Abstract
Nitric oxide (NO), the product of the nitric oxide synthase (NOS) reaction, was previously shown to result in S-nitrosation of the NOS Zn2+-tetrathiolate and inactivation of the enzyme. To probe the potential physiological significance of NOS S-nitrosation, the inactivation timescale of the inducible NOS isoform (iNOS) was determined and found to directly correlate with an increase in iNOS S-nitrosation. A kinetic model of NOS inactivation in which arginine is treated as a suicide substrate was developed. In this model, NO synthesized at the heme cofactor is partitioned between release into solution (NO release pathway) and NOS S-nitrosation followed by NOS inactivation (inactivation pathway). Experimentally determined progress curves of NO formation were fit to the model. The NO release pathway was perturbed through addition of the NO traps oxymyoglobin (MbO2) and β2 H-NOX, which yielded partition ratios between NO release and inactivation of ~100 at 4 μM MbO2 and ~22,000 at saturating trap concentrations. The results suggest that a portion of the NO synthesized at the heme cofactor reacts with the Zn2+-tetrathiolate without being released into solution. Perturbation of the inactivation pathway through addition of the reducing agents GSH or TCEP resulted in a concentration-dependent decrease in iNOS S-nitrosation that directly correlated with protection from iNOS inactivation. iNOS inactivation was most responsive to physiological concentrations of GSH with an apparent Km value of 13 mM. NOS turnover that leads to NOS S-nitrosation might be a mechanism to control NOS activity, and NOS S-nitrosation could play a role in the physiological generation of nitrosothiols.
Nitric oxide (NO) plays essential and disparate roles in mammalian physiology, causing vasodilation in the cardiovascular system, neurotransmission in nervous tissue, and cytotoxicity against pathogens in the immune response. NO is synthesized by nitric oxide synthase (NOS), which catalyzes the conversion of arginine to citrulline and NO, using oxygen (O2) and NADPH as cosubstrates (1). Mammals possess three NOS isoforms: neuronal (nNOS), inducible (iNOS), and endothelial (eNOS). As a signaling agent, low nanomolar NO concentrations are synthesized by eNOS or nNOS. As a cytotoxin, micromolar NO concentrations are generated by iNOS at sites of infection or inflammation (2).
All three NOS isoforms are dimeric and share a common architecture, composed of two domains: a reductase and an oxidase domain. The reductase domain contains the flavin cofactors FAD and FMN and a binding site for co-substrate NADPH, which shuttle electrons from NADPH to the oxidase domain of the opposite monomer in the dimer during catalysis. Therefore, NOS is only active as a homodimer. The oxidase domain binds the substrate arginine as well as the heme and tetrahydrobiopterin (H4B) cofactors. The interface of the oxidase domain homodimer contains a Zn2+-tetrathiolate that is comprised of two cysteines from each monomer.
In its classical signaling role, NO is captured by the heme cofactor of soluble guanylate cyclase (sGC), activating sGC to produce the secondary messenger cyclic GMP (cGMP) (3). However, mounting evidence points toward an alternative, cGMP-independent NO signaling pathway where the S-nitrosation of cysteine residues regulates protein structure and activity (4). S-nitrosation has been implicated in a broad spectrum of diseases including cancer, diabetes, and other cardiovascular, pulmonary, and neurological disorders (4). Yet the mechanism by which nitrosothiols are formed in vivo is unknown.
The direct transfer of a nitrosyl group from a nitrosothiol to a free thiol (i.e. transnitrosation) is a simple bimolecular reaction. However, the initial formation of a nitrosothiol from NO and a free thiol requires a one-electron oxidation. Transnitrosation reactions therefore necessitate an “initiating” nitrosothiol formed by a protein that uses NO and a free thiol to synthesize a nitrosothiol with the aid of an oxidant. The mitochondrial hemoprotein cytochrome c was recently shown to satisfy these criteria using NO, glutathione (GSH), and ferric heme to form S-nitrosoglutathione (GSNO) (5, 6). However, the high Km value of NO and the high protein concentration utilized to observe this reaction suggests that another protein is responsible for the physiological generation of nitrosothiols at the lower NO concentrations commonly found in vivo. Cytochrome c may play a more significant role under the high NO levels encountered during nitrosative stress.
NOS is a potential candidate for the initial formation of nitrosothiols as all three mammalian NOS isoforms selectively form nitrosothiols at their Zn2+-tetrathiolate cysteines (7–11). iNOS S-nitrosation dissociates the iNOS dimer, inactivates the enzyme, and may expose the nitrosothiols for transnitrosation reactions (8). If involved in S-nitrosation, NOS must balance participation in this pathway against enzyme inactivation. Of the three NOS isoforms, only iNOS has been shown to participate in protein–protein interaction mediated S-nitrosation reactions. Kim et al. showed that formation of an iNOS-COX-2 complex was required for S-nitrosation of cyclooxygenase-2 (COX-2) (12). Furthermore, procaspase-3 and iNOS participate in an NO-dependent protein-protein interaction (13). As caspase-3 is known to be nitrosated on its active-site cysteine (14), iNOS might directly transnitrosate caspase-3. Additionally, a protein–protein complex between iNOS and arginase 1 was shown to be necessary for arginase 1 S-nitrosation (15).
Once formed, nitrosothiols can be transferred to downstream targets by formation of specific protein-protein or protein-small molecule interactions in a process analogous to phosphoryl transfer (16). Mirroring the role of ATP in phosphorylation, transnitrosation reactions may involve a small molecule nitrosothiol donor. GSH is the most abundant thiol in mammalian cells (physiological concentrations range from 0.5 to 10 mM) (17), and GSNO has been detected both intra- and extracellularly (18–20). Furthermore, GSNO can specifically transnitrosate thioredoxin (21, 22) and GSNO reductase knockout mice have markedly increased levels of S-nitrosated proteins (4). As NOS is S-nitrosated and can participate in transnitrosation reactions, NOS may also be responsible, at least in part, for the physiological generation of GSNO.
Here, we performed a detailed kinetic analysis of iNOS S-nitrosation and inactivation by treating arginine as a suicide substrate. This model allowed direct determination of the partition ratio between NO release into solution and iNOS S-nitrosation followed by inactivation. Further insight was gained by perturbing this partition ratio through varying the concentration of arginine, NO traps, or reductants. Our results indicate that the main mechanism of iNOS inactivation is S-nitrosation of the Zn2+-tetrathiolate. We hypothesize that a tunnel, present in all three NOS isoforms, acts as a conduit between the heme and Zn2+-tetrathiolate and facilitates this S-nitrosation. Our kinetic results have implications on iNOS S-nitrosation as an initial source of nitrosothiols such as GSNO.
Experimental Procedures
General materials
All chemicals used were of the highest purity commercially available and were purchased from Sigma (St. Louis, MO, USA), Aldrich (Milwaukee, WI, USA), or Fisher Scientific (Pittsburgh, PA, USA) unless otherwise noted below.
Protein expression, purification, and preparation
Expression and purification of murine iNOS (coexpressed with calmodulin) (8, 23) and β2 H-NOX (24) were carried out as described previously. iNOS concentrations were determined using the method of Bradford with bovine serum albumin as the standard (25). Horse heart myoglobin was purchased from Sigma (St. Louis, MO). Horse heart oxymyoglobin (MbO2) was generated through dithionite reduction and desalting as described previously for oxyhemoglobin (26). Oxymyoglobin concentrations were determined using a heme extinction coefficient of 13.9 mM−1cm−1 at 542 nm (27). Generation of horse heart apomyoglobin by acidification to pH 2 with 0.1 N HCl and heme extraction with methyl ethyl ketone was performed as described previously (28). Apomyoglobin concentrations were determined using an extinction coefficient of 15.5 mM−1cm−1 at 280 nm (29).
iNOS activity assays
Horse heart MbO2 assays were carried out as previously described for NOS oxyhemoglobin assays (26) with the following modifications. This assay monitors the absorbance change upon reaction of MbO2 with NO to form nitrate and metmyoglobin (metMb). Assays were monitored at either 401, 540, or 581 nm in 300 μL total volumes in clear 96-well microplates. Due to the strong absorbance of myoglobin at 401 nm, assays with MbO2 concentrations greater than 15 μM were monitored at 540 or 581 nm. Activities were determined using extinction coefficients of 45.5 mM−1cm−1 for the increase in absorbance at 401 nm, 7.5 mM−1cm−1 for the decrease in absorbance at 540 nm, or 10.5 mM−1cm−1 for the increase in absorbance at 581 nm. These extinction coefficients were determined by the difference spectrum of horse heart MbO2 and metMb (formed by reaction of MbO2 with NO) at known initial MbO2 concentrations.
Biotin switch method
The biotin switch method (11) was employed to assay S-nitrosation of iNOS. To avoid nitrosothiol decomposition, samples were protected from direct sunlight as much as possible. Assays contained 1–4 μM iNOS, 5 mM Arg, 2 mM NADPH, 50 mM NaCl, with or without GSH/TCEP in either 50 mM HEPES or HEN buffer (250 mM HEPES, 2 mM EDTA, 0.2 mM neocuproine) at pH 7.5 in 100 μL total volumes. iNOS S-nitrosation was initiated by NADPH addition and quenched by addition of 100 μL blocking buffer (10% SDS and 60 mM N-ethylmaleimide in HEN buffer) and incubated at 55 °C for 30 minutes. Protein was then precipitated with 1.2 mL acetone pre-cooled to −20 °C and pelleted at 14,000 rpm for 10 minutes at 4 °C. The supernatant was removed and the pellet was washed with 0.9 mL more acetone, and the pellets were air-dried. Dried pellets were resuspended in PBS with 5% SDS, 30 mM ascorbate, and 500 μM biotin-maleimide or biotin-iodoacetamide (Thermo Fisher Scientific; Rockford, IL) and incubated at 37 °C for 30 minutes. Biotin labeling was quenched with 50 mM DTT or gel loading buffer containing 10 mM DTT. Relative iNOS S-nitrosation levels were determined by immunoblotting for biotin labeling using the Vectastain ABC Kit (Vector laboratories; Burlingame, CA) and SuperSignal West pico or femto maximum sensitivity substrate (Thermo Fisher Scientific; Rockford, IL). To ensure equal protein loading between samples, the blots were stained with Ponceau stain after imaging.
Results
Nitrosation and inactivation of iNOS during enzymatic turnover
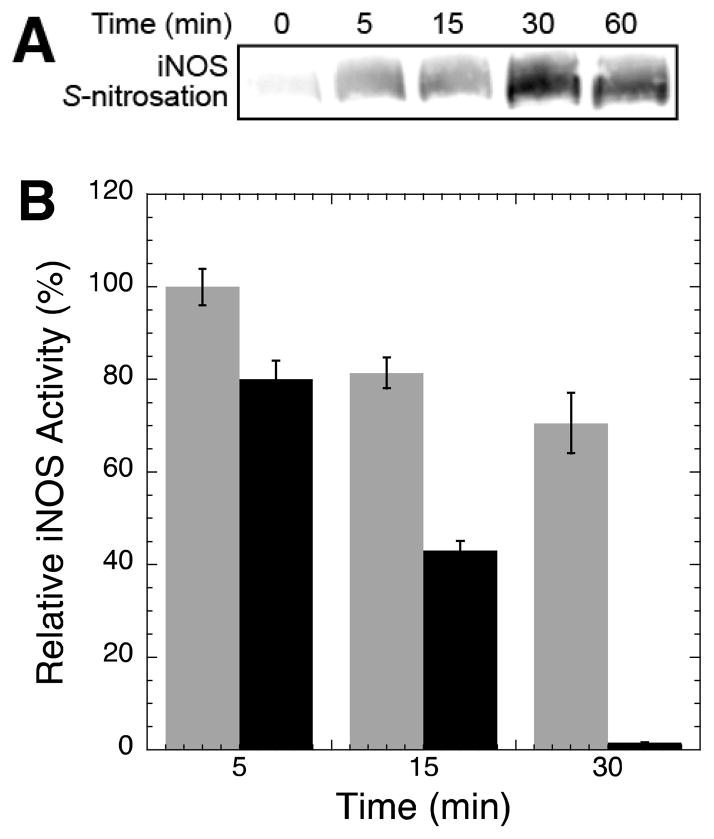
To gain insight into the physiological role of NOS S-nitrosation, we first determined if there was a direct temporal correlation between iNOS S-nitrosation and inactivation. While iNOS was previously shown to be S-nitrosated by exogenously added NO (8) and endogenously generated NO at the heme cofactor (30), the timescale of auto-S-nitrosation were not investigated. Here, iNOS was rapidly S-nitrosated by NO generated during the catalytic reaction (Figure 1A), and the level of iNOS auto-S-nitrosation peaked at ~30 minutes and did not increase at 60 minutes. S-nitrosation levels were determined using the biotin switch assay (11) which selectively and covalently labels sites of S-nitrosation with biotin for detection by immunoblotting.
Figure 1.
iNOS S-nitrosation and inactivation during enzymatic turnover. (A) iNOS S-nitrosation during enzymatic turnover. Each reaction contained 4 μM iNOS, 5 mM Arg, 2 mM NADPH, 50 mM NaCl, and 50 mM HEPES pH 7.5. iNOS activity was initiated by NADPH addition and 100 μL aliquots were taken at the indicated timepoints and quenched with an equal volume of 250 mM HEPES at pH 7.4 containing 2 mM EDTA, 0.2 mM neocuproine, 60 mM N-ethylmaleimide, and 10% SDS. Relative levels of iNOS S-nitrosation were determined by the biotin switch method (see Experimental Procedures). (B) iNOS inactivation during enzymatic turnover. Two separate reactions were initiated containing 90 nM iNOS, 15 mM Arg, 150 mM NaCl, and 100 mM HEPES pH 7.5. To one reaction iNOS activity was initiated at time zero by NADPH (600 μM) addition (black bars), to the other reaction NADPH was only added immediately prior to rate determination (grey bars). At the given timepoints, 100 μL aliquots of each reaction were added to 200 μL of 45 μM MbO2 in 100 mM HEPES pH 7.5 to obtain final concentrations of 30 nM iNOS, 5 mM Arg, 50 mM NaCl, 30 μM MbO2, and 100 mM HEPES pH 7.5.
Exogenous addition of NO was previously shown to inactivate NOS in vitro (8, 31–33). This inactivation correlated with iNOS dimer dissociation due to S-nitrosation of the Zn2+-tetrathiolate (8). However, the timescale of inactivation by endogenously generated NO at the heme cofactor had not been investigated. To determine the timescale of iNOS auto-inactivation, iNOS-catalyzed NO formation (and thus auto-S-nitrosation) was initiated by addition of both Arg and NADPH at time zero and the remaining iNOS activity at 5, 15, and 30 minutes was determined using the increase in absorbance at 401 nm upon reaction of NO with oxymyoglobin (MbO2) to form nitrate and metmyoglobin (metMb). Indeed, iNOS catalyzed NO formation resulted in rapid loss of NOS activity (Figure 1B black bars) that correlated with an increase in iNOS auto-S-nitrosation (Figure 1A). A similar rapid decline in nNOS activity was previously observed with near complete inactivation after 30–90 minutes (34, 35). To ensure the observed iNOS inactivation was a direct result of NOS activity and not general protein instability at room temperature, only Arg was added at time zero and NADPH was not added until immediately prior to assaying the rate at each timepoint. A relatively modest loss in iNOS activity was observed under these conditions (Figure 1B grey bars) indicating that iNOS auto-inactivation was a direct result of NOS activity and auto-S-nitrosation.
Importantly, addition of superoxide dismutase (SOD) (50 units) and catalase (50 units) failed to protect iNOS from inactivation and instead resulted in a slight (~15%) increase in iNOS inactivation (data not shown). A similar slight increase in inactivation rate was previously observed upon addition of SOD to iNOS (36) and eNOS (31) activity assays. This result indicated that iNOS auto-inactivation was not due to generation of reactive oxygen species from uncoupling of the iNOS reductase and oxidase domains.
Kinetic model of iNOS auto-inactivation
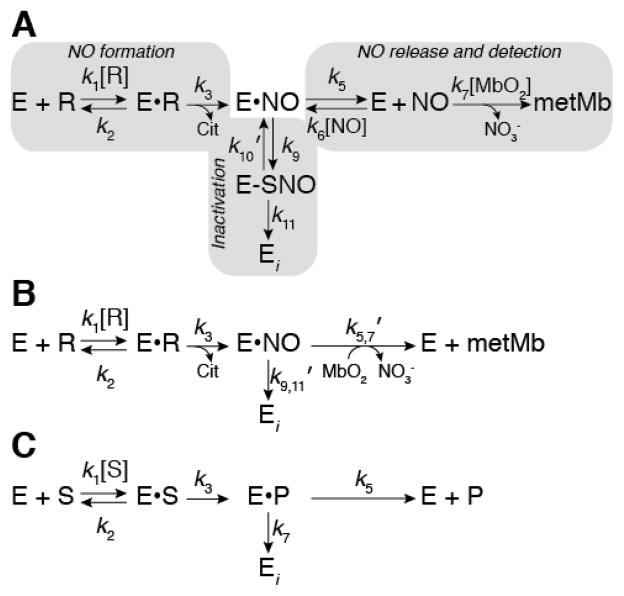
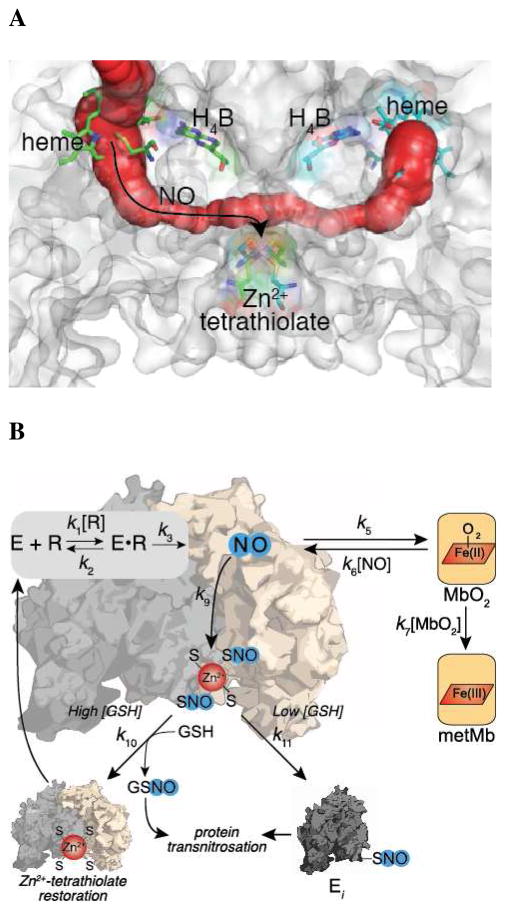
The observation that iNOS auto-inactivation directly correlated with an increase in iNOS auto-S-nitrosation led us to develop a kinetic model for iNOS inactivation by S-nitrosation (Figure 2A). In this model Arg binding to iNOS (k1 and k2) is followed by irreversible formation of NO and citrulline (k3) (NO formation pathway). The NO formed at the heme cofactor is then released into solution (k5 and k6) where NO then reacts with MbO2 to form nitrate and metMb (k7) (NO release/detection pathway). Alternatively, NO may react with the Zn2+-tetrathiolate resulting in S-nitrosation (k9) followed by dimer dissociation and inactivation (k11) (S-nitrosation/inactivation pathway). Prior to dimer dissociation, S-nitrosation can be reversed (k10′) through addition of exogenous reductants (see below). Note that E•NO represents NO sequestered within iNOS such that the NO is inaccessible to MbO2, but not necessarily bound to the heme iron. Therefore, k6 represents the diffusion of NO into the iNOS protein environment, not NO binding to the heme iron. The kinetic model in Figure 2A can be further simplified by replacing the NO release/detection and inactivation pathways with net rate constants (Figure 2B):
| (1) |
| (2) |
Figure 2.
Kinetic model of iNOS inactivation. (A) Complete kinetic model of iNOS auto-inactivation by S-nitrosation and dimer dissociation where the kinetic readout is reaction of NO with MbO2. Gray boxes indicate the NO formation, inactivation, and NO release/detection pathways. E represents the enzyme (i.e. iNOS), R represents arginine, E•R represents arginine bound within the iNOS active-site, E•NO represents nitric oxide sequestered within iNOS but not necessarily bound to the heme iron, E-SNO represents iNOS S-nitrosated at the Zn2+-tetrathiolate cysteines, and Ei represents inactivated iNOS. (B) A simplified kinetic model in which the inactivation and NO release/detection pathways are represented by net rate constants. (C) General kinetic model of a suicide substrate where S represents the substrate and P represents the product.
The kinetic model in Figure 2B is identical to that of a suicide substrate (mechanism-based inhibitor) (Figure 2C). Therefore, using suicide substrate analysis (37–43), plots of NO formation over time may be fit to equation 3:
| (3) |
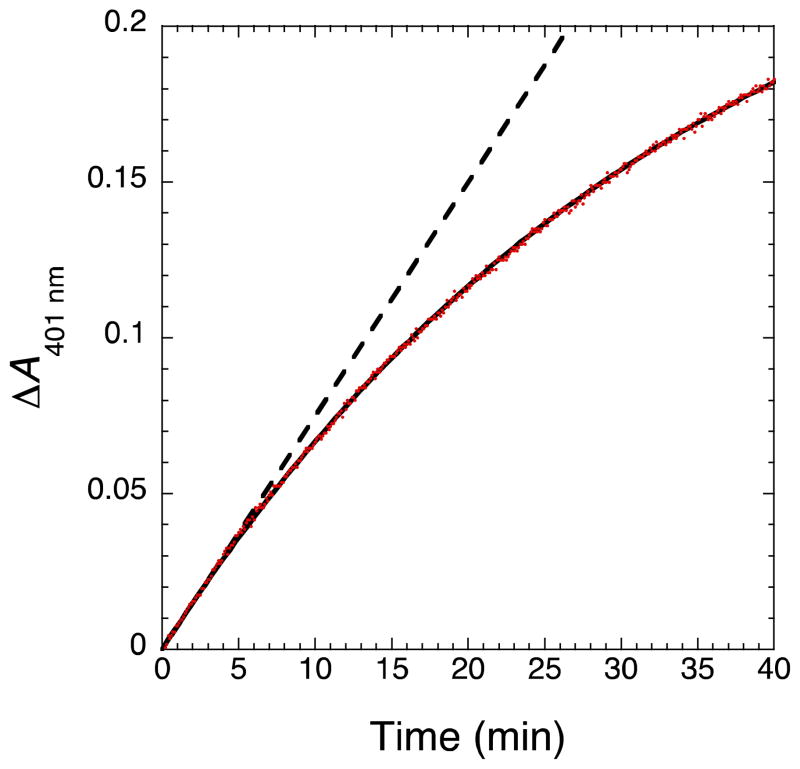
where [NO]∞ is the total concentration of NO formed at infinite time and λ is the NOS inactivation rate. Equation 3 provided excellent fits to progress curves of iNOS catalyzed NO formation obtained using the MbO2 assay (Figure 3). MbO2 rapidly reacts with NO to form nitrate and metMb at a rate of 3.4 × 107 M−1s−1 (44), which may be followed spectrally by a change in absorbance at 401, 540, or 581 nm. The timescale of inactivation in the MbO2 assay (Figure 3) was slower than without MbO2 (Figure 1B), since MbO2 reacts with NO to form nitrate and metmyoglobin (metMb) further indicating that NO is the primary source of iNOS inactivation.
Figure 3.
Progress curve of NO formation exhibits inactivation over time. The reaction contained 15 nM iNOS, 5 mM Arg, 200 μM NADPH, 50 mM NaCl, 4 μM MbO2, and 100 mM HEPES pH 7.5 in 300 μL total volume in a 96-well microplate. NO formation was determined by the increase in absorbance at 401 nm observed upon reaction of NO with MbO2 to form metMb and nitrite. Data was fitted to equation 3 and the resulting fit is shown as a solid black line. The dashed line represents the linear fit to the first five minutes of the progress curve.
Using suicide substrate analysis (37–43) [NO]∞ and λ can be expressed in terms of the kinetic model rate constants:
| (4) |
| (5) |
where r is the partition ratio between the NO release/detection and S-nitrosation/inactivation pathways. This partition ratio can easily be determined for a given set of conditions via a plot of [NO]∞ versus the initial iNOS concentration. If Arg is a suicide substrate, then this plot is predicted to give a straight line that intersects the origin with a slope equal to r. Indeed, fitting a plot of [NO]∞ versus iNOS concentration at saturating substrate concentrations and 4 μM MbO2 yielded a straight line that passed through the origin with a slope of 106 ± 2 (Figure S1). This indicated that ~100 molecules of NO were released into solution and reacted with MbO2 for every one molecule of NO that led to S-nitrosation of the Zn2+-tetrathiolate. However, it should be noted that the [NO]∞ value and therefore the r-value greatly depend on the MbO2 concentration utilized in the assay. This ability of MbO2 to “trap” solution NO and alter the inactivation rate will be exploited below.
Reductants protect iNOS from S-nitrosation and inactivation
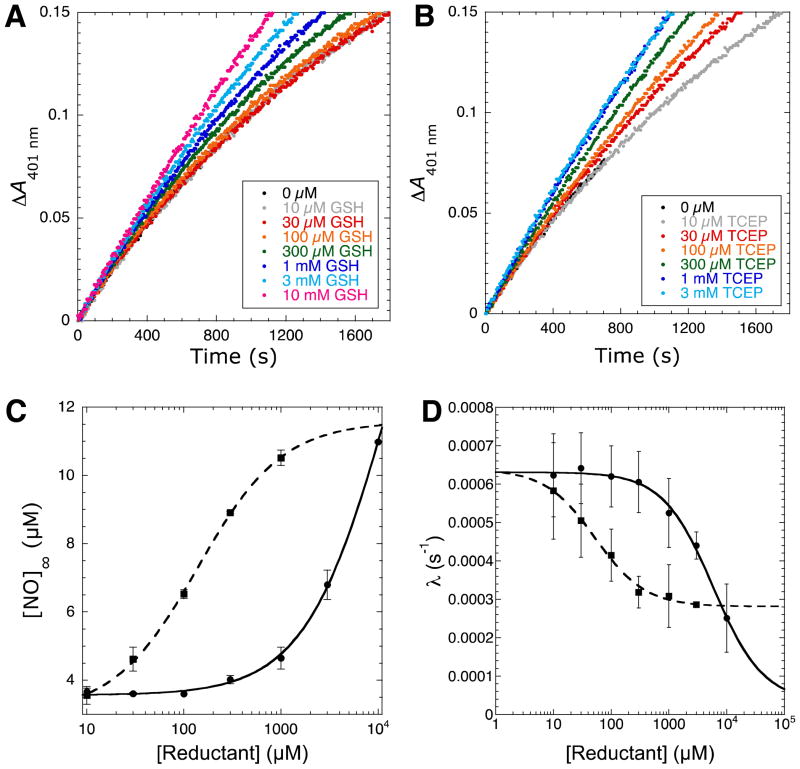
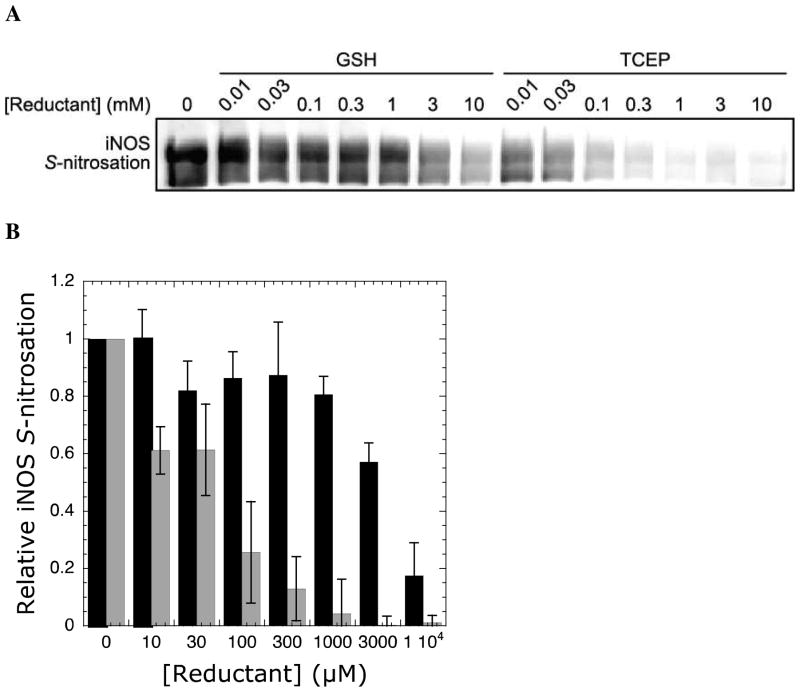
Thiols like glutathione (GSH) and other reducing agents such as TCEP are capable of reducing nitrosothiols to free thiols. GSH and other thiol reducing agents were shown to prevent iNOS (45, 46) and nNOS inactivation (35, 47). Therefore, the inactivation pathway of the kinetic model (Figure 2A) can be perturbed through addition of reducing agents. Indeed, both GSH (Figure 4A) and TCEP (Figure 4B) reduced the inactivation rate, though TCEP required approximately two orders of magnitude lower concentration compared to GSH for a similar effect.
Figure 4.
Protection of iNOS inactivation by reductants. Progress curves of iNOS catalyzed NO formation in the presence of increasing concentrations of (A) GSH and (B) TCEP. Individual reactions contained 15 nM iNOS, 5 mM Arg, 200 μM NADPH, 50 mM NaCl, 4 μM MbO2, 100 mM HEPES pH 7.5, and varying concentrations of either GSH or TCEP in 300 μL total volumes in a 96-well microplate. NO formation was measured via the change in absorbance at 401 nm upon reaction of MbO2 with NO. (C) Plot of [NO]∞ versus GSH (circles, solid line) or TCEP (squares, dashed line) concentration. Data was fitted using equations 2, 5, and 6. (D) Plot of inactivation rate (λ) versus GSH (circles, solid line) or TCEP (squares, dashed line) concentration. Data was fitted using equations 2, 4, and 6.
As only a finite concentration of S-nitrosated iNOS (E-SNO in Figure 2A) exists, we hypothesized that reducing agent addition would exhibit saturation kinetics and alter the rate of nitrosothiol reduction (k10′) through the following relationship:
| (6) |
where k10 is the maximum rate of nitrosothiol reduction, [reductant] is the GSH or TCEP concentration, and is the apparent Km of nitrosothiol reduction. Therefore, when no reductant is added to the assay mixture, k10′ = 0 and nitrosothiol formation is irreversible and committed to enzyme inactivation. Plots of [NO]∞ versus [reductant] yielded excellent fits to the relationship between [NO]∞ and [reductant] shown in equation 7 derived from equations 2, 5, and 6 (Figure 4C).
| (7) |
Similarly, plots of λ versus [reductant] concentration also yielded excellent fits to the relationship derived from equations 2, 4, and 6 (Figure 4D). These experiments and those below allowed estimation of values for the various kinetic constants in the kinetic model. In general, data plots were fitted using non-linear regression in Kaleidagraph (Synergy Software; Reading, PA) and the resulting kinetic constants are displayed in Table 1.
Table 1.
Rate constants derived from the kinetic model of inactivation.
| Rate constant | Kinetic Model Step | Reductant titration | Varying MbO2 | |
|---|---|---|---|---|
| k3 (s−1) | NO formation | NA | ~2.2a | |
| k5 (s−1) | NO diffusion from NOS | NA | 16.1 ± 5.6b | |
| k6[NO] (s−1) | NO diffusion into NOS | NA | (1.6 ± 0.6) × 103 b | |
| k7 (M−1s−1) | MbO2 reaction with NO | 3.4 × 107 c | 3.4 × 107 c | |
|
|
Inactivation pathway without reductant | (6.7 ± 0.2) ± 10−4 d | (7.4 ± 2.5) × 10−4 b | |
|
|
Inactivation pathway with saturating reductant | (1.1 ± 0.1) ± 10−4 d | NA | |
| k10GSH/k10TCEP | S-nitrosothiol reduction | 2.0 ± 0.2d | NA | |
| Km, appGSH (μM) | (1.3 ± 0.2) × 104 d | NA | ||
| Km, appTCEP (μM) | (1.4 ± 0.1) × 102 d | NA |
Rate was approximated from the single turnover rate of NHA formation from arginine, the single turnover rate of NO, citrulline formation from NHA, and the rate of NO release from iNOS ferric heme (50–54).
Rate was determined from fit in Figure 7A.
Rate from reference (44).
Rate was determined from fit in Figure 5C.
If iNOS auto-S-nitrosation is the main source of auto-inactivation, protection from inactivation by reductants should correlate with a decrease in S-nitrosation. Indeed, using the biotin switch assay, a decrease in iNOS S-nitrosation was observed with increasing concentrations of both GSH and TCEP (Figure 5). In particular, a concentration-dependent decrease in iNOS auto-S-nitrosation was observed at GSH concentrations greater than 1 mM and TCEP concentrations greater than 30 μM. Importantly, the effect of both reductants on iNOS S-nitrosation correlated well with the observed decrease in inactivation rate under varying reductant concentrations (Figure 4D).
Figure 5.
Protection of iNOS auto-S-nitrosation by reductants. (A) The anti-biotin blot shows a dose-dependent decrease in iNOS S-nitrosation upon addition of GSH and TCEP. (B) Quantitation of anti-biotin blots with increasing concentrations of GSH (black bars) and TCEP (grey bars). Levels of iNOS S-nitrosation were normalized to the level observed with no added reductant. Error bars represent standard deviation from the mean. Each reaction contained 2 μM iNOS, 5 mM Arg, 2 mM NADPH, 50 mM NaCl, and varying concentrations of either GSH or TCEP in HEN buffer (250 mM HEPES, 2 mM EDTA, 0.2 mM neocuproine, pH 7.5) in 100 μL total volume. iNOS activity was initiated by NADPH addition and quenched after 20 min in HEN buffer with 60 mM N-ethylmaleimide and 10% SDS. Relative levels of iNOS S-nitrosation were determined by the biotin switch method (see Experimental Procedures).
The kinetic model described above (Figure 2) indicates iNOS is only inactivated upon dimer dissociation (k11) following S-nitrosation (k9). If this model is correct, then added reductants (k10′) should protect S-nitrosated iNOS (E-SNO) from inactivation, but not recover activity from inactivated iNOS (Ei) as k11 is irreversible. To determine if the activity of inactivated iNOS could be recovered by reductant addition, iNOS was fully inactivated as in Figure 1B at which point 3 mM GSH or 1 mM TCEP was added and the resulting iNOS activity was assayed. iNOS activity was not recovered by this method (data not shown) indicating that iNOS inactivation is irreversible. This result was consistent with a previous study in which GSH was added to fully inactivated nNOS and no recovery of nNOS activity was observed (35). Therefore, k11 is a necessary irreversible rate constant as part of the kinetic model (Figure 2A). The inability to recover inactivated iNOS is also consistent with the hypothesis that dimer dissociation following S-nitrosation and Zn2+ loss, not S-nitrosation or Zn2+ loss itself, is responsible for inactivation.
iNOS inactivation rate depends on the rate of NO formation
Further insight into the kinetic model was obtained through perturbation of the NO formation pathway (Figure 2A). This was done by utilization of subsaturating Arg concentrations. When Arg is not saturating [NO]∞ is governed by equation 8 (37):
| (8) |
where Km is the Michaelis constant for Arg previously determined to be 8.6 μM (48). Note that when Arg is saturating equation 9 reduces to equation 5. Also, note that the apparent Km value for Arg (Km,app) is larger than the actual Km value by a factor of (k3 + k5,7′ + k9,11′)/k3. Progress curves at varying Arg concentrations from 6.25 to 100 μM were fitted to equation 3 to determine [NO]∞ and then plots of [NO]∞ versus Arg concentration (Figure S3) were fitted to equation 9 by non-linear regression using Kaleidagraph (Synergy Software; Reading, PA) to obtain the k5,7′/k9,11′ ratio and apparent Km value. Using 8 μM MbO2, an apparent Km value of 42 ± 2 μM and k5,7′/k9,11′ ratio of 690 ± 20 was determined. Alternatively, equation 8 may be rearranged to give equation 9:
| (9) |
Therefore, the plot of [iNOS]/[NO]∞ versus 1/[Arg] was predicted to yield a straight line with an ordinate intercept of k9,11′/k5,7′ and a slope of k9,11′Km/k5,7′. Indeed, the plot of [iNOS]/[NO]∞ versus 1/[Arg] yielded a straight line (Figure S3 inset) with a k9,11′/k5,7′ value of 0.0011 ± 0.0002 and a k9,11′Km/k5,7′ value of 0.069 ± 0.002. The positive slope observed highlighted the difference between Arg and a classical suicide substrate as the plot of [E]/[P]∞ versus 1/[S] for a classical suicide substrate would yield horizontal line since [P]∞ is independent of substrate concentration. A classical suicide substrate binds the enzyme and then reacts within the active site in a manner that prevents subsequent substrate binding and turnover. In contrast, NO inactivates iNOS at a site distal to the active site (i.e. the Zn2+-tetrathiolate). Thus, Arg binding and turnover proceeds until iNOS S-nitrosation, dimer dissociation, and enzyme inactivation occurs and [NO]∞ depends on the relative flux between the NO formation and inactivation pathways at subsaturating Arg concentrations.
NO traps decrease the iNOS inactivation rate
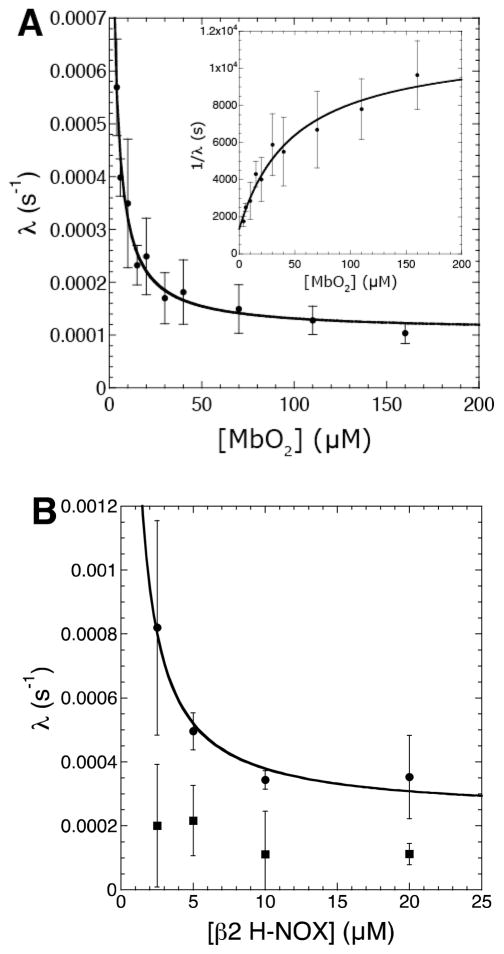
Finally, varying concentrations of solution NO traps were used to perturb the NO release/detection pathway (Figure 2A). Previously, oxyhemoglobin (HbO2) was utilized as a solution NO trap to protect nNOS (32, 33) and eNOS (31, 49) from auto-inactivation in activity assays. Here, in place of hemoglobin, horse heart myoglobin was utilized since it lacks cysteine residues and thus avoids potential assay artifacts due to reaction of NO or nitrosothiols with cysteines. Using saturating Arg, the inactivation rate was determined at varying concentrations of MbO2 from 4–160 μM. The resulting plot of λ versus MbO2 concentration exhibited a dependence on the amount of trap at low MbO2 concentrations but was independent at high MbO2. The trap-dependent portion of the plot is likely due to competition between MbO2 and the Zn2+-tetrathiolate for reaction with NO in solution; the trap-independent portion likely indicates that a portion of NO reacts with the Zn2+-tetrathiolate without first being released into solution (see Discussion). This concentration-dependent trap result was predicted from the kinetic model, and plots of λ versus MbO2 concentration yielded excellent fits to the relationship between λ and MbO2 derived from equations 1 and 4 (Figure 6A). To simplify the determination of individual kinetic constants in the kinetic model, the plot of 1/λ versus MbO2 concentration (Figure 6A inset) was fitted to equation 10 using Kaleidagraph (Synergy Software; Reading, PA).
Figure 6.
Plots of iNOS inactivation rate (λ) versus (A) MbO2 concentration and (B) β2 H-NOX concentration. Individual reactions contained 15 nM iNOS, 5 mM Arg, 200 μM NADPH, 50 mM NaCl, 100 mM HEPES pH 7.5, and varying concentrations of either MbO2 or β2 H-NOX in 300 μL total volumes in a 96-well microplate. NO formation was measured as described under Experimental Procedures. The resulting progress curves were fitted to equation 3 to determine the inactivation rate (λ). The plots of inactivation rate (λ) versus NO trap concentration were fitted using equations 1 and 4. The inset in (A) was fitted to equation 8.
| (10) |
Since k3 could be estimated from published single-turnover rates of heme reduction (50), Arg oxidation (51), and NO formation from N-hydroxyarginine (52–54), the plot of 1/λ versus [MbO2] allowed direct determination of k5, k6, and k9,11′ (Table 1).
To ensure that the effect of MbO2 on the inactivation rate was not due to some property of MbO2 other than the ability to trap solution NO, the ability of apomyoglobin and metmyoglobin (metMb) to protect iNOS from inactivation was tested. The addition of increasing concentrations of apomyoglobin or metMb to 5 μM MbO21 both failed to decrease the inactivation rate (data not shown). Similarly, addition of metHb had no effect on nNOS (33) or eNOS (31) inactivation in previous studies. In addition, inhibition of iNOS by the nitrate generated from the reaction of NO with MbO2 could not account for the decrease in inactivation rate, as nitrate concentrations up to 1 mM did not alter iNOS activity (data not shown). The lack of iNOS inhibition by nitrate was in line with previous reports examining nitrite and nitrate inhibition of nNOS (33, 34). Taken together, these observations indicated that the ability of increasing MbO2 concentrations to decrease the inactivation rate was solely due to an increased ability to trap solution NO.
To assess whether the trap-independent inactivation rate observed at high MbO2 concentrations was due to a thiol-dependent process (i.e. S-nitrosation) or due to other potential, independent inactivation processes, the concentration of NO traps were varied in the presence of 10 mM GSH. If trap-dependent and trap-independent inactivation were both a result of iNOS S-nitrosation, then addition of GSH would result in a horizontal line lying below the trap-independent inactivation rate in the plot of λ versus MbO2 concentration. However, 10 mM GSH interfered with the MbO2 assay at high MbO2 concentrations2. Therefore, an alternative method to trap and detect NO without generating a ferric heme species was sought. The heme-domain (residues 1–217) of the β2 subunit of soluble guanylate cyclase (β2 H-NOX) is ideal in this respect as it is a native NO receptor, does not bind oxygen, is stable to oxidation, and binds NO with a characteristic spectral shift (Soret λmax 433 nm to 399 nm) (24). Therefore, the β2 H-NOX concentration was varied in a similar fashion to MbO2 and inactivation rates were determined in the presence and absence of 10 mM GSH. Plots of λ versus β2 H-NOX concentration were similar to those using MbO2 (Figure 6B) with one important distinction: the concentration of β2 H-NOX necessary to reach the trap-independence was lower than with MbO23. This correlates with the more rapid binding of NO to β2 H-NOX (>1.4 × 108 M−1s−1) compared to the reaction of NO with MbO2 (3.4 × 107 M−1s−1) (44, 55) and further indicates that the observed decreases in inactivation rate were due to trapping solution NO. As hypothesized above, addition of 10 mM GSH resulted in constant inactivation rates over the entire range of β2 H-NOX concentrations, with an inactivation rate below the trap-independent inactivation rate (Figure 6B). This indicated the trap-dependent and trap-independent inactivations are both thiol dependent processes (e.g. S-nitrosation). However, the inactivation rate with GSH was not zero since 10 mM GSH is not saturating for nitrosothiol reduction (the apparent Km value for GSH is 13 mM; Table 1), NO binding to β2 H-NOX is reversible, other minor processes besides S-nitrosation likely also result in inactivation (e.g. protein instability). Therefore, our data is consistent with both trap-dependent and trap-independent iNOS auto-inactivation resulting from Zn2+-tetrathiolate S-nitrosation.
Discussion
Kinetic model of iNOS auto-inactivation
Exposure of NOS to NO lowers the activity of NOS both in vitro (8, 10, 31–33) and in cells (31, 56–58). NO is also capable of S-nitrosating the NOS Zn2+-tetrathiolate (7–11, 30, 59), which (at least for iNOS) results in zinc loss followed by irreversible dimer dissociation and inactivation (7, 8). Here, we present a detailed kinetic model of NOS S-nitrosation and inactivation by treating arginine as a suicide substrate (Figure 2). This kinetic model has allowed for the direct and quantitative determination of the inactivation rate (λ), the NO concentration formed at infinite time ([NO]∞), and the partition ratio between NO release and NOS S-nitrosation/inactivation. As discussed below, perturbation of this kinetic model by varying the concentration of NO traps and reductants provided insight into the role of NOS S-nitrosation in NOS auto-inactivation, the mechanism of NOS S-nitrosation, and the potential role of NOS S-nitrosation in the initial formation of nitrosothiols such as GSNO.
S-nitrosation and iNOS auto-inactivation
The majority of the evidence previously reported and expanded upon here points to S-nitrosation of the Zn2+-tetrathiolate as the primary mechanism of NOS auto-inactivation, although some alternative mechanisms have been described including modification of the pterin cofactor (see Supporting Information). The timescale of iNOS S-nitrosation (Figure 1A) correlated exactly with the observed loss in iNOS activity (Figure 1B). Furthermore, the ability of reducing agents (i.e. GSH and TCEP) to protect iNOS from auto-inactivation (Figure 4) also directly correlated with a decrease in iNOS S-nitrosation (Figure 5). Previous studies showed that treatment of activated macrophages with GSNO or S-nitroso-N-acetyl-penicillamine (SNAP) inactivated iNOS in a concentration-dependent manner, consistent with inactivation occurring through transnitrosation of the Zn2+-tetrathiolate (56). SNAP addition also inhibited nNOS activity in cytosolic extracts from rat brain (56). Furthermore, SNAP inhibition of nNOS was irreversible consistent with nNOS dimer dissociation following S-nitrosation of the Zn2+-tetrathiolate (56) as the cause of inactivation.
Mechanism of iNOS S-nitrosation
Having determined that iNOS S-nitrosation is the primary mechanism of iNOS auto-inactivation, we sought insight into the mechanism of iNOS S-nitrosation. As mentioned above, S-nitrosation is not a simple addition of NO to thiols; the conversion of a thiol to a nitrosothiol involves a 1-electron oxidation. There are two main mechanisms of S-nitrosation (60). An aerobic mechanism (with O2 as the oxidant, forming N2O3 as the nitrosating agent) in which N2O3 reacts with free thiols to form nitrosothiols at a rate of ~6.6 × 107 M−1s−1 (61). Alternately, thiyl radicals of cellular thiols (e.g. GS•) can react with NO at a rate of ~3 × 109 M−1s−1 (62) to produce nitrosothiols. For iNOS, O2 appears to be the oxidant for S-nitrosation, as treatment of iNOS with exogenous NO under anaerobic conditions yielded no S-nitrosation (8). This requirement of O2 for S-nitrosation is consistent with N2O3 as the nitrosating agent for iNOS, but other aerobic oxidation mechanisms cannot be ruled out.
In cells, NO concentrations are in the nanomolar to low micromolar range, but the concentrations of species that bind, react with, and destroy NO are several orders of magnitude higher (heme proteins, reductants, oxygen, superoxide, iron-sulfur clusters, etc.). Lim et al. estimated using a kinetic model that, under NO concentrations representative of an inflammatory response (1 μM), solution N2O3 concentrations are limited to the femtomolar range (63). These low estimated N2O3 concentrations were primarily due to the ability of GSH to scavenge NO and react with N2O3. Regardless of the exact mechanism of Zn2+-tetrathiolate S-nitrosation, the results presented here are consistent with iNOS forming the relevant nitrosating agent sequestered within the protein environment, thus avoiding reactions with GSH and other species that destroy NO.
The trap-dependent and trap-independent components observed upon fitting our kinetic model (Figure 2) at varying trap concentrations (Figure 6) support the hypothesis that NO sequestered within the protein environment is responsible for iNOS S-nitrosation. The trap-independent component suggests that NO generated at the heme cofactor can S-nitrosate iNOS without being released into solution. Importantly, assays varying the β2 H-NOX concentration in the presence of 10 mM GSH (Figure 6B) showed that the majority of the trap-independent component was thiol-dependent consistent with inactivation due to S-nitrosation. Interestingly, examination of iNOS crystal structures revealed a tunnel between the two heme-binding sites that passes directly by the Zn2+-tetrathiolate (Figure 7A). Similar tunnels were also found in eNOS (pdb 1DOC) and nNOS (pdb 1OM4) crystal structures. It is important to note that the Zn2+-tetrathiolate is positioned directly between this tunnel and the solvent exposed surface of NOS. Therefore, S-nitrosation of the Zn2+-tetrathiolate might occur via this conduit tunnel resulting in surface exposed nitrosothiols that may participate in downstream transnitrosation reactions with GSH or other proteins. The hypothesis that the NOS nitrosothiols are surface exposed is supported by the ability of both GSH and TCEP (which is structurally unrelated to GSH) to efficiently reduce iNOS S-nitrosation (Figure 5). This use of a protein tunnel represents a novel mechanism to direct NO reactivity.
Figure 7.
(A) Potential tunnel for NO diffusion from the heme to the Zn2+-tetrathiolate within iNOS. The tunnel within a structure of the iNOS heme domain (pdb 1DWV) (85) was generated using CAVER (86). (B) Summary model of iNOS auto-inactivation through S-nitrosation of the Zn2+-tetrathiolate and protection from inactivation by reductants. The illustrated scheme is superimposed on the kinetic model that was developed by treating the substrate arginine as a mechanism-based inhibitor. Rescue by cellular reductants (GSH) is also shown as well as trapping of released NO by MbO2.
The trap-dependent component suggests that a component of iNOS inactivation involves competition of the Zn2+-tetrathiolate and MbO2 for solution NO. In the kinetic model (Figure 2), the rate of NOS competition for solution NO is represented by the rate constant k6[NO] (Table 1). From the rate of NO rebinding to the heme upon photodissociation, Slama-Schwok et al. estimated that ~4 non-heme bound NO molecules can reside within the eNOS oxidase domain (64). If we estimate that, like eNOS, iNOS also possesses 4 non-heme NO binding sites per monomer in addition to the heme binding site, then the steady-state NO concentration can be estimated as ~75 nM as 15 nM iNOS was utilized in our assays. Using this analysis, the estimated bimolecular rate of NO sequestration by iNOS (k6 in Figure 2) is ~1010 M−1s−1. As N2O3 nitrosates GSH at a rate of 6.6 × 107 M−1s−1 (61), either the iNOS Zn2+-tetrathiolate is over two orders of magnitude more reactive towards N2O3 compared to GSH, N2O3 is not the relevant nitrosating agent, or all S-nitrosation occurs through the tunnel (Figure 7A). In the last case, the trap-dependent component would result from competition of NO released into solution between reaction with MbO2 and diffusion back into iNOS (Figure 7B). Furthermore, rates of ~1010 M−1s−1 are within reason for diffusion of gases into protein environments (65).
Several other lines of evidence are consistent with the ability of NOS to sequester NO and generate nitrosating agents within the protein environment. First, non-heme NO binding sites have been observed in NOS crystal structures (66). Second, HbO2 was found much more efficient in protecting iNOS from inactivation by exogenously added NO compared to generated NO at the active site during turnover (36) suggesting that NO does not dissociate from iNOS prior to S-nitrosation of the Zn2+-tetrathiolate. Finally, the interior of NOS is largely hydrophobic and the S-nitrosation of proteins is accelerated in a hydrophobic environment (67). Therefore, a high local concentration of NO and oxygen sequestered within NOS may accelerate the formation of the nitrosating agent (e.g. N2O3) involved in NOS S-nitrosation.
GSH protection of iNOS from S-nitrosation and inactivation
In assays without added reductant iNOS is fully inactivated in ~30 minutes (Figure 1B). Millimolar GSH concentrations were sufficient to protect iNOS from S-nitrosation and inactivation, but significant iNOS S-nitrosation and inactivation was observed at micromolar GSH concentrations (Figure 5 and 6). Therefore, iNOS might only inactivate in vivo once GSH concentrations reach micromolar levels. In particular, the kinetic data presented here suggests that the rate of iNOS auto-inactivation (i.e. λ) and the total concentration of NO synthesized (i.e. [NO]∞) are carefully controlled by the concentration of reduced cellular thiols (e.g. GSH). Additionally, proteins that may be direct targets of NOS transnitrosation (e.g. COX-2, caspase-3, or arginase 1) may protect NOS from auto-inactivation. Intriguingly, iNOS is most responsive to low millimolar concentrations of GSH, which corresponds to the GSH concentration in normal cells (1–5 mM) (17). In cases where GSH levels drop from low millimolar to high micromolar concentrations (e.g. during endotexemia (68, 69) or ischemia/reperfusion (70) in hepatocytes or during macrophage activation (71)), significant iNOS inactivation would be predicted. Indeed, in activated macrophages total glutathione concentrations (GSH and GSSG) decreased by 45% and the GSH:GSSG ratio decreased from 12:1 to 2:1 after 48 hours. This decrease in GSH levels directly correlated with a drop in NOS activity (71). Depletion of cellular GSH levels through chemical means also led to a sharp decrease in iNOS activity in induced macrophages (71, 72) or hepatocytes (46, 73) and eNOS activity in endothelial cells (74–77). Addition of GSH (46, 74) or glutathione ethyl ester (72, 78) concurrently with GSH-depleting small molecules resulted in protection from NOS inactivation. However, addition of GSH to induced macrophage cytosolic extracts failed to recover iNOS activity (72), suggesting that GSH protects iNOS from inactivation in vivo but that GSH is incapable of recovering activity once iNOS is inactivated, an observation that mirrors in vitro results reported here.
Implications for NOS S-nitrosation in the physiological generation of nitrosothiols
To gain insight into the physiological relevance of NOS S-nitrosation as an initial source of nitrosothiols, we determined the partition ratio of NO release versus iNOS S-nitrosation/inactivation (Figure 2) at varying concentrations of NO traps (Figure 6). In vivo, this partition ratio likely resembles the trap-independent component in Figure 6 as the concentrations of species that bind, react with, and destroy NO are high compared to iNOS concentrations. Therefore, NO release is likely irreversible in vivo and the partition ratio is represented by k5/k9,11′, which is ~22,000 (Table 1). The k9,11′ value used in this calculation was the higher value without added reductant since GSH reversal of iNOS S-nitrosation also results in the formation of a nitrosothiol, GSNO. Importantly, the half-life of NO in vivo is on the order of 2 milliseconds to 2 seconds (79, 80) and the half-lives of nitrosothiols in plasma are ~40 minutes (81). Assuming an NO half-life of one second, then a partition ratio of ~2,400 would result in equal steady state NO and nitrosothiol concentrations, which is within an order of magnitude of the determined partition ratio of ~22,000. Since iNOS produces low micromolar NO concentrations and current estimates of cellular nitrosothiol concentrations generally fall within the nanomolar range (20), the determined partition ratio is consistent with iNOS S-nitrosation being responsible, at least in part, for the physiological generation of nitrosothiols. However, future work is needed to determine if the primary function of iNOS S-nitrosation is as a mechanism to control iNOS activity or as an initial source of nitrosothiols. Furthermore, future work is needed to determine what proteins, if any, are direct targets of iNOS transnitrosation.
While the partition ratio between NO release and iNOS S-nitrosation favors NO release for iNOS, eNOS and nNOS might favor S-nitrosation. eNOS and nNOS are more susceptible to inactivation by NO (31, 33, 36) suggesting greater S-nitrosation of the constitutive NOS isoforms. In addition, S-nitrosation of eNOS and nNOS might be less coupled to dimer dissociation allowing transnitrosation reactions to occur without dimer dissociation and inactivation. Consistent with this hypothesis, S-nitrosated eNOS is mostly dimeric and both eNOS S-nitrosation and inactivation were fully reversible within endothelial cells (10). In vitro, the eNOS oxidase domain forms the most stable dimer, followed by nNOS and then iNOS (82). However, further studies are needed to determine the partition ratio between NO release and S-nitrosation/inactivation for the constitutive NOS isoforms.
As intracellular GSH concentrations range from 0.5 mM to 10 mM (17), GSNO is likely a major product of transnitrosation reactions with S-nitrosated NOS. NOS has been shown to produce nanomolar to low micromolar GSNO concentrations both in vitro (83), in cells (19), and plasma (20, 83, 84). Based on the results in Figure 3, an intracellular GSH concentration of ~3 mM approximately doubles [NO]∞ and halves the iNOS auto-inactivation rate. This suggests that approximately equal GSNO and S-nitrosated iNOS concentrations are formed at 3 mM GSH. The balance between GSNO formation and iNOS S-nitrosation will favor iNOS S-nitrosation if GSH levels are decreased and favor GSNO if GSH levels are increased. Thus, protein transnitrosation reactions with S-nitrosated iNOS or GSNO might directly respond to GSH levels as well as the cellular redox state (Figure 7B).
In summary, iNOS S-nitrosation plays an important role in the control of NO concentrations and potentially as an initial source of nitrosothiols in vivo. Whether NOS S-nitrosation initiates protein transnitrosation signaling pathways analogous to phosphoryl transfer (16) is an interesting avenue for future work. A complete understanding of the molecular details involved in the initial formation and transfer of nitrosothiols by NOS might lead to treatments of a broad spectrum of diseases including cancer, diabetes, and other cardiovascular, pulmonary, and neurological disorders (4).
Supplementary Material
Acknowledgments
This project originated from discussion with Dr. Douglas Mitchell and Dr. Joshua Woodward, who made the initial observation of the NOS tunnel. Additionally, we thank members of the Marletta lab for discussions and critical reading of this manuscript.
Abbreviations
- cGMP
cyclic guanosine monophosphate
- GSH
glutathione
- GSNO
S-nitrosoglutathione
- β2
beta 2 subunit of soluble guanylate cyclase
- H4B
tetrahydrobiopterin
- H-NOX
Heme Nitric oxide and/or OXygen binding domain
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HbO2
oxyhemoglobin
- Mb
myoglobin
- MbO2
oxymyoglobin
- metMb
metmyoglobin
- NADPH
β-Nicotinamide adenine dinucleotide 2′-phosphate reduced form
- NO
nitric oxide
- NOS
nitric oxide synthase
- eNOS
endothelial nitric oxide synthase
- iNOS
inducible nitric oxide synthase
- nNOS
neuronal nitric oxide synthase
- sGC
soluble guanylate cyclase
- SNAP
S-nitroso-N-acetyl-penicillamine
- SNO
S-nitrosothiol
- SOD
superoxide dismutase
- TCEP
tris(2-carboxyethyl)phosphine
Footnotes
Since MbO2 serves as the spectral readout for NO production it was necessary to maintain a low concentration of MbO2 in activity assays.
The interference of GSH was likely due to reduction of the metMb formed upon reaction of NO with MbO2. The reduced Mb can then rebind O2 to reform MbO2, which results in an increase in the apparent inactivation rate.
The magnitude of the inactivation rates titrating β2 H-NOX cannot be directly compared to those with MbO2 due to the presence of four cysteines in β2 H-NOX that may alter apparent inactivation rates and the fact that NO binding to β2 H-NOX is reversible whereas NO reaction with MbO2 is irreversible.
Financial support was provided by the Aldo DeBenedictis Fund of the University of California, Berkeley (M.A.M.) and an NIH National Institute of General Medical Sciences postdoctoral fellowship 5F32GM095023 (B.C.S).
Supporting Information Available. Further results and discussion of the relevance of alternative proposed mechanisms of NOS auto-inactivation besides S-nitrosation. Figure S1: Plot of [NO]∞ versus iNOS concentration. Figure S2: Chemical structures of a protein nitrosothiol, GSNO, N5-nitrosated H4B, and N-nitrosomorpholine. Figure S3: Plot of [NO]∞ versus Arg concentration. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Marletta MA, Hurshman AR, Rusche KM. Catalysis by nitric oxide synthase. Curr Opin Chem Biol. 1998;2:656–663. doi: 10.1016/s1367-5931(98)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Aktan F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004;75:639–653. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 3.Derbyshire ER, Marletta MA. Biochemistry of soluble guanylate cyclase. Handb Exp Pharmacol. 2009:17–31. doi: 10.1007/978-3-540-68964-5_2. [DOI] [PubMed] [Google Scholar]
- 4.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basu S, Keszler A, Azarova NA, Nwanze N, Perlegas A, Shiva S, Broniowska KA, Hogg N, Kim-Shapiro DB. A novel role for cytochrome c: Efficient catalysis of S-nitrosothiol formation. Free Radic Biol Med. 2010;48:255–263. doi: 10.1016/j.freeradbiomed.2009.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broniowska KA, Keszler A, Basu S, Kim-Shapiro DB, Hogg N. Cytochrome c-mediated formation of S-nitrosothiol in cells. Biochem J. 2011 doi: 10.1042/BJ20111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravi K, Brennan LA, Levic S, Ross PA, Black SM. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc Natl Acad Sci USA. 2004;101:2619–2624. doi: 10.1073/pnas.0300464101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell DA, Erwin PA, Michel T, Marletta MA. S-Nitrosation and regulation of inducible nitric oxide synthase. Biochemistry. 2005;44:4636–4647. doi: 10.1021/bi0474463. [DOI] [PubMed] [Google Scholar]
- 9.Erwin PA, Mitchell DA, Sartoretto J, Marletta MA, Michel T. Subcellular targeting and differential S-nitrosylation of endothelial nitric-oxide synthase. J Biol Chem. 2006;281:151–157. doi: 10.1074/jbc.M510421200. [DOI] [PubMed] [Google Scholar]
- 10.Erwin PA, Lin AJ, Golan DE, Michel T. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J Biol Chem. 2005;280:19888–19894. doi: 10.1074/jbc.M413058200. [DOI] [PubMed] [Google Scholar]
- 11.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;86:pl1–9. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 12.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto A, Comatas KE, Liu L, Stamler JS. Screening for nitric oxide-dependent protein-protein interactions. Science. 2003;301:657–661. doi: 10.1126/science.1079319. [DOI] [PubMed] [Google Scholar]
- 14.Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 15.Dunn J, Gutbrod S, Webb A, Pak A, Jandu SK, Bhunia A, Berkowitz DE, Santhanam L. S-nitrosation of arginase 1 requires direct interaction with inducible nitric oxide synthase. Mol Cell Biochem. 2011;355:83–89. doi: 10.1007/s11010-011-0841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaller GE, Shiu S-H, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21:R320–30. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 17.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 18.Yap L-P, Sancheti H, Ybanez MD, Garcia J, Cadenas E, Han D. Determination of GSH, GSSG, and GSNO Using HPLC with Electrochemical Detection. Thiol Redox Transitions in Cell Signaling, Part A. 2010;473:137–147. doi: 10.1016/S0076-6879(10)73006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluge I, Gutteck-Amsler U, Zollinger M, Do KQ. S-nitrosoglutathione in rat cerebellum: identification and quantification by liquid chromatography-mass spectrometry. J Neurochem. 1997;69:2599–2607. doi: 10.1046/j.1471-4159.1997.69062599.x. [DOI] [PubMed] [Google Scholar]
- 20.Giustarini D, Milzani A, Dalle-Donne I, Rossi R. Detection of S-nitrosothiols in biological fluids: A comparison among the most widely applied methodologies. Journal of Chromatography B. 2007;851:124–139. doi: 10.1016/j.jchromb.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 22.Barglow KT, Knutson CG, Wishnok JS, Tannenbaum SR, Marletta MA. Site-specific and redox-controlled S-nitrosation of thioredoxin. Proc Natl Acad Sci USA. 2011;108:E600–6. doi: 10.1073/pnas.1110736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusche KM, Spiering MM, Marletta MA. Reactions catalyzed by tetrahydrobiopterin-free nitric oxide synthase. Biochemistry. 1998;37:15503–15512. doi: 10.1021/bi9813936. [DOI] [PubMed] [Google Scholar]
- 24.Karow DS, Pan D, Davis JH, Behrends S, Mathies RA, Marletta MA. Characterization of Functional Heme Domains from Soluble Guanylate Cyclase. Biochemistry. 2005;44:16266–16274. doi: 10.1021/bi051601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Hevel JM, Marletta MA. Nitric-oxide synthase assays. Meth Enzymol. 1994;233:250–258. doi: 10.1016/s0076-6879(94)33028-x. [DOI] [PubMed] [Google Scholar]
- 27.Antonini E, Brunori M. Hemoglobin and Myoglobin in Their Reactions with Ligands. North-Holland, Amsterdam: [Google Scholar]
- 28.Teale F. Cleavage of the haem-protein link by acid methylethylketone. Biochim Biophys Acta. 1959;35:543–543. doi: 10.1016/0006-3002(59)90407-x. [DOI] [PubMed] [Google Scholar]
- 29.Paulson DR, Addison AW, Dolphin D, James BR. Preparation of ruthenium(II) and ruthenium(III) myoglobin and the reaction of dioxygen, and carbon monoxide, with ruthenium(II) myoglobin. J Biol Chem. 1979;254:7002–7006. [PubMed] [Google Scholar]
- 30.Rosenfeld RJ, Bonaventura J, Szymczyna BR, Maccoss MJ, Arvai AS, Yates JR, Tainer JA, Getzoff ED. Nitric-oxide Synthase Forms N-NO-pterin and S-NO-Cys: Implications for activity, allostery, and regulation. J Biol Chem. 2010;285:31581–31589. doi: 10.1074/jbc.M109.072496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buga GM, Griscavage JM, Rogers NE, Ignarro LJ. Negative feedback regulation of endothelial cell function by nitric oxide. Circ Res. 1993;73:808–812. doi: 10.1161/01.res.73.5.808. [DOI] [PubMed] [Google Scholar]
- 32.Rengasamy A, Johns RA. Regulation of nitric oxide synthase by nitric oxide. Mol Pharmacol. 1993;44:124–128. [PubMed] [Google Scholar]
- 33.Rogers NE, Ignarro LJ. Constitutive nitric oxide synthase from cerebellum is reversibly inhibited by nitric oxide formed from L-arginine. Biochem Biophys Res Commun. 1992;189:242–249. doi: 10.1016/0006-291x(92)91550-a. [DOI] [PubMed] [Google Scholar]
- 34.Kotsonis P, Frey A, Fröhlich LG, Hofmann H, Reif A, Wink DA, Feelisch M, Schmidt HH. Autoinhibition of neuronal nitric oxide synthase: distinct effects of reactive nitrogen and oxygen species on enzyme activity. Biochem J. 1999;340(Pt 3):745–752. [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann H, Schmidt HH. Thiol dependence of nitric oxide synthase. Biochemistry. 1995;34:13443–13452. doi: 10.1021/bi00041a023. [DOI] [PubMed] [Google Scholar]
- 36.Griscavage JM, Rogers NE, Sherman MP, Ignarro LJ. Inducible nitric oxide synthase from a rat alveolar macrophage cell line is inhibited by nitric oxide. J Immunol. 1993;151:6329–6337. [PubMed] [Google Scholar]
- 37.Garrido-del Solo C, García-Cánovas F, Havsteen BH, Varón-Castellanos R. Kinetic analysis of a Michaelis-Menten mechanism in which the enzyme is unstable. Biochem J. 1993;294(Pt 2):459–464. doi: 10.1042/bj2940459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varón R, García-Cánovas F, García-Moreno M, Valero E, Molina-Alarcón M, García-Meseguers MJ, Vidal de Labra JA, Garrido-del Sol C. Kinetic analysis of the general modifier mechanism of Botts and Morales involving a suicide substrate. Journal of Theoretical Biology. 2002;218:355–374. doi: 10.1006/jtbi.2002.3083. [DOI] [PubMed] [Google Scholar]
- 39.Duggleby RG. Progress curves of reactions catalyzed by unstable enzymes. A theoretical approach. Journal of Theoretical Biology. 1986;123:67–80. doi: 10.1016/s0022-5193(86)80236-3. [DOI] [PubMed] [Google Scholar]
- 40.Burke MA, Maini PK, Murray JD. On the kinetics of suicide substrates. Biophys Chem. 1990;37:81–90. doi: 10.1016/0301-4622(90)88009-h. [DOI] [PubMed] [Google Scholar]
- 41.Tatsunami S, Yago N, Hosoe M. Kinetics of suicide substrates steady-state treatments and computer-aided exact solutions. Biochimica et Biophysica Acta (BBA) - Enzymology. 1981;662:226–235. doi: 10.1016/0005-2744(81)90034-6. [DOI] [PubMed] [Google Scholar]
- 42.Tudela J, García Cánovas F, Varón R, García Carmona F, Gálvez J, Lozano JA. Transient-phase kinetics of enzyme inactivation induced by suicide substrates. Biochim Biophys Acta. 1987;912:408–416. doi: 10.1016/0167-4838(87)90046-x. [DOI] [PubMed] [Google Scholar]
- 43.Waley SG. The kinetics of substrate-induced inactivation. Biochem J. 1991;279(Pt 1):87–94. doi: 10.1042/bj2790087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eich RF, Li T, Lemon DD, Doherty DH, Curry SR, Aitken JF, Mathews AJ, Johnson KA, Smith RD, Phillips GN, Olson JS. Mechanism of NO-induced oxidation of myoglobin and hemoglobin. Biochemistry. 1996;35:6976–6983. doi: 10.1021/bi960442g. [DOI] [PubMed] [Google Scholar]
- 45.Stuehr DJ, Kwon NS, Nathan CF. FAD and GSH participate in macrophage synthesis of nitric oxide. Biochem Biophys Res Commun. 1990;168:558–565. doi: 10.1016/0006-291x(90)92357-6. [DOI] [PubMed] [Google Scholar]
- 46.Harbrecht BG, Di Silvio M, Chough V, Kim YM, Simmons RL, Billiar TR. Glutathione regulates nitric oxide synthase in cultured hepatocytes. Ann Surg. 1997;225:76–87. doi: 10.1097/00000658-199701000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komori Y, Hyun J, Chiang K, Fukuto JM. The role of thiols in the apparent activation of rat brain nitric oxide synthase (NOS) J Biochem. 1995;117:923–927. doi: 10.1093/oxfordjournals.jbchem.a124797. [DOI] [PubMed] [Google Scholar]
- 48.Martin NI, Woodward JJ, Winter MB, Marletta MA. 4,4-Difluorinated analogues of l-arginine and N(G)-hydroxy-l-arginine as mechanistic probes for nitric oxide synthase. Bioorg Med Chem Lett. 2009;19:1758–1762. doi: 10.1016/j.bmcl.2009.01.076. [DOI] [PubMed] [Google Scholar]
- 49.Ravichandran LV, Johns RA, Rengasamy A. Direct and reversible inhibition of endothelial nitric oxide synthase by nitric oxide. Am J Physiol. 1995;268:H2216–23. doi: 10.1152/ajpheart.1995.268.6.H2216. [DOI] [PubMed] [Google Scholar]
- 50.Roman LJ, Miller RT, La Garza de MA, Kim JJ, Siler Masters BS. The C terminus of mouse macrophage inducible nitric-oxide synthase attenuates electron flow through the flavin domain. J Biol Chem. 2000;275:21914–21919. doi: 10.1074/jbc.M002449200. [DOI] [PubMed] [Google Scholar]
- 51.Wei CC, Wang ZQ, Wang Q, Meade AL, Hille R, Stuehr DJ. Rapid kinetic studies link tetrahydrobiopterin radical formation to heme-dioxy reduction and arginine hydroxylation in inducible nitric-oxide synthase. J Biol Chem. 2001;276:315–319. doi: 10.1074/jbc.M008441200. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z-Q, Wei C-C, Stuehr DJ. A conserved tryptophan 457 modulates the kinetics and extent of N-hydroxy-L-arginine oxidation by inducible nitric-oxide synthase. J Biol Chem. 2002;277:12830–12837. doi: 10.1074/jbc.M111967200. [DOI] [PubMed] [Google Scholar]
- 53.Wang ZQ, Wei CC, Ghosh S, Meade AL, Hille R, Stuehr DJ. A conserved tryptophan in nitric oxide synthase regulates heme-dioxy reduction by tetrahydrobiopterin. Biochemistry. 2001;40:12819–12825. doi: 10.1021/bi011182s. [DOI] [PubMed] [Google Scholar]
- 54.Lefèvre-Groboillot D, Boucher J-L, Mansuy D, Stuehr DJ. Reactivity of the heme-dioxygen complex of the inducible nitric oxide synthase in the presence of alternative substrates. FEBS J. 2006;273:180–191. doi: 10.1111/j.1742-4658.2005.05056.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Y, Brandish PE, Ballou DP, Marletta MA. A molecular basis for nitric oxide sensing by soluble guanylate cyclase. Proc Natl Acad Sci USA. 1999;96:14753–14758. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Assreuy J, Cunha FQ, Liew FY, Moncada S. Feedback inhibition of nitric oxide synthase activity by nitric oxide. Br J Pharmacol. 1993;108:833–837. doi: 10.1111/j.1476-5381.1993.tb12886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheehy AM, Burson MA, Black SM. Nitric oxide exposure inhibits endothelial NOS activity but not gene expression: a role for superoxide. Am J Physiol. 1998;274:L833–41. doi: 10.1152/ajplung.1998.274.5.L833. [DOI] [PubMed] [Google Scholar]
- 58.Ma XL, Lopez BL, Christopher TA, Birenbaum DS, Vinten-Johansen J. Exogenous NO inhibits basal NO release from vascular endothelium in vitro and in vivo. Am J Physiol. 1996;271:H2045–51. doi: 10.1152/ajpheart.1996.271.5.H2045. [DOI] [PubMed] [Google Scholar]
- 59.Taldone FS, Tummala M, Goldstein EJ, Ryzhov V, Ravi K, Black SM. Studying the S-nitrosylation of model peptides and eNOS protein by mass spectrometry. Nitric Oxide. 2005;13:176–187. doi: 10.1016/j.niox.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Keszler A, Zhang Y, Hogg N. Reaction between nitric oxide, glutathione, and oxygen in the presence and absence of protein: How are S-nitrosothiols formed? Free Radic Biol Med. 2010;48:55–64. doi: 10.1016/j.freeradbiomed.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keshive M, Singh S, Wishnok JS, Tannenbaum SR, Deen WM. Kinetics of S-nitrosation of thiols in nitric oxide solutions. Chemical research in toxicology. 1996;9:988–993. doi: 10.1021/tx960036y. [DOI] [PubMed] [Google Scholar]
- 62.Madej E, Folkes LK, Wardman P, Czapski G, Goldstein S. Thiyl radicals react with nitric oxide to form S-nitrosothiols with rate constants near the diffusion-controlled limit. Free Radic Biol Med. 2008;44:2013–2018. doi: 10.1016/j.freeradbiomed.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 63.Lim CH, Dedon PC, Deen WM. Kinetic analysis of intracellular concentrations of reactive nitrogen species. Chemical research in toxicology. 2008;21:2134–2147. doi: 10.1021/tx800213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slama-Schwok A, Négrerie M, Berka V, Lambry JC, Tsai AL, Vos MH, Martin JL. Nitric oxide (NO) traffic in endothelial NO synthase. Evidence for a new NO binding site dependent on tetrahydrobiopterin? J Biol Chem. 2002;277:7581–7586. doi: 10.1074/jbc.M108657200. [DOI] [PubMed] [Google Scholar]
- 65.Lakowicz JR, Weber G. Quenching of protein fluorescence by oxygen. Detection of structural fluctuations in proteins on the nanosecond time scale. Biochemistry. 1973;12:4171–4179. doi: 10.1021/bi00745a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pant K, Crane BR. Nitrosyl-Heme Structures of Bacillus subtilisNitric Oxide Synthase Have Implications for Understanding Substrate Oxidation. Biochemistry. 2006;45:2537–2544. doi: 10.1021/bi0518848. [DOI] [PubMed] [Google Scholar]
- 67.Möller MN, Li Q, Lancaster JR, Denicola A. Acceleration of nitric oxide autoxidation and nitrosation by membranes. IUBMB Life. 2007;59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 68.Keller GA, Barke R, Harty JT, Humphrey E, Simmons RL. Decreased hepatic glutathione levels in septic shock. Predisposition of hepatocytes to oxidative stress: an experimental approach. Arch Surg. 1985;120:941–945. doi: 10.1001/archsurg.1985.01390320065013. [DOI] [PubMed] [Google Scholar]
- 69.Okabe H, Irita K, Taniguchi S, Kurosawa K, Tagawa K, Yoshitake J, Takahashi S. Endotoxin causes early changes in glutathione concentrations in rabbit plasma and liver. J Surg Res. 1994;57:416–419. doi: 10.1006/jsre.1994.1163. [DOI] [PubMed] [Google Scholar]
- 70.McKelvey TG, Höllwarth ME, Granger DN, Engerson TD, Landler U, Jones HP. Mechanisms of conversion of xanthine dehydrogenase to xanthine oxidase in ischemic rat liver and kidney. Am J Physiol. 1988;254:G753–60. doi: 10.1152/ajpgi.1988.254.5.G753. [DOI] [PubMed] [Google Scholar]
- 71.Hothersall JS, Cunha FQ, Neild GH, Norohna-Dutra AA. Induction of nitric oxide synthesis in J774 cells lowers intracellular glutathione: effect of modulated glutathione redox status on nitric oxide synthase induction. Biochem J. 1997;322(Pt 2):477–481. doi: 10.1042/bj3220477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buchmuller-Rouiller Y, Corradin SB, Smith J, Schneider P, Ransijn A, Jongeneel CV, Mauël J. Role of Glutathione in Macrophage Activation: Effect of Cellular Glutathione Depletion on Nitrite Production and Leishmanicidal Activity. Cellular Immunology. 1995;164:73–80. doi: 10.1006/cimm.1995.1144. [DOI] [PubMed] [Google Scholar]
- 73.Tirmenstein MA, Nicholls-Grzemski FA, Schmittgen TD, Zakrajsek BA, Fariss MW. Glutathione-dependent regulation of nitric oxide production in isolated rat hepatocyte suspensions. Antioxid Redox Signal. 2000;2:767–777. doi: 10.1089/ars.2000.2.4-767. [DOI] [PubMed] [Google Scholar]
- 74.Ghigo D, Alessio P, Foco A, Bussolino F, Costamagna C, Heller R, Garbarino G, Pescarmona GP, Bosia A. Nitric oxide synthesis is impaired in glutathione-depleted human umbilical vein endothelial cells. Am J Physiol. 1993;265:C728–32. doi: 10.1152/ajpcell.1993.265.3.C728. [DOI] [PubMed] [Google Scholar]
- 75.Murphy ME, Piper HM, Watanabe H, Sies H. Nitric oxide production by cultured aortic endothelial cells in response to thiol depletion and replenishment. J Biol Chem. 1991;266:19378–19383. [PubMed] [Google Scholar]
- 76.Laursen JB, Boesgaard S, Trautner S, Rubin I, Poulsen HE, Aldershvile J. Endothelium-dependent vasorelaxation in inhibited by in vivo depletion of vascular thiol levels: role of endothelial nitric oxide synthase. Free Radic Res. 2001;35:387–394. doi: 10.1080/10715760100300901. [DOI] [PubMed] [Google Scholar]
- 77.Sugiyama T, Michel T. Thiol-metabolizing proteins and endothelial redox state: differential modulation of eNOS and biopterin pathways. AJP: Heart and Circulatory Physiology. 2010;298:H194–H201. doi: 10.1152/ajpheart.00767.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song M, Kellum JA, Kaldas H, Fink MP. Evidence that glutathione depletion is a mechanism responsible for the anti-inflammatory effects of ethyl pyruvate in cultured lipopolysaccharide-stimulated RAW 264.7 cells. J Pharmacol Exp Ther. 2004;308:307–316. doi: 10.1124/jpet.103.056622. [DOI] [PubMed] [Google Scholar]
- 79.Liu X, Miller MJ, Joshi MS, Sadowska-Krowicka H, Clark DA, Lancaster JR. Diffusion-limited reaction of free nitric oxide with erythrocytes. J Biol Chem. 1998;273:18709–18713. doi: 10.1074/jbc.273.30.18709. [DOI] [PubMed] [Google Scholar]
- 80.Thomas DD, Liu X, Kantrow SP, Lancaster JR. The biological lifetime of nitric oxide: implications for the perivascular dynamics of NO and O2. Proc Natl Acad Sci USA. 2001;98:355–360. doi: 10.1073/pnas.011379598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci USA. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Panda K, Rosenfeld RJ, Ghosh S, Getzoff ED, Stuehr DJ. Distinct dimer interaction and regulation in nitric-oxide synthase types I, II, and III. J Biol Chem. 2002;277:31020–31030. doi: 10.1074/jbc.M203749200. [DOI] [PubMed] [Google Scholar]
- 83.Mayer B, Pfeiffer S, Schrammel A, Koesling D, Schmidt K, Brunner F. A new pathway of nitric oxide/cyclic GMP signaling involving S-nitrosoglutathione. J Biol Chem. 1998;273:3264–3270. doi: 10.1074/jbc.273.6.3264. [DOI] [PubMed] [Google Scholar]
- 84.Tsikas D. Is S-nitrosoglutathione formed in nitric oxide synthase incubates? FEBS Lett. 2000;483:83–84. doi: 10.1016/s0014-5793(00)02078-0. [DOI] [PubMed] [Google Scholar]
- 85.Crane BR, Arvai AS, Ghosh S, Getzoff ED, Stuehr DJ, Tainer JA. Structures of the N(omega)-hydroxy-L-arginine complex of inducible nitric oxide synthase oxygenase dimer with active and inactive pterins. Biochemistry. 2000;39:4608–4621. doi: 10.1021/bi992409a. [DOI] [PubMed] [Google Scholar]
- 86.Beneš P, Chovancová E, Kozlíková B, Pavelka A, Strnad O, Brezovský J, Sustr V, Klva3a M, Szabó T, Gora A, Zamborský M, Biedermannová L, Medek P, Damborský J, Sochor J. CAVER 2.1 2010 January 1; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.