Abstract
As a highly vascularized tissue, the placenta mediates gas and solute exchange between maternal and fetal circulations. In the human placenta, the interface with maternal blood is a unique epithelial structure known as the syncytiotrophoblast. Previously we developed a colloidal-silica based method to generate highly enriched preparations of the apical plasma membrane of the syncytiotrophoblast. Using similar preparations, a proteomics assessment of this important sub-proteome has identified 340 proteins as part of this apical membrane fraction. The expression of 38 of these proteins was previously unknown in the human placental syncytiotrophoblast. Together with previous studies, the current proteomic database expands our knowledge of the proteome of the syncytiotrophoblast apical plasma membrane from normal placentas to include more than 500 proteins. This database is a valuable resource for future comparisons to diseased placentas. Additionally, this dataset provides a basis for further experimental studies of placenta and trophoblast function.
Keywords: Placenta, syncytiotrophoblast, proteomics, placental proteomics, cationic colloidal silica
Introduction
The placenta plays critical roles in many physiological functions including the exchange of nutrients, ions, water, respiratory gases, hormones, vitamins, and other key molecules necessary for fetal development and metabolism. The interface between maternal blood and the placenta is a specialized epithelial structure known as the syncytiotrophoblast (STB). The STB is remarkably unique among mammalian cells in that cell-cell fusion during gestation forms a single cell syncytium containing tens of millions of individual nuclei housed within a single plasma membrane [1,2]. At present, little molecular information is available regarding the STB as it exists in native tissue, and a comprehensive analysis of the proteins present in the apical plasma membrane of the STB does not exist.
Several proteomics studies have been carried out using placental tissue [3], but there have only been two previous studies examining the protein composition of STB membrane fractions [4,5]. In each of these studies, membrane fractions were obtained based upon the preparation of crude microsomal-enriched fractions from placental homogenates from which the apical STB membrane was further enriched following magnesium chloride treatment to remove the basolateral membranes as described by Jimenez et al. [6]. The aim of the present study was to provide a further analysis of the sub-cellular proteome of the apical plasma membrane of the STB of the normal placenta from samples obtained using an alternative sample preparation methodology. To facilitate this analysis, we developed a novel method that utilized cationic colloidal silica (CCS) particles to isolate highly enriched preparations of microvilli containing the apical plasma membrane [7]. Additionally, our method incorporated extraction steps that dissociate protein-protein interactions thus further enriching the isolated apical membrane fraction for integral membrane proteins.
The strategy of isolation of sub-proteomes, specifically plasma membranes, has been an important component in other proteomics studies [8–10]. The utilization of CCS based plasma membrane isolation methods in proteomics studies are limited to applications in which the plasma membrane is readily accessible to allow coating with the silica nanoparticles. To date, proteomics analysis of plasma membranes isolated using CCS have been reported for various cultured cell lines [11–13], isolated primary hepatocytes [14], and endothelial cell membranes [15–18]. In the present study, we expanded the application of CCS methods for the isolation of plasma membranes obtained from intact tissue beyond those reported for endothelial cells to include the syncytiotrophoblast of the human placenta. The proteomics profiling of STB apical plasma membrane identified 340 proteins. Herein, these proteins have been classified in several different ways including the identification of many proteins whose expression was heretofore not known in the STB of the human placenta.
Materials and Methods
Microscopy
The placentas used in this study, were obtained with informed consent from elective caesarean surgeries of normal full term pregnancies following a protocol approved by the Institutional Review Board at The Ohio State University. Freshly obtained placentas were immediately dissected to obtain villi samples. Small pieces of tissue (~2mm) were placed into the primary fixative [2% glutaraldehyde in 100 mM cacodylate buffer, pH 7.2, containing 5% sucrose] for 90 min. Fixed tissue was washed prior to post-fixation with 2% OsO4 in 100 mM cacodylate buffer for 60 min. The samples were then dehydrated in ethanol, infiltrated and embedded in Epon 812 resin as we have previously reported [19].
Preparation and analysis of samples
The handling of placental tissue, coating with CCS, fractionation steps, and immunoblot procedures utilized to prepare and analyze the STB apical plasma membrane was described recently [7]. The method yielded STB apical membrane samples enriched at least 1000 fold as determined using placental alkaline phosphatase as a marker. The enriched membrane fraction (100 μg) was solubilized in boiling 1% SDS in 10 mM Tris buffer, pH 7.4, and separated by SDS-PAGE. Coomassie stained gels were cut into 32 equal pieces, and each slice was processed in an Ettan Spot Handling Workstation Robot following the Ettan Spot Handling Workstation 2.1 User Manual from Amersham Biosciences. Digestion was carried out after the gel bands were exposed to iodoacetamide (prepared as 15 mg/ml iodoacetamide in 100 mM ammonium bicarbonate solution) for 30 min in the dark. Sequencing grade modified trypsin (Promega Biosciences, Madison WI), was prepared at 5 μg/mL in 50 mM ammonium bicarbonate, and protease digestion was carried out for 180 min. The peptides were extracted from the polyacrylamide with 50% acetonitrile and 5% formic acid.
Capillary-liquid chromatography-nanospray tandem mass spectrometry (Nano-LC/MS/MS) was performed on all gel pieces generated from one placental sample as well as a total of 15 additional gel slices obtained from two separate placental preparations for comparison purposes. Samples were analyzed using a Thermo Finnigan LTQ mass spectrometer equipped with a nanospray source operated in positive ion mode. Five microliters of each sample was first injected on to the trapping column (LC-Packings A Dionex Co, Sunnyvale, CA), and washed with 50 mM acetic acid. A 5 cm 75 mm ID ProteoPep II C18 column (New Objective, Inc. Woburn, MA) packed directly in the nanospray tip was used for chromatographic separations. Peptides were eluted directly off the column into the LTQ system using a gradient of acetonitrile (2–80%) in water over 50 min, with a flow rate of 300 nl/min. The MS/MS was acquired using a nanospray source operated with a spray voltage of 3 kV and a capillary temperature of 200°C.
Sequence information from the MS/MS data were searched using Mascot Daemon by Matrix Science version 2.3.2 (Boston, MA) and the database searched against the full SwissProt database version 2010_12 (523,151 sequences; 184,678,199 residues) limited to human. The mass accuracy of the precursor ions were set to 1.8 Da given that the data was acquired on an ion trap mass analyzer and the fragment mass accuracy was set to 0.5 Da. Considered modifications (variable) were methionine oxidation and carbamidomethyl cysteine. Two missed cleavages for the enzyme were permitted. A decoy database was searched to determine the false discovery rate (FDR) and peptides were filtered according to the FDR. Protein identifications were checked manually and proteins with a Mascot score of 40 or higher having a -b or -y ion sequence tag of five residues or better were accepted. The resulting search results were merged using the Mascot merge option.
Database Searches
Predicted protein sub-cellular localization and function were determined by searching the Universal Protein Resource (UniProt) [20], and the Human Protein Reference Database [21] including gene ontology annotations and literature citations contained within. In order to determine if the identified proteins were known to be expressed in the human placenta, we used the protein name coupled with the terms “placenta” and “syncytiotrophoblast”in the PubMed search engine (Library of Medicine, National Institutes of Health). If no positive results were found, or if the studies identified were in species other than human, it was assumed that the particular protein at issue had not been detected in the human placenta. Also, if five or less “citation hits” were obtained then those papers were examined to determine if the actual protein had been detected and not just the mRNA.
Results
Microscopic Examination of Human Placental Villi
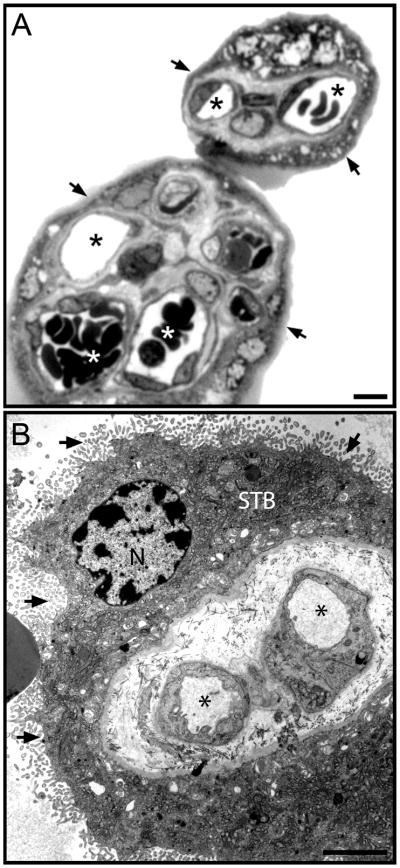
The complexity of the human placenta can be appreciated by examining the microscopic structure of the placental villus, which consists of a variety of cell types (Fig. 1A). Electron microscopy of the villus further illustrates the structural intricacies of this organ at both the cellular and sub-cellular levels, and shows numerous microvilli that project from the apical surface of the STB layer (Fig. 1B). Based upon this structural organization, we modified the Jacobson silica pellicle method to highly enrich for the apical plasma membrane of the STB [7].
Figure 1.
Microscopy of human placental villi. (A) Light micrograph of 1 μm thick section of Epon-embedded placental tissue that was stained with toluidine blue. Exposed surfaces of the STB in cross-sections of villi are indicated with arrows. The lumens of capillaries, some with red blood cells evident, are indicated (*). (B) Low-magnification electron micrograph of a thin section through a terminal villus. Numerous microvilli project from the exposed surface of the STB (arrows). A nucleus (N) in the STB is noted as are capillary lumens (*). Scale bars = 10 μm.
Proteomics Analysis
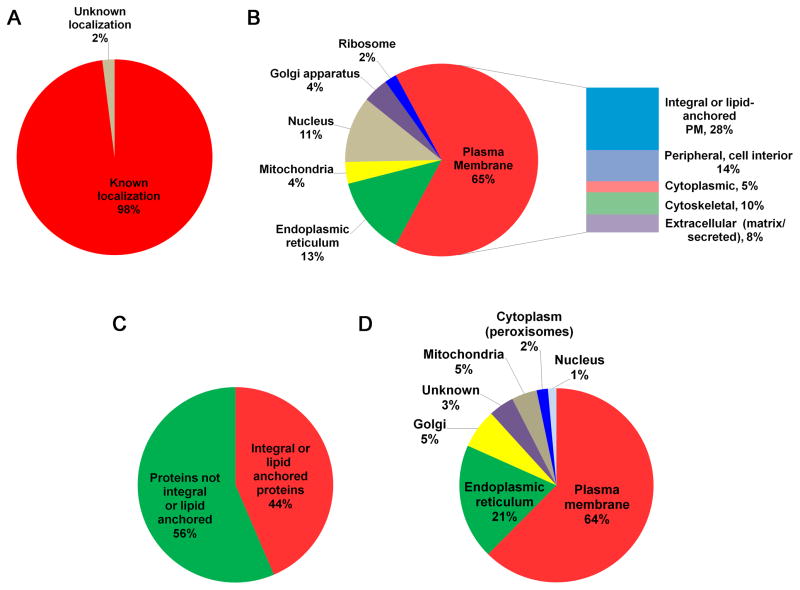
We identified 340 non-redundant proteins in the apical plasma membrane fraction of the STB (see supplemental table 1). The protein dataset includes 14 proteins that were identified based upon a single unique peptide, since these proteins have also been reported in a proteomics analysis of STB apical plasma membranes that were isolated using separate methods [5]. The proteins included in our dataset were classified based upon analysis of reported sub-cellular localization, gene ontology annotations, and literature resources contained within the UniProt and Human Protein Reference databases. Of the proteins identified in the apical membrane fraction obtained from the human STB, only 2% were classified as having an unknown sub-cellular localization (Fig 2A). Of the remaining characterized proteins, 65% could be classified as being associated with the plasma membrane (Fig 2B), while the remaining proteins were localized primarily to the nucleus (11%) or endoplasmic reticulum (13%), with fewer proteins localized to the Golgi apparatus (4%), mitochondria (4%), or ribosome (2%). Of the 220 proteins classified as associated with the plasma membrane, 95 of these (43%) could be further defined as being integral or lipid anchored membrane proteins, while 22% were considered to be peripherally associated with the interior of the plasma membrane (Fig 2B). When considering all 340 proteins identified in this analysis, (44%) could be classified as integral or lipid anchored membrane proteins (Fig 2C). Most of these transmembrane or membrane anchored proteins were localized to the plasma membrane (64%), endoplasmic reticulum (21%), or Golgi apparatus (5%), with the remaining 11% distributed to other sub-cellular organelles or having unknown localization (Fig 2D).
Figure 2.
Classification of proteins identified in the apical plasma membrane of human placental syncytiotrophoblasts. (A) Pie chart of all 340 identified proteins classified according to known localization. (B) Pie chart of proteins with known localization classified according to primary subcellular localization. The side bar shows a breakdown of the distributions of proteins potentially association with the plasma membrane. Note that the numbers in the side bar reflect the percentages with respect to total proteins with known localization. (C) Pie chart of all identified proteins classified according to membrane association (integral/lipid anchored, or not). (D) Pie chart of integral or lipid-anchored proteins classified according to reported primary sub-cellular localization.
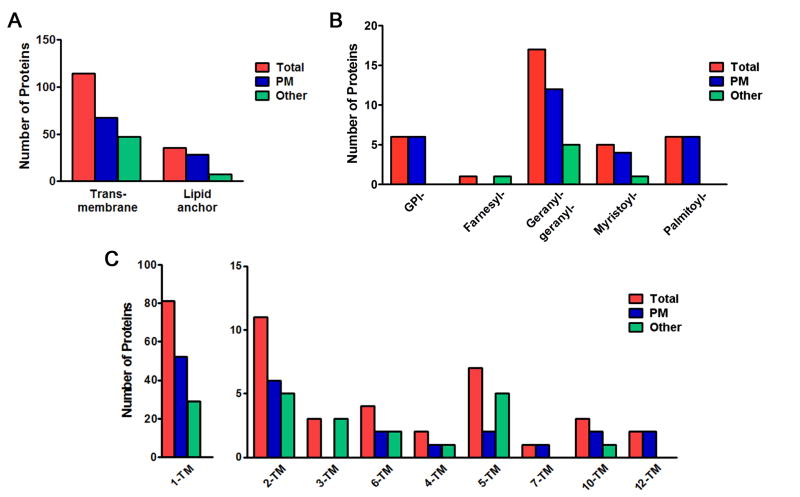
Further analysis of those proteins classified as being membrane proteins revealed that most were integral membrane proteins, which contained from 1 to 13 transmembrane domains (Figure 3A and 3C). Examination of the distribution of proteins containing lipid anchors (Fig 3B), showed that most of these proteins were associated with the plasma membrane as would be predicted.
Figure 3.
Analysis of identified integral plasma membrane proteins in human placental syncytiotrophoblast.microvilli. (A) Graph of proteins with transmembrane domains or lipid anchors, and localization to the plasma membrane (PM) or other sub-cellular organelle membranes. (B) Distribution of proteins with lipid anchors by type of linkage to the membrane. (C) Distribution of integral membrane proteins by the number of transmembrane (TM) domains.
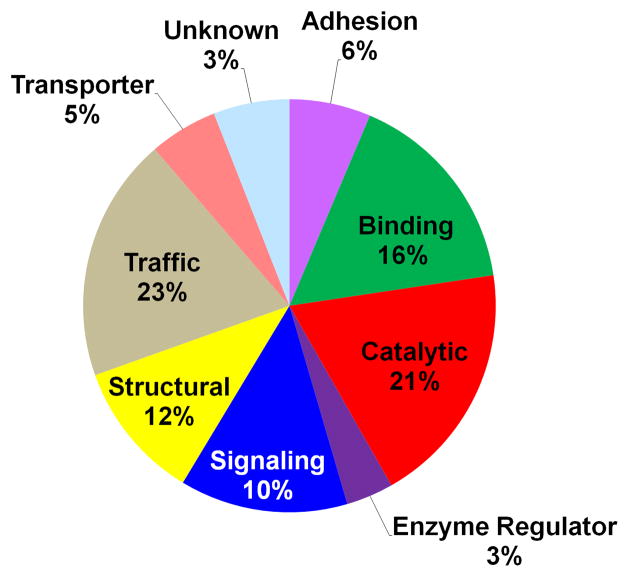
The molecular functions of the proteins identified in this study were classified into nine general categories according to gene ontology annotations and literature reports (Fig 4). It is of interest to point out that 44% of the proteins identified could be specifically classified as having potential functional roles often attributed to plasma membrane proteins including: vesicular trafficking (23%), cell signaling (10%), cell adhesion (6%), or membrane transport (5%). Many of the structural and binding proteins are also likely to have functional roles in either membrane stability or chaperone activity associated with protein folding and unfolding within the membrane.
Figure 4.
Classification of syncytiotrophoblast proteins according to the reported function.
Ultrastructural examination of the STB reveals a cytoplasm rich in membrane bound vesicles (Fig. 1). Considering the spatial orientation of the STB at the interface between the maternal blood and the fetus, and the role played by the STB in transporting nutrients necessary for fetal development, these ultrastructural characteristics are to be expected. Of particular relevance in the STB sub-proteome data set are proteins that are functionally classified as being involved in vesicular trafficking (Table 1). Among these proteins are dysferlin and myoferlin, whose expression was previously unknown in the placenta prior to the initiation of our proteomics analysis of the STB [22,23].
Table 1.
STB Proteins Associated With Vesicular Trafficking
| Protein name | UniProt# | Gene Symbol | Mol. Weight |
|---|---|---|---|
| ADP-ribosylation factor 4 | P18085 | ARF4 | 20,511 |
| ADP-ribosylation factor-like protein 8A | Q96BM9 | ARL8A | 21,416 |
| Annexin A1 | P04083 | ANXA1 | 38,714 |
| Annexin A2 | P07355 | ANXA2 | 38,604 |
| Annexin A4 | P09525 | ANXA4 | 35,883 |
| AP-1 complex subunit beta-1 | Q10567 | AP1B1 | 104,607 |
| AP-2 complex subunit alpha-1 | O95782 | AP2A1 | 107,555 |
| AP-2 complex subunit alpha-2 | O94973 | AP2A2 | 103,960 |
| AP-2 complex subunit beta | P63010 | AP2B1 | 104,553 |
| AP-2 complex subunit mu | Q96CW1 | AP2M1 | 49,655 |
| AP-3 complex subunit mu-1 | Q9Y2T2 | AP3M1 | 46,939 |
| B-cell receptor-associated protein 31 | P51572 | BCAP31 | 27,992 |
| Calnexin | P27824 | CANX | 67,568 |
| Calreticulin | P27797 | CALR | 48,142 |
| CAP-Gly domain-containing linker protein 1 | P30622 | CLIP1 | 160,990 |
| Charged multivesicular body protein 4b | Q9H444 | CHMP4B | 24,950 |
| Clathrin heavy chain 1 | Q00610 | CLTC | 191,615 |
| Coatomer subunit alpha | P53621 | COPA | 138,332 |
| Coatomer subunit beta | P53618 | COPB1 | 107,142 |
| Coatomer subunit gamma | Q9Y678 | COPG | 97,718 |
| Cytoplasmic dynein 1 heavy chain 1 | Q14204 | DYNC1H1 | 532,406 |
| Cytoskeleton-associated protein 4 | Q07065 | CKAP4 | 66,022 |
| Dysferlin | O75923 | DYSF | 237,295 |
| Exocyst complex component 2 | Q96KP1 | EXOC2 | 104,066 |
| Exocyst complex component 3 | O60645 | EXOC3 | 86,845 |
| Exocyst complex component 8 | Q8IYI6 | EXOC8 | 81,799 |
| Extended synaptotagmin-1 | Q9BSJ8 | ESYT1 | 122,856 |
| Flotillin-1 | O75955 | FLOT1 | 47,355 |
| General vesicular transport factor p115 | O60763 | USO1 | 107,895 |
| Glyceraldyde-3-phosphate dehydrogenase | P04406 | GAPDH | 36,053 |
| Heat shock cognate 71 kDa protein | P11142 | HSPA8 | 70,898 |
| Kinesin-1 heavy chain | P33176 | KIF5B | 109,685 |
| Kinesin-like protein KIF2A | O00139 | KIF2A | 84,089 |
| Lysosome-associated membrane glycoprotein 1 | P11279 | LAMP1 | 44,773 |
| Myoferlin | Q9NZM1 | MYOF | 234,709 |
| Myosin-14 | Q7Z406 | MYH14 | 228,889 |
| Myosin-Ib | O43795 | MYO1B | 131,985 |
| Myosin-Ic | O00159 | MYO1C | 118,038 |
| Myosin-Id | O94832 | MYO1D | 116,202 |
| Myosin-Ie | Q12965 | MYO1E | 127,041 |
| Myosin regulatory light polypeptide 9 | P24844 | MYL9 | 19,871 |
| PRA1 family protein 3 | O75915 | ARL6IP5 | 21,615 |
| Protein kinase C and casein kinase substrate in neurons protein 3 | Q9UKS6 | PACSIN3 | 48,487 |
| Protein RER1 | O15258 | RER1 | 22,958 |
| PTB domain-containing engulfment adapter protein 1 | Q9UBP9 | GULP1 | 34,490 |
| Ras-related C3 botulinum toxin substrate 1 | P63000 | RAC1 | 21,450 |
| Ras-related protein Rab-10 | P61026 | RAB10 | 22,541 |
| Ras-related protein Rab-11B | Q15907 | RAB11B | 24,489 |
| Ras-related protein Rab-14 | P61106 | RAB14 | 23,897 |
| Ras-related protein Rab-15 | P59190 | RAB15 | 24,660 |
| Ras-related protein Rab-18 | Q9NP72 | RAB18 | 22,977 |
| Ras-related protein Rab-1A | P62820 | RAB1A | 22,678 |
| Ras-related protein Rab-1B | Q9H0U4 | RAB1B | 22,171 |
| Ras-related protein Rab-2A | P61019 | RAB2A | 23,546 |
| Ras-related protein Rab-35 | Q15286 | RAB35 | 23,296 |
| Ras-related protein Rab-5A | P20339 | RAB5A | 23,659 |
| Ras-related protein Rab-5B | P61020 | RAB5B | 23,707 |
| Ras-related protein Rab-5C | P51148 | RAB5C | 23,483 |
| Ras-related protein Rab-6A | P20340 | RAB6A | 23,593 |
| Ras-related protein Rab-7a | P51149 | RAB7A | 23,490 |
| Ras-related protein R-Ras2 | P62070 | RRAS2 | 23,613 |
| Sorting nexin-9 | Q9Y5X1 | SNX9 | 66,592 |
| Surfeit locus protein 4 | O15260 | SURF4 | 30,394 |
| Synaptosomal-associated protein 23 | O00161 | SNAP23 | 23,354 |
| Syntaxin-11 | O75558 | STX11 | 33,196 |
| Syntaxin-12 | Q86Y82 | STX12 | 31,642 |
| Syntaxin-7 | O15400 | STX7 | 29,816 |
| Trans-Golgi network integral membrane protein 2 | O43493 | TGOLN2 | 51,007 |
| Transmembrane emp24 domain-containing protein 10 | P49755 | TMED10 | 24,976 |
| Transmembrane emp24 domain-containing protein 9 | Q9BVK6 | TMED9 | 25,105 |
| Tubulointerstitial nephritis antigen-like | Q9GZM7 | TINAGL1 | 52,387 |
| Vacuolar protein sorting-associated protein 45 | Q9NRW7 | VPS45 | 65,077 |
| Vesicle transport protein GOT1B | Q9Y3E0 | GOLT1B | 15,426 |
| Vesicle-associated membrane protein-associated protein A | Q9P0L0 | VAPA | 27,893 |
| Vesicle-associated membrane protein-associated protein B/C | O95292 | VAPB | 27,228 |
| Vesicle-trafficking protein SEC22b | O75396 | SEC22B | 24,741 |
| Vesicular integral-membrane protein VIP36 | Q12907 | LMAN2 | 40,229 |
There have been two previous proteomics data sets generated from analysis of the STB apical plasma membrane that was prepared using standard biochemical approaches [4,5]. In the first of these studies by Paradela and coworkers, only 57 proteins were identified, and most of these were based on single unique peptide identifications [4]. A more recent study by Zhang and coworkers [5], identified 292 proteins. A comparison of the proteins identified in each of the studies shows that a total of 506 proteins have been identified in the proteome of the STB apical plasma membrane identified (Fig. 5). Our dataset includes 132 proteins that were found in at least one of the previous proteomics studies, and an additional 208 proteins not previously identified in the STB apical membrane sub-proteome. A review of the literature showed that for 38 proteins associated with the plasma membrane, this was the first report of their presence in the STB of the human placenta (Table 2).
Figure 5.
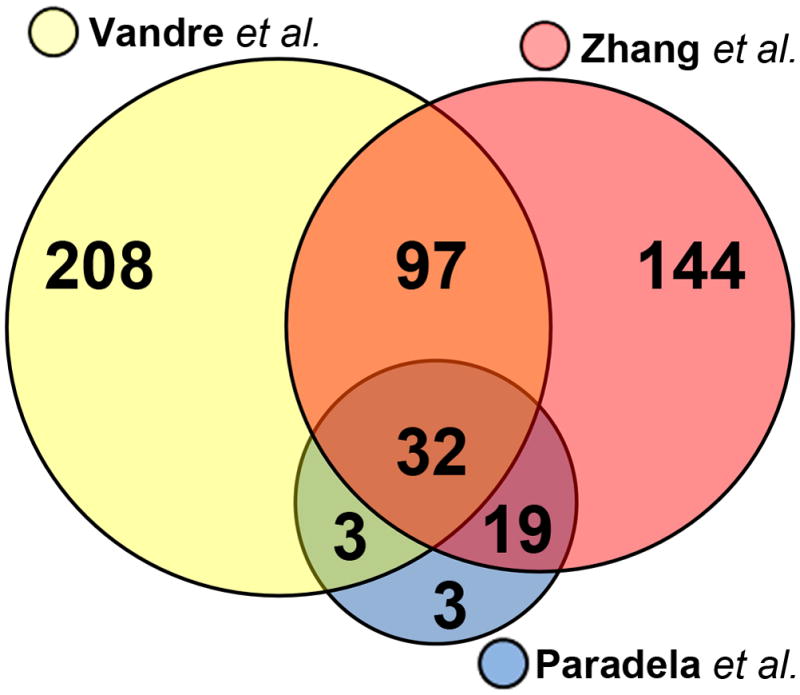
Comparison of the sub-proteome data set in this study with the STB apical membrane data set of Paradela and coworkers [4] and Zhang and coworkers [5].
Table 2.
Proteins Associated with the Plasma Membrane Not Previously Known to be in the STB of the Human
| Protein name | UniProt# | Gene Symbol | Mol. Weight |
|---|---|---|---|
| Adipocyte plasma membrane-associated protein | Q9HfDC9 | APMAP | 46,480 |
| Ankycorbin | Q9P0K7 | RAI14 | 110,041 |
| CAP Gly-domain containing linker protein 1 | P30622 | CLIP1 | 160,990 |
| Cytoskeleton-associated protein 4 | Q07065 | CKAP4 | 66,022 |
| DDB1 and CUL4-associated factor 7 | P61962 | DCAF7 | 38,926 |
| Developmentally regulated GTP-binding protein 1 | Q9Y295 | DRG1 | 40,542 |
| Developmentally regulated GTP-binding protein 2 | P55039 | DRG2 | 40,746 |
| Disintegrin and metalloproteinase domain-containing protein 10 | O14672 | ADAM10 | 84,142 |
| DnaJ homology subfamily A member 1 | P31689 | DNAJA1 | 44,868 |
| Exocyst complex component 2 | Q96KP1 | EXOC2 | 104,066 |
| Exocyst complex component 3 | O60645 | EXOC3 | 86,845 |
| Exocyst complex component 8 | Q8IYI6 | EXOC8 | 81,799 |
| Extended synaptotagmin | Q9BSJ8 | ESYT1 | 122,856 |
| Flotillin-1 | O75955 | FLOT1 | 47,355 |
| Kinesin-like protein KIF2A | O00139 | KIF2A | 84,089 |
| Metalloreductase STEAP4 | Q8NFT2 | STEAP4 | 52,518 |
| Myosin 11 | Q3MNF1 | MYH11 | 228,090 |
| Myosin-14 | Q7Z406 | MYH14 | 228,889 |
| Myosin-Ib | O43795 | MYO1B | 131,985 |
| Myosin-Id | O94832 | MYO1D | 116,202 |
| Myosin-Ie | Q12965 | MYO1E | 127,041 |
| Myosin-regulatory light polypeptide 9 | P24844 | MYL9 | 19,871 |
| Nck-associated protein 1-like | P55160 | NCKAP1L | 128,215 |
| Nicalin | Q969V3 | NCLN | 62,974 |
| PRA1 family protein 3 | O75915 | ARL6IP5 | 21,615 |
| Prohibitin 2 | Q99623 | PHB2 | 33,296 |
| PTB domain-containing engulfment adapter protein 1 | Q9UBP9 | GULP1 | 34,490 |
| Ras suppressor protein 1 | Q15404 | RSU1 | 25,545 |
| Receptor expression enhancing protein 5 | Q00765 | REEP5 | 21,493 |
| Receptor-type tyrosine protein phosphatase delta | P23468 | PTPRD | 215,651 |
| Septin 7 | Q16181 | SEPT7 | 50,680 |
| Sialic acid-binding Ig-like lectin 6 | O43699 | SIGLEC6 | 48,823 |
| Sorting nexin-9 | Q9Y5X1 | SNX9 | 66,592 |
| Synaptophysin-like protein 1 | Q16563 | SYPL1 | 28,565 |
| Synaptosomal-associated protein 23 | O00161 | SNAP23 | 23,354 |
| Syntaxin 7 | O15400 | STX7 | 29,816 |
| Syntaxin-12 | Q86Y82 | STX12 | 31,642 |
| WD repeat-containing protein 37 | Q9Y2I8 | WDR37 | 54,665 |
| Vesicle-associated membrane protein-associated protein A | Q9P0L0 | VAPA | 27,893 |
| Vesicle-associated membrane protein-associated protein B/C | O95292 | VAPB | 27,228 |
| 78 kDa glucose-regulated protein | P11021 | HSPA5 | 72,333 |
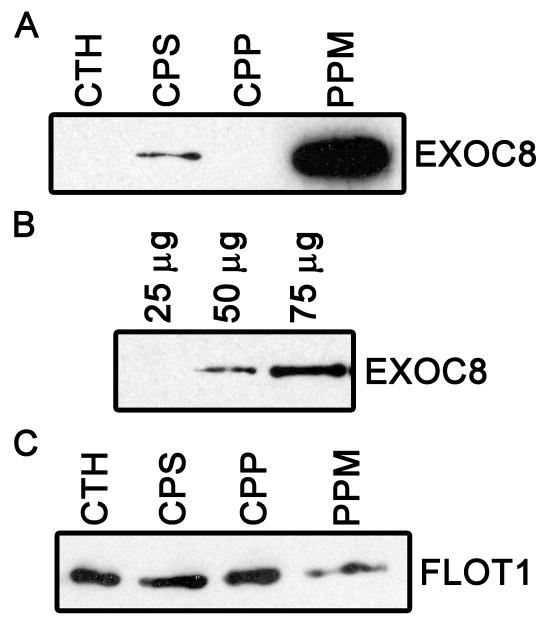
We examined the placental fractions used to generate the enriched STB apical membrane fractions by immunoblot to verify the presence of some of these previously unidentified proteins (Figure 6). Exocyst complex 8 and flotillin 1 were both detected in the placental fractions and enriched STB apical membrane fractions. The exocyst complex 8 protein was highly enriched in the final STB membrane fraction in comparison to flotillin 1. These results indicate that exocyst 8 expression is not widely distributed in other cell types within placental tissue, but is more restricted to the STB. In contrast, flotillin is likely present in other placental cell types, and may not be as highly expressed in the STB in comparison (manuscript in preparation).
Figure 6.
Immunoblots validating the presence of exocyst 8 (EXOC8) and flotillin 1 (FLOT1) in the human placenta. (A) Detection of EXOC8 in different fractions of the placenta used to prepare the STB apical plasma membranes as described [4]. Each lane contained 25 μg of protein from the crude tissue homogenate (CTH), low speed supernatant of the CTH (CPS), low speed pellet of the CTH (CPP), and the final apical plasma membrane fraction of the STB (PPM). These data indicate that EXOC8 is highly enriched in the PPM compared to other fractions including the starting material, the CTH. (B) Exocyst 8 appears to be a relatively low abundance protein in the placenta. The indicated concentrations of CTH protein were loaded onto the gel. As in panel A, 25 μg of CTH protein contained insufficient amounts of Exocyst 8 to be detected using this assay, and could onlt be detected at higher protein loads. (C) Each lane contained 25 μg of protein from the same fractions used in panel A. FLOT1 was readily detected in all fractions including the final PPM fraction, but was not highly enriched in the PPM fraction.
Discussion
Coating of exposed cell surface membranes with CCS is a useful method for isolating highly enriched plasma membrane fractions suitable for detailed proteomics analysis [11–18]. Our refinement of the CCS isolation method for the enrichment of the STB apical plasma membrane [7], and the sub-proteome dataset reported here, expands the applications of this methodology to additional tissues.
While we identified 340 proteins in this proteomics survey of the STB apical membrane, this should not be taken as a comprehensive analysis of the apical STB microvillus brush border membranome for a variety of reasons. First, following isolation of the CCS coated STB membranes, the samples were exposed to a series of extraction procedures designed to disrupt protein-protein interactions in order to strip the membranes of associated proteins that were not integral to the membrane [7]. This treatment effectively removed ~80% of the protein content associated with CCS coated STB microvilli. Proteomics analyses of the STB apical membranes that were prepared from a microsomal membrane fraction [4,5], identified 166 additional proteins that were not found in the present study. It is possible that some of the proteins detected in these previous studies may have been removed from our membrane samples as the result of the sequential extractions prior to our proteomic analysis. Secondly, it has been reported that at least 4–5 replicate mass spectrometry analyses are required to achieve 95% coverage of the proteins present in 1D-gel separated samples [24], and we were unable to fully analyze this number of replicate samples. We predict that the STB apical membrane proteome will be further refined upon additional proteomics analysis of the CCS coated STB microvillus membranes and the associated proteins that were extracted from the samples as prepared for the present analysis.
A proteomics comparison of human placenta with the mouse placental labyrinth was carried out by Cox and colleagues [25]. While complementary, their study and the present one are not directly comparable, since different sub-proteomes were analyzed. We focused on the apical plasma membrane of a single cell type, the STB. In contrast Cox and colleagues generated four sub-proteomes (i.e., nuclei, mitochondria, microsomes, and cytosol) simultaneously taken from all cell types within the tissues [25], and while they can conclude that the identified proteins were expressed in their placental samples they cannot, at present, know what cell type(s) express any given protein in their dataset. While we identified far fewer proteins than were identified by Cox and colleagues, this is to be expected since we addressed the restricted STB sub-proteome.
Electron microscopy reveals that the cytoplasm of the STB is very complex; numerous membrane vesicle profiles are observed suggestive of high levels of vesicular trafficking in the STB. The proteomics screen identified several proteins that participate in various aspects of membrane trafficking. Many of these trafficking proteins were not known to be expressed in the human placenta. We identified several proteins associated with clathrin-dependent and –independent endocytosis [26–30], as well as the exocyst complex [29], and exosomes [30]. Proteins of the exocyst complex have never been studied in the placenta, but may be important in maintenance of the microvilli and the apical plasma membrane of the STB. It has been reported that the STB of placentas from pregnancies complicated by intrauterine growth restriction (IUGR) have less elaborate microvilli that in normal placentas [31]. It is interesting to speculate that that expression of exocyst complex proteins may be modulated in IUGR. In addition, we identified several non-muscle myosins and small GTPases of the Rab family that resisted the extraction protocol applied to the isolated STB microvilli prior to the proteomics analysis [7], and thus are considered as membrane associated in the placenta. These myosin molecules were previously not known to be expressed in the STB of the human placenta, and are likely to play a role in vesicular transport. Similarly, the Rab family of small GTPases are known to regulate vesicular trafficking events, and likely play a role in the tethering of vesicles to target membranes such as the plasma membrane [32].
As we have previously reported [3,22,23], in our preliminary proteomics analysis we identified dysferlin and myoferlin as novel components of the STB apical plasma membrane. Dysferlin has been shown to function as a plasma membrane repair protein in the sarcolemma of skeletal muscle, and participates in forming a “patch” for damaged muscle fibers [33, 34]. Additionally, myoferlin has been implicated in membrane repair in cultured endothelial cells [35]. Since the STB exists as a syncytium, injury resulting from a breach in the plasma membrane might be widely propagated since there are no intervening plasma membranes between nuclei to retard the movement of injurious agents. The need for membrane repair may also be of particular significance in the placenta as a result of the shedding of aggregated nuclei from the apical STB plasma membrane in the form of syntcytial knots or as a result of ischemic injury or necrosis [36]. Our data suggests that the placenta may express more dysferlin than skeletal muscle; furthermore dysferlin is strategically located in the apical plasma membrane of the STB where it could function in membrane repair [22].
A major value of the present study is that it opens several avenues for future research. Firstly, as an extensive, though not exhaustive, proteomics analysis of the apical plasma membrane fraction of the STB it provides a basis for comparison of normal placenta and placental disorders leading to complications of pregnancy. For example, we recently carried out a pilot study to compare the expression of dysferlin in placentas from normal pregnancies and severe preeclampsia. Pre-eclamptic placentas expressed significantly less dysferlin, at both the mRNA and protein levels, than did normal placentas [37]. Two recent proteomics studies comparing normal and pre-eclamptic tissue extracts prepared from whole placental lysates separated by 2D-polyacrylamide gel electrophoresis have reported an increase in apolipoprotein A1 expression in the pre-eclamptic placenta [38,39]. Soluble forms of endoglin secreted by the placenta have also been studied as a potential marker for pre-eclampsia [40]. Interestingly, we identified both apolipoprotein A1 and endoglin as components of the STB apical membrane sub-proteome. Whether, the expression levels of these proteins, dysferlin, apolipoprotein A1, and endoglin consistently vary in the STB isolated from pre-eclamptic placenta will require further proteomic analysis. The clinical relevance of these observations have not been determined, and will need additional study with much larger sample sizes, but further serve to illustrate the importance of defining the composition of this important sub-proteome.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants HD058084 (JMR) and K08 HD49628 (WEA), and The Ohio State University Perinatal Research and Development Fund. We wish to thank Dr. Kari Green-Church from the Campus Chemical Instrument Center – Mass Spectrometry & Proteomics Facility for providing assistance with mass spectroscopy and data analysis, and the Campus Microscopy and Imaging Facility for assistance with electron microscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abumaree MH, Stone PR, Chamley LW. An in vitro model of human placental trophoblast deportation/shedding. Mol Hum Reprod. 2006;12:687–94. doi: 10.1093/molehr/gal073. [DOI] [PubMed] [Google Scholar]
- 2.Benirschke K, Kaufmann P. Pathology of the human placenta. 5. New York: Springer-Verlag; 2006. [Google Scholar]
- 3.Robinson JM, Ackerman WE, IV, Kniss DA, Takizawa T, Vandré DD. Proteomics of the human placenta: Promises and realities. Placenta. 2008;29:135–43. doi: 10.1016/j.placenta.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paradela A, Bravo SB, Henríquez M, Riquelme G, Gavilanes F, González-Ros JM, et al. Proteomic analysis of apical microvillous membranes of syncytiotrophoblast cells reveals a high degree of similarity with lipid rafts. J Proteome Res. 2005;4:2435–41. doi: 10.1021/pr050308v. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Q, Schulenborg T, Tan T, Lang B, Friauf E, Fecher-Trost C. Proteome analysis of a plasma membrane-enriched fraction at the placental feto-maternal barrier. Proteomics Clin Appl. 2010;4:538–49. doi: 10.1002/prca.200900048. [DOI] [PubMed] [Google Scholar]
- 6.Jimenez V, Henriquez M, Llanos P, Riquelme G. Isolation and purification of human placental membranes from normal and pre-eclamptic pregnancies. A comparative study. Placenta. 2004;25:422–37. doi: 10.1016/j.placenta.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Robinson JM, Ackerman WE, IV, Tewari AK, Kniss DA, Vandré DD. Isolation of highly enriched apical plasma membranes of the placental syncytiotrophoblast. Anal Biochem. 2009;387:87–94. doi: 10.1016/j.ab.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu B, McClathchy DB, Kim JY, Yates III., JR Strategies for shotgun identification of integral membrane proteins by tandem mass spectrometry. Proteomics. 2008;8:3947–55. doi: 10.1002/pmic.200800120. [DOI] [PubMed] [Google Scholar]
- 9.Codwell SJ, Thingholm TE. Technologies for plasma membrane proteomics. Proteomics. 2010;10:1–17. doi: 10.1002/pmic.200900521. [DOI] [PubMed] [Google Scholar]
- 10.Elschenbroich S, Kim Y, Medin JA, Kislinger T. Isolation of cell surface proteins for mass spectrometry-based proteomics. Expert Rev Proteomics. 2010;7:141–54. doi: 10.1586/epr.09.97. [DOI] [PubMed] [Google Scholar]
- 11.Harvey S, Zhang Y, Landry F, Miller C, Smith JW. Insights into a plasma membrane signature. Physiol Genomics. 2001;5:129–36. doi: 10.1152/physiolgenomics.2001.5.3.129. [DOI] [PubMed] [Google Scholar]
- 12.Rahbar AM, Fenselau C. Integration of Jacobson’s pellicle method into proteomic strategies for plasma membrane proteins. J Proteome Res. 2004;3:1267–77. doi: 10.1021/pr040004t. [DOI] [PubMed] [Google Scholar]
- 13.Rahbar AM, Fenselau C. Unbiased examination of changes in plasma membrane proteins in drug resistant cancer cells. J Proteome Res. 2005;4:2148–53. doi: 10.1021/pr0502370. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Jin Q, Cao J, Xie C, Cao R, Liu Z, et al. Evaluation of two cell surface modification methods for proteomic analysis of plasma membrane from isolated mouse hepatocytes. Biochim Biophys Acta. 2009;1794:32–41. doi: 10.1016/j.bbapap.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Durr E, Yu J, Krasinska KM, Carver LA, Yates III, JR, Testa JE, et al. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nature Biotech. 2004;22:985–92. doi: 10.1038/nbt993. [DOI] [PubMed] [Google Scholar]
- 16.Oh P, Li Y, Yu J, Durr E, Krasinska KM, Carver LA, et al. Subtractive proteomic mapping of the endothelial surface in lung and solid tumors for tissue specific therapy. Nature. 2004;429:629–35. doi: 10.1038/nature02580. [DOI] [PubMed] [Google Scholar]
- 17.Arjunan S, Reinartz M, Emde B, Zanger K, Schrader J. Limitations of the colloidal silica method in mapping the endothelial plasma membrane proteome of the mouse heart. Cell Biochem Biophys. 2009;53:135–43. doi: 10.1007/s12013-009-9045-8. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Xie C, Cao J, He Q, Cao R, Lin Y, et al. An in vivo membrane density perturbation strategy for identification of liver sinusoidal surface proteome accessible from the vasculature. J Proteome Res. 2009;8:123–32. doi: 10.1021/pr8006683. [DOI] [PubMed] [Google Scholar]
- 19.Lyden TW, Anderson CL, Robinson JM. The endothelium but not the syncytiotrophoblast of human placenta expresses caveolae. Placenta. 2002;23:640–52. doi: 10.1053/plac.2002.0847. [DOI] [PubMed] [Google Scholar]
- 20.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, et al. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, et al. Human Protein Reference Database – 2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandré DD, Ackerman WE, IV, Kniss DA, Tewari A, Mori M, Takizawa T, Robinson JM. Dysferlin is expressed in human placenta but does not associate with caveolin. Biol Reprod. 2007;77:533–42. doi: 10.1095/biolreprod.107.062190. [DOI] [PubMed] [Google Scholar]
- 23.Robinson JM, Ackerman WE, IV, Behrendt NJ, Vandré DD. While dysferlin and myoferlin are coexpressed in the human placenta, only dysferlin expression is responsive to trophoblast fusion in model systems. Biol Reprod. 2009;81:33–9. doi: 10.1095/biolreprod.108.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Yu J, Wang Y, Griffin NM, Long F, Shore S, et al. Enhancing identifications of lipid-embedded proteins by mass spectrometry for improved mapping of endothelial plasma membranes in vivo. Mol Cell Proteomics. 2009;8:1219–35. doi: 10.1074/mcp.M800215-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox B, Kotlyar M, Evangelou AI, Ignatchenko V, Whiteley K, Jurisica I, et al. Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology. Mol Systems Biol. 2009;5:279. doi: 10.1038/msb.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mousavi SA, Malerød L, Berg T, Kjeken R. Clathrin-dependent endocytosis. Biochem J. 2004;377:1–16. doi: 10.1042/BJ20031000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol. 2007;8:603–12. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donaldson JG, Porat-Shiliom N, Cohen LA. Clathrin-independent endocytosis: a unique platform for cell signaling and PM remodeling. Cell Signal. 2009;21:1–6. doi: 10.1016/j.cellsig.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–42. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons M, Raposo G. Exosomes – vesicular carriers for intracellular communication. Curr Opin Cell Biol. 2009;21:575–81. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Battistelli M, Burattini S, Pomini F, Scavo M, Caruso A, Falcieri E. Ultrastructural study on human placenta from intrauterine growth retardation cases. Microsc Res Tech. 2004;65:150–8. doi: 10.1002/jemt.20120. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Rossi G, Brennwald P. The ghost in the machine: small GTPases as spatial regulators of exocytosis. Trends Cell Biol. 2008;18:397–404. doi: 10.1016/j.tcb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lennon NJ, Kho A, Backai BJ, Perlmutter SL, Hyman BT, Brown RH., Jr Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J Biol Chem. 2003;278:50466–73. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 34.Bansal D, Miyake K, Vogel SS, Groh S, Chen CC, Williamson R, et al. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–72. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 35.Bernatchez PN, Sharma A, Kodaman P, Sessa WC. Myoferlin is critical for endocytosis in endothelial cells. Am J Physiol Cell Physiol. 2009;297:C484–92. doi: 10.1152/ajpcell.00498.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burton GJ, Jones CJ. Syncytial knots, sprouts, apoptosis, and trophoblast deportation from the human placenta. Taiwan J Obstet Gynecol. 2009;48:28–37. doi: 10.1016/S1028-4559(09)60032-2. [DOI] [PubMed] [Google Scholar]
- 37.Lang CT, Markham KB, Behrendt NJ, Suarez AA, Samules R, Vandré DD, et al. Placental dysferlin expression is reduced in severe preeclampsia. Placenta. 2009;30:711–18. doi: 10.1016/j.placenta.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Centlow M, Hansson SR, Welinder C. Differential proteome analysis of the preeclamptic placenta using optimized protein extraction. J Biomed Biotech. 2010;2010:458748. doi: 10.1155/2010/458748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gharesi-Fard B, Zolghardi J, Kamali-Sarvestani E. Proteome differences of placenta between pre-eclampsia and normal pregnancy. Placenta. 2010;31:121–5. doi: 10.1016/j.placenta.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Steinberg G, Khankin EV, Karumanchi SA. Angiogenic factors and preeclampsia. Thromb Res. 2009;123:S93–9. doi: 10.1016/S0049-3848(09)70020-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.