Abstract
We have achieved enhanced lipid imaging to a ~10 μm spatial resolution using negative ion mode matrix assisted laser desorption ionization (MALDI) imaging mass spectrometry, sublimation of 2,5-dihydroxybenzoic acid as the MALDI matrix and a sample preparation protocol that uses aqueous washes. We report on the effect of treating tissue sections by washing with volatile buffers at different pHs prior to negative ion mode lipid imaging. The results show that washing with ammonium formate, pH 6.4, or ammonium acetate, pH 6.7, significantly increases signal intensity and number of analytes recorded from adult mouse brain tissue sections. Major lipid species measured were glycerophosphoinositols, glycerophosphates, glycerolphosphoglycerols, glycerophosphoethanolamines, glycerophospho-serines, sulfatides, and gangliosides. Ion images from adult mouse brain sections that compare washed and unwashed sections are presented and show up to fivefold increases in ion intensity for washed tissue. The sample preparation protocol has been found to be applicable across numerous organ types and significantly expands the number of lipid species detectable by imaging mass spectrometry at high spatial resolution.
Keywords: lipids, imaging, glycerophospholipid, sulfatide, brain, negative ion, MALDI IMS
Introduction
Lipids are essential components of all tissues as constituents of the cell membrane and participate in cellular functions ranging from modulation of membrane protein signaling, signal transduction, cell motility and adhesion, and apoptosis.1,2 The diversity of lipid function is matched by a diversity of structure, with over a hundred thousand potential lipid structures possible from what appears to be simple combinations of headgroups and fatty acid constituents.1,2 Changes in regulated expression patterns of this complex molecular population have been correlated to numerous disease processes including cancer, neurological disorders, cardiovascular disease and immune response.3–7 Defining the synthesis, metabolism and regulation of complex lipid populations in relation to the biological status of an organism is an ongoing analytical challenge in lipidomics.
In recent years, matrix-assisted laser desorption ionization (MALDI) imaging mass spectrometry (IMS) has emerged as a powerful tool for detecting the spatial localization of lipids in relation to disease states or developmental biology investigations. 8,9 IMS employs a laser to probe matrix coated thin tissue sections at discrete and ordered points across the tissue, generating a lipid signature from the cellular composition of the tissue at each x and y coordinate.8 IMS is used together with histological studies to produce lipid patterns from clusters of cells, cellular microenvironments and single cells.10 Examples of recent studies utilizing IMS for lipid mapping include examination of lipid distributions in evaluation of ovarian cancer,3 embryonic implantation,11 brain tissue,12,13 and eye lens deterioration.14
Sample preparation is critical for achieving sensitive, reproducible and high spatial resolution images using IMS.15 Methods have been reported for the optimization of sample preparation procedures for positive ion mode lipid imaging.13,16–19 However, imaging by negative mode ionization has been limited by a lack of sensitivity,9 even though many lipids, including all classes of glycerophospholipids (GPL), acidic glycosphingolipids (sulfatides, ST) and sialylated glycosphingolipids (gangliosides), preferentially ionize in negative mode.20–22 Efforts to increase the sensitivity in negative ion mode have involved the development of new matrices and matrix application techniques for lipid extraction.13,17,18,23 Matrices such as 2,5-dihydroxyacetophenone (2,5-DHA) and 4-nitroaniline produce intense lipid signals in negative ion mode but are not ideal since they are readily removed from the sample due to sublimation of the matrix under the high vacuum condition of MALDI TOF instruments. Ionic matrices overcome this problem and the use of 2,5-DHA coupled with aniline has allowed improvements in peak detection for negative ion mode imaging.11 Similarly, the addition of butylamine to 2,5-dihydroxybenzoic acid (DHB) increased peak intensity in negative ion mode allowing improved analysis of phospholipids in the mouse brain cerebellum,13 and the preparation of a mixture of 2,6-dihydroxyacetophenone/ammonium sulfate/heptafluorbutyric acid enhanced imaging of sialylated sphingoglycolipids in the negative ion mode.23 In an elegant study, the matrix 9-aminoacridine (9-AA) was used to illustrate micrometer-scale gradients of phospholipids in the drosophila melanogaster egg chamber,24 yet 9-AA has been reported to result in erroneous assignments of glycerophosphocholines (PC) and glycerophosphoethanolamines (PE).25 There is a continued need to explore new approaches that increase both the sensitivity and the detection of lipids, particularly for negative ion mode imaging.
In this work we have investigated sample preparation protocols to enhance lipid detection in the negative ion mode. Washing the tissue with ammonium formate or ammonium acetate significantly improves sensitivity for negative ion mode lipid imaging. Spatial localization after treatment is retained to ~10 μm spatial resolution. Lipids are identified directly from the tissue using accurate mass collected by MALDI FT-ICR MS and fragmentation by MALDI ion trap MS/MS. Lipid signatures from washed and unwashed tissues across several organ tissues are compared. This sample preparation protocol produces a nearly fivefold increase in total ion current, allowing imaging and identification of glycerophospholipids as well as sulfatides and gangliosides in negative ion mode.
Experimental Section
Materials
Ammonium acetate, ammonium bicarbonate, ammonium formate, ammonium phosphate and formic acid were purchased from Sigma-Aldrich (St. Louis, MO, USA). 2,5-dihydroxybenzoic acid was purchased from Acros-Organics (Morris Plains, NJ, USA). Fresh frozen adult mouse brain was purchased from Pel-Freez Biologicals (Rogers, AR, USA).
Animal Care
All animal use and handling was in accordance with protocols approved by the Vanderbilt University Institutional Animal Care and Use Committee (IACUC). Postnatal day zero (P0) pups were euthanized by CO2 asphyxiation and immediately flash frozen by submerging in 1,1,1,2-tetrafluorethane and stored at −80°C until analysis.
Tissue Preparation
Tissue was cryosectioned at a 12 μm thickness, thaw mounted on gold coated targets, and vacuum dried in a desiccator for ≤ 4 hours. For washing studies, serial tissue sections were collected from the same brain tissue block. All comparative studies were performed by mounting four consecutive sections on the four edges of a gold target, with an unwashed section mounted in the center as section #3. Buffer concentrations of over 75 mM frequently resulted in cracking of tissue, and subsequently all buffers for washing were prepared to 50 mM concentration, chilled to 4°C, and the pH was measured.
The following buffers were examined:
Ammonium bicarbonate, pH 8.4
Ammonium bicarbonate, pH 7.2
Ammonium acetate, pH 6.7
Ammonium formate, pH 6.4
Ammonium phosphate, pH 4.6
Ammonium formate, pH 3.0
Formic acid, pH 2.5
Tissue sections were washed by submerging the plate edge with the mounted section into chilled (4°C) 50 mM buffer, holding the sample plate stationary for 5 seconds. Special care was taken not to agitate the mounted sample during washing. After each 5 second wash, the tissue was withdrawn completely from the buffer, and placed in fresh buffer for the next washing increment. After washing, excess liquid was blotted away and the sample was placed back into the desiccator. For ammonium formate (pH 6.4), washing length was examined at 5 s, 10 s, 15 s, and 30 s, evaluating for signal intensity increase and retention of lipid localization. After drying the tissue for ≤15 minutes under vacuum, matrix was applied by sublimation26 as previously described.10 Briefly, 363.6 ± 1.09 mg DHB was sublimated at 120°C/35 mTorr for 5.5 minutes resulting in a coating of 293.24 ± 39.75 μg/cm2. For all studies, the imaging experiments, accurate mass measurement and fragmentation experiments were performed on the same tissue section.
Imaging Mass Spectrometry and Data Analysis
Imaging experiments were performed using a reflectron geometry MALDI TOF-TOF mass spectrometer (Ultraflextreme, Bruker Daltonics, Bremen, Germany). For all analyses, the Nd:YAG/355 nm laser spot size was focused to a 8.40 ± 0.84 μm spot size and imaging data were acquired by summing up 100 shots per array position without intraspot rastering at a laser repetition rate of 1000 Hz. For evaluation of washing, image data from same brain tissue sections was collected by analyzing at an 80 μm lateral resolution in the negative ion mode, offsetting x and y by 40 μm and imaging in the positive ion mode. All other imaging was performed in negative ion mode: Sagittal brain sections were collected at a 60 μm lateral resolution, high spatial resolution images were collected at 10 μm step intervals and whole body mouse pup sections were imaged at a 125 μm lateral resolution. Variations in spatial resolution were performed in order to limit data size of the collected image. Imaging data were processed using FlexAnalysis v3.3. Images were visualized using the FlexImaging software v2.1. ClinProTools v2.2 was used to calculate individual peak intensities and standard deviations from total ion current.
Fragmentation
Negative and positive mode lipid fragmentation was performed on a MALDI-LTQ-XL hybrid linear ion trap instrument (Thermo Scientific, Waltham, MA, USA), further information in supplemental data. Lipid fragmentation patterns were manually interpreted using tools and standard spectra available at lipidmaps.org.
Accurate Mass Measurements
Accurate masses were determined using a 9.4 Tesla Apex-Qe FT-ICR MS (Bruker Daltonics, Bremen, Germany) equipped with a MALDI source on same sections used in previous experiments. Data were collected at a 200 μm lateral resolution using 50 shots by an Nd:YAG laser operating at 200 Hz, offsetting by 100 μm for positive ion mode imaging. The instrument was tuned to obtain high mass accuracy for broadband measurement over m/z 450–1600 with a 1 M transient (~ 200,00 resolution) averaging four scans per spectra.
Results and Discussion
The main objective of this study was to investigate sample preparations procedures that enhance on-tissue detection of lipid species in the negative ion mode. We present data showing which steps in sample preparation are critical to lipid analysis by IMS and report an approach that dramatically enhances negative ion mode detection of multiple lipid classes at a spatial resolution of ~10 μm using sublimated 2,5-DHB as the MALDI matrix.
Aqueous washing increases sensitivity for negative and positive ion mode imaging
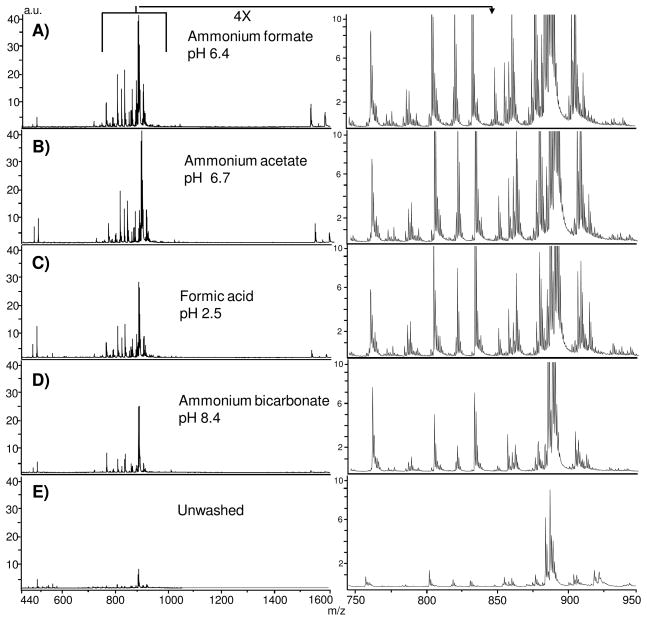
Aqueous buffers should retain lipid localization within the tissue while enhancing detection, either by formation of adducts, as observed in liquid chromatography electrospray ionization tandem mass spectrometry (LC/ESI-MS/MS) analysis of lipids,27 or by removing sample constituents that interfere with ionization of lipids. We investigated several volatile buffers at different pH values for their influence on lipid signal intensity. Adult mouse brain has been well characterized by positive ion mode IMS of lipids, 13, 26,28 and was used as the test tissue in our experiments. Figure 1 shows the effects of selected buffer composition on total ion current across thin tissue sections of adult mouse brain. All washes improved signal intensity in comparison to unwashed tissue. Washes using additional buffers are shown in (Supplemental Figure 1). We did not observe a shift in m/z values for any buffer, indicating that buffer components do not form adducts with the analytes during laser ablation, unlike LC/ESI-MS/MS analysis where buffer components readily adduct with lipid species.29 The use of ammonium formate (pH 6.4) and ammonium acetate (pH 6.7) produced the largest increase in signal intensity across the recorded mass range. A physiologically neutral pH (ammonium bicarbonate 7.2) produced intense signals, but with a total ion count less than that of ammonium formate or acetate. Altering the pH using ammonium bicarbonate (pH 8.4) or adjusting ammonium formate to pH 3.0 decreased the signal intensity and resulted in fewer detectable analytes. Likewise, washing with 0.1% formic acid (pH 2.5) yielded a reduced signal intensity. The highly acidic and basic washes (<pH 3.0; >pH 8.0) resulted in cellular content being released from the MALDI sample target. At these extreme pH values, the main functional groups of lipids become charged and/or hydrated,30 disrupting the cellular lipid membrane and this results in loss of tissue from the plate.
Figure 1.
Comparison of total ion current after buffer washes on thin tissue sections from adult mouse brain tissue coated with 2,5-DHB analyzed by MALDI TOF. Each spectra are reported relative to the unwashed tissue and are collected with the laser spot size focused to an 8.4 μm spot size. A) ammonium formate, pH 6.4; B) ammonium acetate, pH 6.7; C) formic acid, pH 2.5; D) ammonium bicarbonate, pH 8.4; E) Unwashed tissue.
Although our objective was to improve sensitivity for negative ion mode imaging, we simultaneously examined signal intensity in positive ion mode. We observed that washing significantly reduced the sodium and potassium adducted species, resulting in up to seven fold increase in M+H signal for glycerophosphocholines (Supplemental Figure 2 and 3), using ammonium formate (pH 6.4) or ammonium acetate (pH 6.7). These results correlate with previous studies reporting an increase in signal intensity due after aqueous washing for direct profiling of tissue.19 We observe that in positive ion mode removal of sodium and potassium adducts reduces the complexity of statistical comparison between salt adducted and M+H peaks and simplifies investigation of the spatial references between lipid species.
Characterization of the lipid signature after washing with ammonium formate
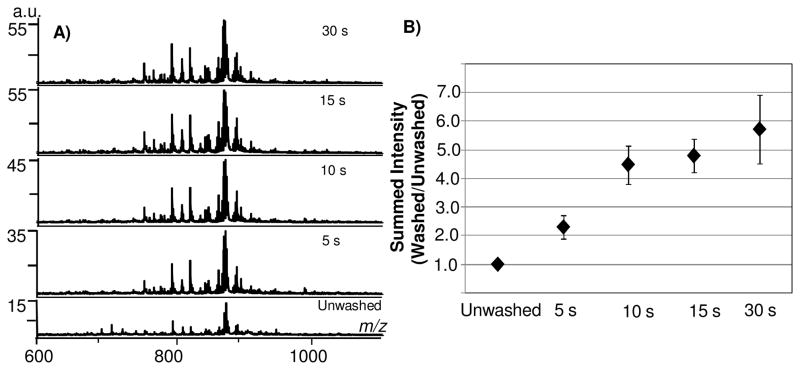
We defined the optimal washing conditions for maximized signal intensity using ammonium formate. After washing for 15 seconds, many peaks were observed that were not detected in the unwashed tissue sections resulting in more than doubling of the number of peaks detected with a S/N > 5 compared to unwashed tissue (excluding matrix-clusters). A nearly fivefold increase in total ion current was consistently observed relative to unwashed tissue (Figure 2A, B). Relative standard deviation was low at brief wash times (16% at 10 seconds, and 12% after a 15 second wash), showing good reproducibility in signal after washing the tissue. At 30 seconds, there was a larger variability in signal in comparison to earlier washing times (21% RSD). At 45 seconds (data not shown), there was frequently an observable loss of tissue from the sample target; the variability observed at 30 seconds is likely a sign of early tissue loss from the sample target. Therefore, lengthy incubations in aqueous solution must be avoided to reduce loss of tissue from the MALDI sample target. We have since found that adding a drying step after 15 seconds washing assists in maintaining the integrity of tissues that require more washing. This is particularly helpful for tissues embedded in optimal cutting temperature (OCT) media, a polyvinyl alcohol based medium. OCT embedding allows long term preservation of frozen tissue, facilitates the sectioning of small pieces of tissue, such as biopsies, and permits specific anatomical orientation of tissue sections.31 However, IMS lipid analysis of tissue sections embedded in OCT show a series of polymeric peaks overlaying the lipid region and inhibiting interpretation of lipid species. Careful washing of the tissue in the buffer solution with incorporation of a brief drying step between washes significantly decreases the OCT signature (Supplemental Figure 4) and retains the cellular content of the tissue.
Figure 2.
Aqueous washing on adult mouse brain tissue sections by MALDI TOF. A) Increased detection of analytes in total ion current during incrementally increased wash times of 5 seconds. B) Washing for 15 seconds results in nearly fivefold increase in total ion current.
We identified the primary analytes observed by negative ion mode in the overall average mass spectra from unwashed and washed tissue using accurate mass and fragmentation analysis. We found that signal from unwashed tissue is dominated by matrix clusters. Previous work has thoroughly characterized the formation of matrix-clusters observed by negative ion mode MALDI analysis of solution based mixtures.32,33 In these reports, mathematically predictable combinations of matrix molecules, water, sodium and potassium suppressed signal from in-solution analyses using the matrices α-cyano-4-hydroxycinnamic acid (CHCA) or 2,5-DHB. Factors that influence matrix cluster formation include high salt conditions, laser fluency, and high matrix:analyte ratios.32,33 In studies of unwashed tissue using the low laser fluencies necessary to obtain high spatial resolution, the 2,5-DHB matrix-cluster peaks dominated the spectrum within the range m/z 450–700 and lipid species appeared with low signal intensity (Supplemental Figure 5). On washed tissue, the matrix cluster peaks were either removed or significantly minimized. Fragmentation of the matrix-cluster peaks produced losses of 136.02 (DHB-H2O), 153.02 (DHB-H), 154.02 (DHB), 176.01 (DHB-H+Na) or 191.99 (DHB-H+K), following past reports of DHB cluster composition.32 DHB-adducted PC species were not identified from washed tissue, thus washing appears to eliminate the potential for incorrect assignments of phosphatidylcholines species due to DHB.34 We observed that the matrix-cluster peaks produced distinctive images. From studies that examined trace metals in tissue, sodium and potassium were shown to form distinctive distributions in brain tissue and other organs.35 Distinctive patterns of matrix-clusters could be due to components of the complex sample matrix of the tissue (e.g., native salts, minerals and metabolites) that are removed upon aqueous treatment. Matrix-cluster patterns potentially lead to false or skewed images through peak overlap with lipids species and may interfere with subsequent statistical analyses.
Identification of lipids from adult mouse brain tissue
An advantage of negative ion mode imaging is that subsequent identification, including the fatty acid content of glycerophospholipids, may be obtained directly from the same tissue section and matrix application. Supplemental Table 1 lists the main lipids from total ion current collected of adult mouse brain tissue, identified by fragmentation and accurate mass profiling after washing the tissue with ammonium formate. The major fatty acid chain composition of identified glycerophospholipids was octadecanoic acid (18:0) found in 51% of the fragmentation patterns and hexadecanoic acid (16:0) found in 32% of the fragmentation patterns. Main polyunsaturated fatty acids found were eicosatetraenoic acid (20:4, 19% of fragmentation patterns) and docosahexaenoic acid (22:6, 13.5% of fragmentation patterns). When using an ion trap mass spectrometer for fragmentation, we were able to readily detect the shorter chain fatty acids by the presence of the lysophosphatidic acid (LPA) fragments or by performing MS3 on peaks resulting from headgroup loss.
An abundant species in brain tissue are the acidic sulfoglycosphingolipids (sulfatides, ST). Sulfatide fragmentation by MALDI LTQ using pulsed q-dissociation produced characteristic headgroup ions at m/z 97 (sulfate group) and m/z 241 (dehydrated galactose-sulfate). We consistently observed m/z 522, 540, or 568 resulting from α-hydroxy-directed cleavage of the fatty acid side-chain from the carbonyl carbon, followed by elimination of CO (m/z 568) and double water loss (m/z 540 and 522).36 We identified sixteen sulfatides from brain tissue that in imaging experiments aligned to the white matter of the brain. This is consistent with reports utilizing LC/ESI-MS/MS and multiple reaction monitoring, where seventeen sulfatides were identified from adult mouse brain,21 and shows that IMS operated in the negative ion mode correlates well with LC/ESI-MS studies.
Finally, we identified sialylated glycolipids (gangliosides) in brain tissue.15,22 For example, m/z 1544.88 increased from negligible levels (≤3) to a signal to noise ratio of 27 by washing. MS3 allowed characterization of the structural moieties of GM1 including fatty acid fragments. Supplemental Figures 6–8 shows example fragmentation patterns of glycerophospholipids, sulfatides, and gangliosides detected from mouse brain tissue.
Aqueous washing increases sensitivity for tissue imaging while retaining high spatial resolution
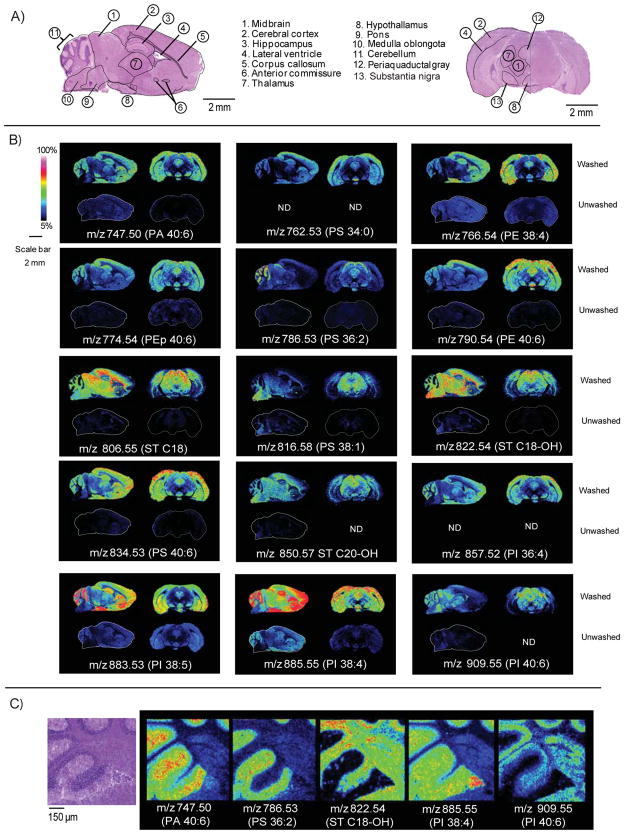
Figure 3 compares imaged ion intensities from unwashed brain tissue to those utilizing ammonium formate wash. Upon washing, all lipid images displayed an increase in ion intensity with no evidence of delocalization at a 10 μm lateral resolution. Sagittal and coronal sections showed excellent correlation in lipid expression between anatomical regions of the brain from different mice illustrating a high level of reproducibility of the method. In some cases such as for PI 38:4, a well characterized and abundant lipid observed in gray matter of the brain, 16,37,38 lipid patterns observed in unwashed tissue followed the same spatial patterns when washed, but with a significant increase in intensity. Further examples of signal enhancement following same spatial patterns in both unwashed and washed tissue are PA 40:6, PS 36:2, PS 38:1 and the sulfatide C18-OH.
Figure 3.
Comparison of washed or unwashed tissue analyzed by MALDI TOF. A) Hematoxylin and eosin stain showing anatomical features of adult mouse brain tissue sections. B) Comparison of signal across sagittal and coronal sections of adult mouse brain tissue. Images were collected with the laser spot focused to 8.4 μm, spatial resolution was 60 μm. For each lipid, the top figure represents the washed tissue; the bottom figure represents unwashed tissue analyzed under the exact same conditions. C) Image results showing adult mouse brain cerebellum collected at a 10 μm lateral resolution with the laser spot size focused to 8.4 μm. PI- glycerophosphoinositol, PA-glycerophosphatidic acid, PS- glycerophosphoserine, PG- glycerophosphoglycerol, ST- sulfatide (acidic glycosphingolipid).
The washing protocol also allowed detection of new species and ion image patterns in the negative ion mode. For example, the lipid PS 34:0 was not detected in unwashed tissue, but was detected in the cortex and hypothalamus of both sagittal and coronal sections of the brain after washing. Also, the PI 36:4 was not observed in unwashed tissue, but appeared within the gray matter of the cerebellum, cortex and hypothalamus in both the sagittal and coronal sections. Other examples of increased sensitivity allowing detection of species were the PEp 40:6 in the gray matter of the cerebellum, cortex and hypocampus, and ST d18:0/C20-OH in the white matter of the cerebellum and midbrain sections.
We examined the ability of the method to produce high spatial resolution images by imaging a portion of the cerebellum at a ~10 μm spatial resolution (Figure 3C). The ions m/z 747.50 (PA 40:6), and m/z 885.55 (PI 38:4), mapped to the gray matter and granular cells, features as small as 25 μm. The ion m/z 822.54, identified as sulfatide C18-OH, was expressed in both white matter and the granular cell layer. An example of granular cell layer distribution not observed in gray matter was seen in patterns of m/z 909.55 (PI 40:6).
Negative ion mode imaging across different organs
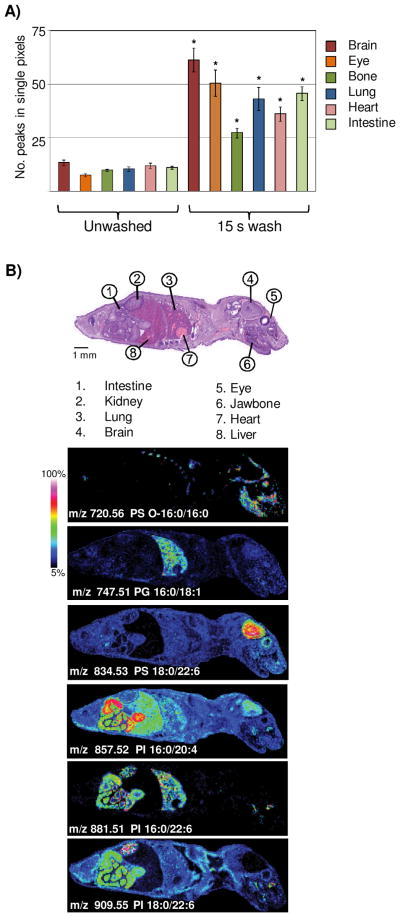
To test if washing with ammonium formate buffer enhances signal over many different tissue types, we collected and washed a thin tissue section from a whole body mouse pup at postnatal day zero. Figure 4A shows the average increase in peak numbers (excluding matrix-cluster peaks) from various organs after treatment of the tissue. Peak number increase was different for each tissue type, potentially indicating that differing organ tissue may benefit from sample preparation techniques directed at the specific tissue type, as has been shown for protein analysis by IMS.39 Examination of these spectra show unique lipid signatures that are distinctive to tissue types (Figure 4B). We found that like adult brain, PS (18:0/22:6), m/z 834.53, was abundant within the brain of the postnatal day zero mouse. Interestingly, the bone from the jawbone and rib cage appeared to have unique lipid expression, and we identified PS (O-16:0/16:0), m/z 720.56, as a lipid distinguishing these structures from other tissue types. Lung tissue has high level of surfactants and phospholipids40 and many lipid species were observed with high expression in the lung. Unique to lung was the PG (16:0/18:1), m/z 747.51. Several GPIs appeared with high abundance within the intestinal muscle. The PI (16:0/20:0), m/z 857.52, was found with intense expression in the intestinal wall and within the liver, but low abundance within lung, whereas PI (16:0/22:6), m/z 881.51, was found in the intestinal muscle, lung tissue, and kidney. The PI (18:0/22:6), m/z 909.55, was observed with highest expression in the kidney and also observed in the neck muscle and intestinal tract. This analysis shows that the aqueous wash enhances lipid signal across all examined tissue types and allows high spatial resolution analysis of lipid signatures from a variety of organs.
Figure 4.
Aqueous washing illustrated on a whole body mouse pup by MALDI TOF. A) Comparison of peak number between unwashed and washed tissue. * = student’s t-test p-value <2.0 E-4. B) Examples of images obtained across a whole body mouse pup, illustrating organ specific lipid expression in the negative ion mode.
Conclusions
The use of a simple aqueous wash was shown to produce a fivefold improvement in total ion current and up to eight fold improvement in number of analytes detected during negative ion mode imaging of lipids by IMS. All aqueous washes increased intensity of signal, but basic washes and acidic washes resulted in tissue loss from the MALDI target. This method was demonstrated utilizing 2,5-DHB, a matrix known to be stable under the high vacuum conditions within a mass spectrometer source, but sensitive to ion suppression through matrix-cluster formation in negative ion mode. Matrix-cluster formation was significantly decreased using the sample preparation protocol.
Aqueous washing has several advantages. There are obvious advantages in increased peak intensity towards robust detection of lipid species for imaging. This facilitated the use of a focused laser for high spatial resolution negative mode imaging, reported here with a 8.4 μm spot size, a level near or at single cell resolution for many tissues. The increase in peak intensity allows a greater number of lipids to be selected for MSn identification. Negative ion mode images included glycerophospholipids, sulfatides and gangliosides, illustrating that negative ion imaging can be used for a diverse array of lipid species. Last, the aqueous wash protocol is compatible with the use of the optimal cutting temperature polymer as an embedding medium. This facilitates access to lipid imaging of small pieces of tissue, such as human biopsies, and enables the use of tissue microarrays in studies aimed at determining lipid markers of disease.
Supplementary Material
Acknowledgments
The authors thank Fabian Deutskens for expertise in identifying brain anatomy and Erin Seeley for review of the material. P.M. A. was supported by the Interdisciplinary Postdoctoral Fellowship through the Systems-based Consortium for Organ Design and Engineering, 5RL1 HL092551-02 and by the NIH grants 5R01GM058008 and 1P41RR031461.
Abbreviations
- PI
glycerophosphoinositol
- PS
glycerophosphoserine
- PE
glycerophosphoethanolamine
- PA
glycerophosphate
- PG
glycerophosphoglycerol
- CerP
ceramide phosphate
- PIP
glycerolphopshoinositol monophosphate
- PIP2
glycerophosphoinositol biphosphate
- ST
sulfatide (acidic glycosphingolipid)
- FA
fatty acid
- LPA
lysophosphatidic acid
References
- 1.Brown HA, Murphy RC. Nature Chemical Biology. 2009;5:602–606. doi: 10.1038/nchembio0909-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shevchenko A, Simons K. Nature Reviews Molecular Cell Biology. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Chen Y, Momin A, Shaner R, Wang E, Bowen NJ, Matyunina LV, Walker LDE, McDonald JF, Sullards MC. Molecular Cancer. 2010;9:186. doi: 10.1186/1476-4598-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farooqui AA. The Neuroscientist. 2009;15:392–407. doi: 10.1177/1073858409337035. [DOI] [PubMed] [Google Scholar]
- 5.Lin ME, Herr DR, Chun J. Prostaglandins & Other Lipid Mediators. 2009;91:130– 138. doi: 10.1016/j.prostaglandins.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu C, Van der Heijden R, Wang M, Van der Greef J, Hankemeier T, Xu G. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2836–2846. doi: 10.1016/j.jchromb.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 7.Watson AD. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Murphy RC, Hankin JA, Barkley RM. J Lipid Res. 2009;50:S317. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs B, Süß R, Schiller J. Prog Lipid Res. 2010;49:450–475. doi: 10.1016/j.plipres.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Chaurand P, Cornett DS, Angel PM, Caprioli RM. Mol Cell Proteomics. 2011;10(2):O110.004259, 1–11. doi: 10.1074/mcp.O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnum KE, Cornett DS, Puolitaival SM, Milne SB, Myers DS, Tranguch S, Brown HA, Dey SK, Caprioli RM. J Lipid Res. 2009;50:2290–2298. doi: 10.1194/jlr.M900100-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugiura Y, Konishi Y, Zaima N, Kajihara S, Nakanishi H, Taguchi R, Setou M. J Lipid Res. 2009;50:1776–1788. doi: 10.1194/jlr.M900047-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrivas K, Hayasaka T, Goto-Inoue N, Sugiura Y, Zaima N, Setou M. Anal Chem. 2010;82:8800–8806. doi: 10.1021/ac102422b. [DOI] [PubMed] [Google Scholar]
- 14.Deeley JM, Hankin JA, Friedrich MG, Murphy RC, Truscott RJW, Mitchell TW, Blanksby SJ. J Lipid Res. 2010;51:2753–2760. doi: 10.1194/jlr.M007716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson SN, Woods AS. J Chrom B. 2009;877:2822–2829. doi: 10.1016/j.jchromb.2008.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang HYJ, Post SN, Woods AS. Int J Mass Spectrom. 2008;278:143–149. doi: 10.1016/j.ijms.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugiura Y, Setou M. Rapid Commun Mass Spectrom. 2009;23:3269–3278. doi: 10.1002/rcm.4242. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Allegood J, Liu Y, Wang E, Cachon-Gonzalez B, Cox TM, Merrill AH, Jr, Sullards MC. Anal Chem. 2008;80:2780–2788. doi: 10.1021/ac702350g. [DOI] [PubMed] [Google Scholar]
- 19.Wang HJ, Liu CB, Wu H. J Lipid Res. 2011;52:840–849. doi: 10.1194/jlr.D013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova PT, Milne SB, Myers DS, Brown HA. Curr Opin Chem Biol. 2009;13:526–531. doi: 10.1016/j.cbpa.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marbois BN, Faull KF, Fluharty AL, Raval-Fernandes S, Rome LH. Biochim Biophys Acta. 2000;1484:59–70. doi: 10.1016/s1388-1981(99)00201-2. [DOI] [PubMed] [Google Scholar]
- 22.Sugiura Y, Shimma S, Konishi Y, Yamada MK, Setou M. PLoS One. 2008;3:e3232. doi: 10.1371/journal.pone.0003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colsch B, Woods AS. Glycobiology. 2010;20:661–667. doi: 10.1093/glycob/cwq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urban PL, Chang CH, Wu JT, Chen YC. Anal Chem. 2011;83:3918–3925. doi: 10.1021/ac200481j. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs B, Bischoff A, Süß R, Teuber K, Schürenberg M, Suckau D, Schiller J. Anal Bioanal Chem. 2009;395:2479–2487. doi: 10.1007/s00216-009-3032-1. [DOI] [PubMed] [Google Scholar]
- 26.Hankin JA, Barkley RM, Murphy RC. J Am Soc Mass Spectrom. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng L, T’Kind R, Decuypere S, Von Freyend SJ, Coombs GH, Watson DGR. Rapid Commun Mass Spectrom. 2010;24:2074–2082. doi: 10.1002/rcm.4618. [DOI] [PubMed] [Google Scholar]
- 28.Sjövall P, Lausmaa J, Johansson B. Anal Chem. 2004;76:4271–4278. doi: 10.1021/ac049389p. [DOI] [PubMed] [Google Scholar]
- 29.Pulfer M, Murphy RC. Mass Spectrom Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 30.Boggs JM. Biochim Biophysic Acta. 1987;906:353–404. doi: 10.1016/0304-4157(87)90017-7. [DOI] [PubMed] [Google Scholar]
- 31.Seeley EH, Caprioli RM. Trends in Biotech. 2011;29:136–143. doi: 10.1016/j.tibtech.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller BO, Li L. J Am Soc Mass Spectrom. 2000;11:88–93. doi: 10.1016/S1044-0305(99)00126-9. [DOI] [PubMed] [Google Scholar]
- 33.Knochenmuss R, Karbach V, Wiesli U, Breuker K, Zenobi R. Rapid Commun Mass Spectrom. 1998;12:529–534. [Google Scholar]
- 34.Schiller J, SuB R, Petković M, Zschörnig O, Arnold K. Analytical Biochem. 2002;309:311–314. doi: 10.1016/s0003-2697(02)00308-1. [DOI] [PubMed] [Google Scholar]
- 35.Qin Z, Caruso JA, Lai B, Matusch A, Becker JS. Metallomics. 2011;3:28–37. doi: 10.1039/c0mt00048e. [DOI] [PubMed] [Google Scholar]
- 36.Hsu FF, Bohrer A, Turk J. Biochim Biophys Acta. 1998;1392:202–216. doi: 10.1016/s0005-2760(98)00034-4. [DOI] [PubMed] [Google Scholar]
- 37.Jackson SN, Wang HYJ, Woods AS. Anal Chem. 2005;77:4523–4527. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- 38.Asakawa D, Yoshimura K, Takeda S, Hiraoka K. J Mass Spectrom. 2010;45:437–443. doi: 10.1002/jms.1729. [DOI] [PubMed] [Google Scholar]
- 39.Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. J Am Soc Mass Spectrom. 2008;19:1069–1077. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berry KAZ, Li B, Reynolds SD, Barkley RM, Gijon MA, Hankin JA, Henson PM, Murphy RC. J Lipid Res. 2011;52:1551–1560. doi: 10.1194/jlr.M015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.