFT or the LogR.E-value method. Therefore, we expect that the DS-score methodology will be incorporated into the analysis of large-scale sequencing projects to identify both novel and rare variants associated with disease development. The most significant feature of the DS-score methodology, however, will be to provide the critical functional explanation for how a variant contributes to disease, essential for the development of future personalized treatment strategies.
Acknowledgments
We would like to thank Yana Bromberg for supplying the Protein Mutant Database dataset used to benchmark the performance of the methods in predicting the effect of non-synonymous mutations on protein function.
Footnotes
Funding: This work was supported by the National Institutes of Health (NIH) 1K22CA143148 to MGK (PI). NLN is funded by the Research Participation Program administered by ORISE through an interagency agreement between DOE and FDA.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Relevant files to this submission are publicly available via our FTP site (http://bioinf.umbc.edu/ds-score/ftp).
References
- 1.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet 2010;375:1525–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong Q, Simonis N, Li QR, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol 2009;5:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diella F, Haslam N, Chica C, et al. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci 2008;13:6580–603 [DOI] [PubMed] [Google Scholar]
- 4.Peterson TA, Adadey A, Santana-Cruz I, et al. DMDM: domain mapping of disease mutations. Bioinformatics 2010;26:2458–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong S, Blundell TL. Structural and functional restraints on the occurrence of single amino acid variations in human proteins. PLoS One;5:e9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stehr H, Jang SH, Duarte JM, et al. The structural impact of cancer-associated missense mutations in oncogenes and tumor suppressors. Mol Cancer 2011;10:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res 2001;11:863–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002;30:3894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003;13:2129–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res 2005;15:978–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao L, Zhou M, Cui Y. nsSNPAnalyzer: identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res 2005;33(Web server issue):W480–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrer-Costa C, Gelpi JL, Zamakola L, et al. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 2005;21:3176–8 [DOI] [PubMed] [Google Scholar]
- 13.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics 2006;7:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavtigian SV, Deffenbaugh AM, Yin L, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 2006;43:295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res 2006;34(Web server issue):W239–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006;22:2729–34 [DOI] [PubMed] [Google Scholar]
- 17.Conde L, Vaquerizas JM, Santoyo J, et al. PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res 2004;32(Web server issue):W242–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminker JS, Zhang Y, Waugh A, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res 2007;67:465–73 [DOI] [PubMed] [Google Scholar]
- 19.Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol 2007;8:R232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res 2007;35:3823–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res 2009;69:6660–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Krishnan VG, Mort ME, et al. Automated Inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009:2744–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calabrese R, Capriotti E, Fariselli P, et al. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat 2009;30:1237–44 [DOI] [PubMed] [Google Scholar]
- 24.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clifford RJ, Edmonson MN, Nguyen C, et al. Large-scale analysis of non-synonymous coding region single nucleotide polymorphisms. Bioinformatics 2004;20:1006–14 [DOI] [PubMed] [Google Scholar]
- 26.Thusberg J, Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum Mutat 2009;30:703–14 [DOI] [PubMed] [Google Scholar]
- 27.Mooney SD, Krishnan VG, Evani US. Bioinformatic tools for identifying disease gene and SNP candidates. Methods Mol Biol 2010;628:307–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cline MS, Karchin R. Using bioinformatics to predict the functional impact of SNVs. Bioinformatics 2011;27:441–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan DM, Ramensky VE, Sunyaev SR. Human allelic variation: perspective from protein function, structure, and evolution. Curr Opin Struct Biol 2010;20:342–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 2007;35(Database issue):D61–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeckmann B, Bairoch A, Apweiler R, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 2003;31:365–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, Anderson JB, Derbyshire MK, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 2007;35(Database issue):D237–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letunic I, Copley RR, Pils B, et al. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 2006;34(Database issue):D257–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 2003;4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Res 2008;36(Database issue):D281–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKusick VA. Mendelian Inheritance in man and its online version, OMIM. Am J Hum Genet 2007;80:588–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eddy SR. Hidden Markov models. Curr Opin Struct Biol 1996;6:361–5 [DOI] [PubMed] [Google Scholar]
- 38.Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics 2001;17:700–12 [DOI] [PubMed] [Google Scholar]
- 39.Kawabata T, Ota M, Nishikawa K. The protein mutant database. Nucleic Acids Res 1999;27:355–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215:403–10 [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, Iversen ES, Parmigiani G. Classification of missense mutations of disease genes. J Am Stat Assoc 2005;100:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue P, Forrest WF, Kaminker JS, et al. Inferring the functional effects of mutation through clusters of mutations in homologous proteins. Hum mutation 2010;31:264–71 [DOI] [PubMed] [Google Scholar]
- 43.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of somatic Mutations in cancer) database and website. Br J cancer 2004;91:355–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye J, Pavlicek A, Lunney EA, et al. Statistical method on nonrandom clustering with application to somatic mutations in cancer. BMC Bioinformatics 2010;11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talavera D, Taylor MS, Thornton JM. The (non)malignancy of cancerous amino acidic substitutions. Proteins 2009;78:518–29 [DOI] [PubMed] [Google Scholar]
- 46.Dixit A, Yi L, Gowthaman R, et al. Sequence and structure signatures of cancer mutation hotspots in protein kinases. PLoS One 2009;4:e7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenflo J, Stenberg Y, Muranyi A. Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim Biophys Acta 2000;1477:51–63 [DOI] [PubMed] [Google Scholar]
- 48.Ekman D, Bjorklund AK, Frey-Skott J, et al. Multi-domain proteins in the three kingdoms of life: orphan domains and other unassigned regions. J Mol Biol 2005;348:231–43 [DOI] [PubMed] [Google Scholar]
- 49.Rao Z, Handford P, Mayhew M, et al. The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell 1995;82:131–41 [DOI] [PubMed] [Google Scholar]
- 50.Capriotti E, Nehrt N, Kann M, et al. Bioinformatics for personal genome interpretation. Brief Bioinform 2012. [Epub ahead of print Jan 13]. http://www.ncbi.nlm.nih.gov/pubmed/22247263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annu Rev Genet 2010;44:293–308 [DOI] [PubMed] [Google Scholar]
- 52.Doughty E, Kertesz-Farkas A, Bodenreider O, et al. Toward an automatic method for extracting cancer- and other disease-related point mutations from the biomedical literature. Bioinformatics 2011;27:408–15 [DOI] [PMC free article] [PubMed] [Google Scholar]