Abstract
The transcription factor SOX10 has essential roles in neural crest-derived cell populations, including myelinating Schwann cells—specialized glial cells responsible for ensheathing axons in the peripheral nervous system. Importantly, SOX10 directly regulates the expression of genes essential for proper myelin function. To date, only a handful of SOX10 target loci have been characterized in Schwann cells. Addressing this lack of knowledge will provide a better understanding of Schwann cell biology and candidate loci for relevant diseases such as demyelinating peripheral neuropathies. We have identified a highly-conserved SOX10 binding site within an alternative promoter at the SH3-domain kinase binding protein 1 (Sh3kbp1) locus. The genomic segment identified at Sh3kbp1 binds to SOX10 and displays strong promoter activity in Schwann cells in vitro and in vivo. Mutation of the SOX10 binding site ablates promoter activity, and ectopic expression of SOX10 in SOX10-negative cells promotes the expression of endogenous Sh3kbp1. Combined, these data reveal Sh3kbp1 as a novel target of SOX10 and raise important questions regarding the function of SH3KBP1 isoforms in Schwann cells.
Keywords: SOX10, Schwann cells, Comparative sequence analysis, transcriptional regulation, SH3KBP1, cin85
Introduction
Myelinating Schwann cells are a critical component of the peripheral nervous system, providing trophic factors to motor and sensory axons and allowing rapid communication to and from the periphery. One gene with an essential role in proper Schwann cell development and function encodes the SRY-box containing gene 10 (SOX10) transcription factor (Stolt and Wegner, 2010). SOX10 is expressed at all stages of Schwann cell development, and is required for Schwann cell specification (Britsch et al., 2001; Kuhlbrodt et al., 1998)—ablation of SOX10 function during Schwann cell development results in a complete loss of these cells (Finzsch et al., 2010). The importance of SOX10 for Schwann cells is further underscored by data showing that SOX10 works both independently and synergistically with other transcription factors (e.g., EGR2, POU3F1, and POU3F2) to transcriptionally regulate Schwann-cell specific loci (Stolt and Wegner, 2010; Svaren and Meijer, 2008)—SOX10 has a similar and equally important role in myelinating oligodendrocytes in the central nervous system (Kuhlbrodt et al., 1998). Of note, mutations in the human SOX10 gene are associated with peripheral nerve demyelination in the multi-syndrome disorder PCWH (Peripheral demyelinating neuropathy, Central dysmyelinating leukodystrophy, Waardenburg syndrome and Hirschsprung disease) (Inoue et al., 1999), and SOX10 directly regulates loci frequently mutated in patients with demyelinating Charcot-Marie-Tooth disease (CMT1); specifically, Peripheral Myelin Protein 22 (PMP22), Gap Junction Beta 1 (GJB1), and myelin protein zero (MPZ)(Bondurand et al., 2001; Jones et al., 2007; Jones et al., 2011; LeBlanc et al., 2006; Peirano et al., 2000). Furthermore, SOX10 directly regulates the locus encoding the key Schwann cell transcription factor EGR2 (Ghislain and Charnay, 2006; Reiprich et al., 2010; Werner et al., 2007). Thus, SOX10 directs a transcriptional hierarchy critical for Schwann cell development and function.
To date, SOX10 is known to regulate only a small set of loci in Schwann cells (Svaren and Meijer, 2008). The identification of additional SOX10 target genes will provide insight into Schwann cell biology as well as candidate genes for myelin-related pathologies. Here, we show that SOX10 directly regulates an alternative promoter at the SH3-domain kinase binding protein 1 (Sh3kbp1) locus in Schwann cells. Interestingly, SOX10 is believed to transcriptionally regulate another locus that encodes an SH3 domain-containing protein (SH3BP4) in Schwann cells (Lee et al., 2008), and yet another (SH3TC2) is mutated in patients with demyelinating peripheral neuropathy (Senderek et al., 2003). The data presented here expand our knowledge of SOX10 target genes and raise important questions regarding the role of specific SH3KBP1 isoforms in Schwann cells.
Results
The Sh3kbp1 locus harbors highly-conserved transcription factor binding site predictions
Sh3kbp1 encodes three major mRNAs (mRNA-1, mRNA-2, and mRNA-3 in Fig. 1A) expressed from three alternative promoters (P1, P2, and P3 in Fig. 1A) (Buchman et al., 2002). However, nothing is known regarding the transcriptional regulation of Sh3kbp1. To address this, we performed stringent comparative sequence analysis and transcription factor binding site predictions at Sh3kbp1. Briefly, we aligned genomic sequences at the ~350 kb Sh3kbp1 locus from Human, Mouse, Rat, Dog, Cow, and Chicken using MultiPipMaker software (Elnitski et al., 2003). Next, we searched for non-coding regions within the alignment that are 100% identical among all six species and at least 6 base pairs long (Antonellis et al., 2006). This revealed 12 non-coding ‘EP hits’ ranging from 6 to 15 base pairs (Table 1). Interestingly, seven of the EP hits fall into two clusters—a cluster of four EP hits was identified in P2, while another cluster of three EP hits was identified in P3 (boldfaced text in Table 1). Based on these data, we considered the seven clustering EP hits as potential transcription factor binding sites. To assess this, we identified potential transcription factor binding sites within each clustering EP hit (see Methods for details) (Matys et al., 2003). Three of the seven clustering EP hits matched with a known transcription factor binding site (Table 2). Two of these three EP hits harbor consensus sequences for transcription factors implicated in Schwann cell development—EP04 contains a POU3F2 binding site (Table 2 and Fig. 1B) and EP10 contains a SOX10 binding site (Table 2 and Fig. 1C). Importantly, the predicted SOX10 binding site within EP10 overlaps with the consensus sequence identified in our preliminary screen (Antonellis et al., 2008).
Fig. 1.
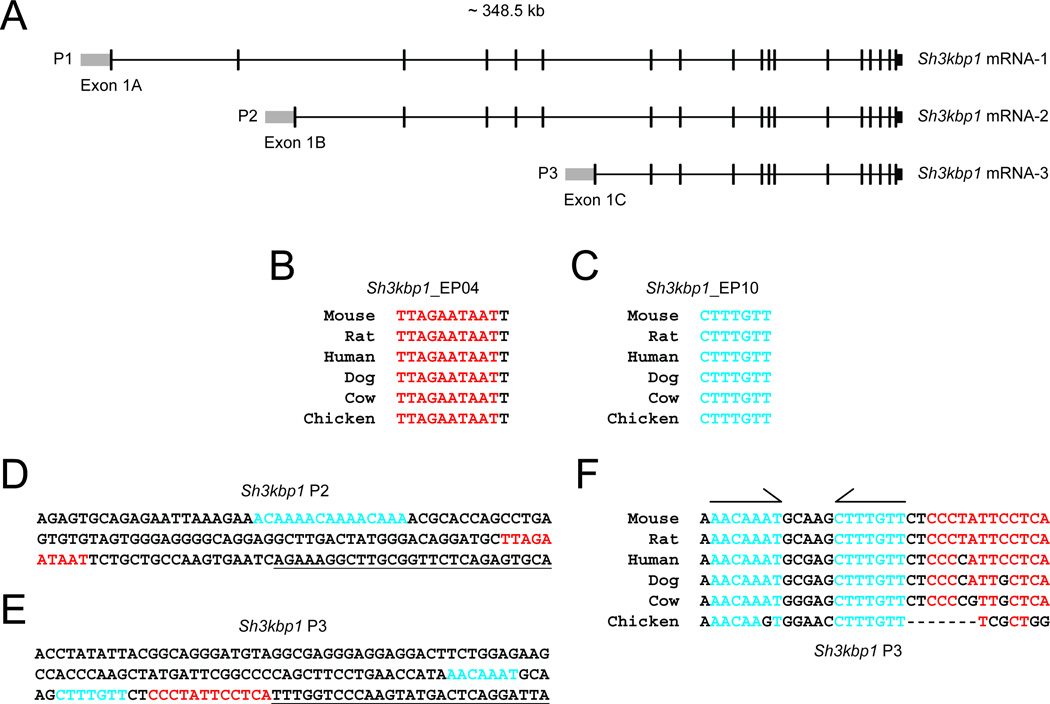
Conserved transcription factor binding sites at the Sh3kbp1 locus. (A) Organization of the mouse Sh3kbp1 locus. Note the three major mRNA variants (mRNA-1, -2, and -3), the associated promoters (P1, P2, and P3), and first exons (Exon 1A, 1B, and 1C). (B and C) Individual ExactPlus (EP) hits were submitted to TRANSFAC to identify potential transcription factor binding sites. POU3F2 and SOX10 binding sites are noted in red (B) and blue (C), respectively. (D and E) Mouse genomic sequences within Sh3kbp1 P2 and P3 were submitted to TRANSFAC to identify binding sites for transcription factors with a role in Schwann cell biology. POU3F2 and SOX10 binding sites are noted in red and blue, respectively. Note that the blue text in (D) represents three consecutive SOX10 binding site predictions. Underlined text indicates mRNA sequences, which were not included in computational analyses. (F) Genomic sequences at Sh3kbp1 P3 from six vertebrate species were extracted from the MultiPipMaker alignment file. The POU3F2 binding site is indicated in red. The dimeric, head-to-head SOX10 binding site is indicated in blue and by half-arrows. Note that each SOX10 half site is fully-conserved among vertebrate species with the exception of one nucleotide difference in one half site in chicken.
Table 1.
Position of ExactPlus (EP) hits at Sh3kbp1
| Element ID |
Browser Position1 |
Size (bp) |
|---|---|---|
| Sh3kbp1_EP01 | chrX:156126771-156126777 | 7 |
| Sh3kbp1_EP02 | chrX:156146226-156146235 | 10 |
| Sh3kbp1_EP03 | chrX:156146461-156146466 | 6 |
| Sh3kbp1_EP04 | chrX:156146505-156146515 | 11 |
| Sh3kbp1_EP05 | chrX:156146518-156146525 | 8 |
| Sh3kbp1_EP06 | chrX:156265867-156265873 | 7 |
| Sh3kbp1_EP07 | chrX:156265938-156265947 | 10 |
| Sh3kbp1_EP08 | chrX:156278354-156278359 | 6 |
| Sh3kbp1_EP09 | chrX:156278366-156278371 | 6 |
| Sh3kbp1_EP10 | chrX:156278379-156278385 | 7 |
| Sh3kbp1_EP11 | chrX:156283747-156283761 | 15 |
| Sh3kbp1_EP12 | chrX:156396255-156396260 | 6 |
Coordinates are from the July 2007 UCSC Genome Browser Mouse assembly (mm9)
Bold text indicates elements that fall within the two clusters (see text)
Table 2.
TRANSFAC predictions within clustered ExactPlus (EP) hits at Sh3kbp1
| Element ID |
Factor ID |
Factor Name |
Core Score |
Matrix Score |
Sequence |
Relative Position1 |
|---|---|---|---|---|---|---|
| Sh3kbp1_EP02 | GEN_INI | NA - General Initiation Site | 0.997 | 0.969 | ctcCATTT | 299 bp upstream of TSS2 |
| HOXA4 | homeobox A4 | 0.939 | 0.856 | CCATTttg | 299 bp upstream of TSS2 | |
| Nkx2-5 | NK2 transcription factor related, locus 5 | 0.665 | 0.708 | cCATTTtg | 299 bp upstream of TSS2 | |
| Sh3kbp1_EP04 | POU3F2 | POU class 3 homeobox 2 | 0.566 | 0.743 | TTAGAataat | 17 bp upstream of TSS2 |
| Sh3kbp1_EP10 | SOX10 | SRY box-containing 10 | 1.000 | 0.995 | cTTTGTt | 15 bp upstream of TSS3 |
TSS refers to the transcription start sites of Sh3kbp1
The SOX10, POU3F1, POU3F2, and EGR2 transcription factors work synergistically to regulate gene expression in Schwann cells (Svaren and Meijer, 2008). Our computational analyses identified highly-conserved consensus sequences for POU3F2 and SOX10 in two promoter regions at Sh3kbp1—P2 and P3. To more broadly assess Sh3kbp1 P2 and P3 for a role in Schwann cells, we analyzed the corresponding mouse genomic sequences for additional, relevant binding sites (see Methods for details). This revealed three SOX10 consensus sequences in Sh3kbp1 P2 (Fig. 1D) and one consensus sequence each for SOX10 and POU3F2 in Sh3kbp1 P3 (Fig. 1E). Furthermore, the two SOX10 consensus sequences in P3 are oriented in a head-to-head manner and are highly-conserved among vertebrate species (Fig. 1F). Similarly conserved and oriented SOX10 binding sites have proven to be functional at SOX10 target loci (Jang and Svaren, 2009; Jones et al., 2007; LeBlanc et al., 2006; Peirano and Wegner, 2000). Combined, these data raise the possibility that POU3F2 and SOX10 transcriptionally regulate the Sh3kbp1 locus in Schwann cells.
Sh3kbp1 is expressed in Schwann cells in vitro and in vivo
If POU3F2 and SOX10 directly regulate the transcription of Sh3kbp1, one expectation is that Pou3f2, Sox10, and Sh3kbp1 mRNAs are expressed in Schwann cells. To confirm this, we performed RT-PCR on cDNA derived from immortalized rat Schwann cells (S16), mouse sciatic nerve (mSN), and immortalized mouse motor neurons (MN-1). Importantly, sciatic nerve tissue contains mRNA mainly from Schwann cell bodies and provides information regarding mRNA expression in vivo. Using gene-specific RT-PCR assays, we detected Sox10 and Pou3f2 mRNA in cultured S16 cells and sciatic nerve tissue, and not in MN-1 cells (Fig. 2A) consistent with previous studies (Jaegle et al., 2003; Kuhlbrodt et al., 1998)—SOX10 protein was also detected in S16 cells and sciatic nerve, and not in MN-1 cells (Supplementary Fig. 1A). To test the expression of Sh3kbp1 in these cells, we developed RT-PCR assays specific to the first exon of each mRNA (Fig. 1A). Exon 1A was detected in S16, sciatic nerve, and MN-1 cells (Fig. 2B). In contrast, exon 1B was only detected in MN-1 cells, and exon 1C was only detected in S16 cells and sciatic nerve (Fig. 2B). SOX10 also directs the expression of loci in oligodendrocytes, raising the possibility that SOX10 transcriptionally regulates Sh3kbp1 in these cells; however, Sh3kbp1 exon 1C was not detected in oligodendrocytes upon RT-PCR analysis (Supplementary Fig. 1B). Combined, these data are consistent with POU3F2 and SOX10 regulating Sh3kbp1 promoter 3 (P3) [but not promoter 2 (P2)] in Schwann cells.
Fig. 2.
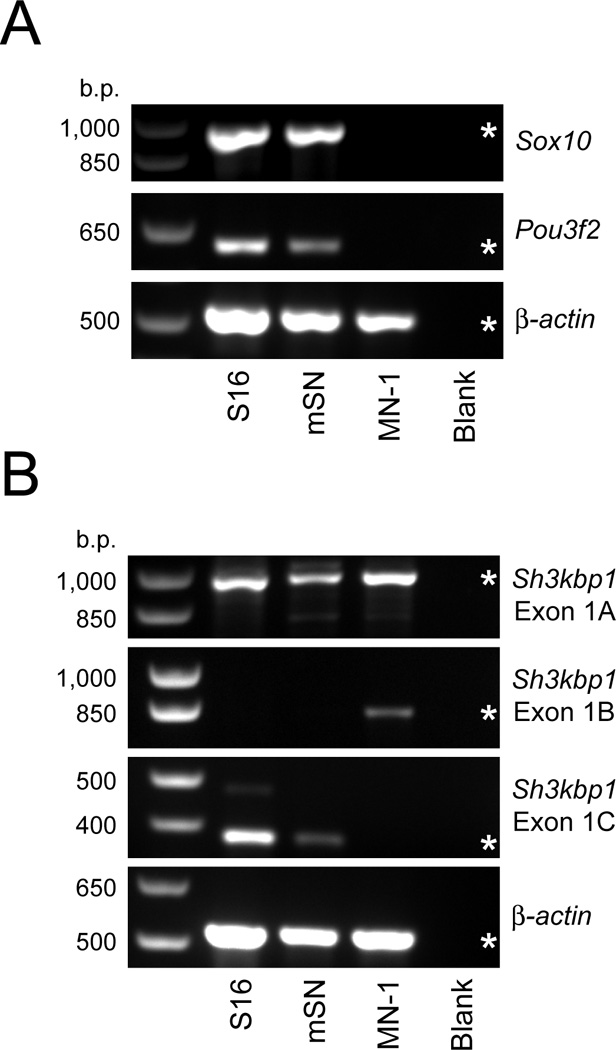
Sh3kbp1 mRNA-3 is expressed in Schwann cells in vitro and in vivo. (A) RT-PCR was performed on cDNA generated from cultured Schwann cells (S16), mouse sciatic nerve (mSN), and cultured motor neurons (MN-1) to test for the presence of Sox10 and Pou3f2 transcripts. (B) Similar RT-PCR assays as described in (A) to detect Sh3kbp1 transcripts harboring exon 1A, exon 1B, and exon 1C. Negative controls without cDNA [‘Blank’ in (A) and (B)] were included for each reaction and primers for β-actin mRNA were used as a positive control. Base pair (b.p.) sizes of markers are provided on the left, and expected band sizes for each product are indicated with an asterisk.
Sh3kbp1 P3 displays strong promoter activity in cultured Schwann cells
Combined, our computational and expression studies deem Sh3kbp1 P3 an excellent candidate for activity in Schwann cells. However, other Schwann cell-specific factors may bind to P1 and P2, and exon 1A is expressed in Schwann cells, suggesting that P1 may be active in these cells. To determine the activity of each Sh3kbp1 promoter region (Fig. 1A), we cloned the corresponding mouse genomic segments directly upstream of a luciferase reporter gene and transfected each resulting construct into cultured Schwann (S16) cells and motor neurons (MN-1). Subsequently, we performed luciferase assays using a 10-fold increase in luciferase activity as an indicator of “strong” promoter activity (red lines in Fig. 3A and 3B). Sh3kbp1 P1 directed luciferase expression in a manner ~2-fold higher than the control vector in both S16 and MN-1 cells, and Sh3kbp1 P2 did not increase luciferase expression over that observed in the control vector in either cell line (Fig. 3A and 3B). In contrast, Sh3kbp1 P3 directed luciferase expression in a manner ~50-fold higher than the control vector in S16 cells, and ~3.5-fold higher in MN-1 cells (Fig. 3A and 3B).
Fig. 3.
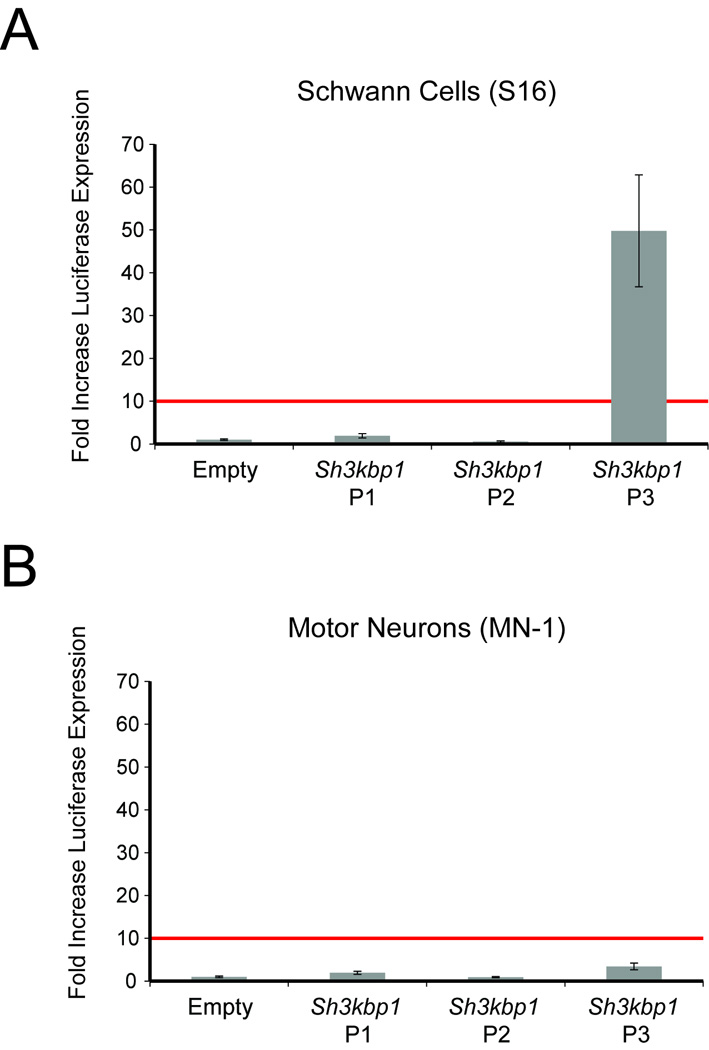
Sh3kbp1 P3 directs reporter gene expression in cultured Schwann cells. Sh3kbp1 P1, P2, and P3 cloned upstream of a luciferase reporter gene were assayed for enhancer activity in S16 (A) and MN-1 (B) cells relative to a control vector [‘Empty’ in (A) and (B)]. The red lines demarcate the 10-fold cut-off for enhancer activity (see Results) and error bars indicate standard deviations.
Sh3kbp1 P3 directs reporter gene expression in the Schwann cells of developing zebrafish
To explore the function of Sh3kbp1 P3 in vivo we employed a zebrafish model system. Zebrafish are a tractable organism for studying reporter gene expression in Schwann cells based on observations that zebrafish embryos are transparent, develop externally, and harbor Schwann cells along peripheral nerves by 48 hours post fertilization (hpf) (Brosamle and Halpern, 2002; Westerfield, 2000). Furthermore, zebrafish have been successfully employed to study Schwann-cell-specific activities of mammalian enhancers (Antonellis et al., 2010; Antonellis et al., 2008; Fisher et al., 2006a). Briefly, mouse Sh3kbp1 P3 was cloned upstream of an enhanced green fluorescent protein (EGFP) reporter gene, and the resulting construct was injected into wild-type (AB) zebrafish embryos. EGFP-positive fish were selected at 24 hpf, allowed to develop to sexual maturity, and mated with wild-type fish to identify EGFP-positive, transgene-stable F1 progeny. Subsequently, EGFP expression was examined in F1 zebrafish at 24 and 48 hpf. Sh3kbp1 P3 directs EGFP expression in the forebrain (white arrows in Fig. 4A and 4B), presumptive fin bud (arrowhead in Fig. 4A), and caudal region (red arrows in Fig. 4A and 4B) at 24 hpf. By 48 hpf, EGFP expression continues in the fin bud (arrowheads in Fig. 4C and 4D) and hindbrain (asterisks in Fig. 4C and 4D). Sh3kbp1 P3 directs reporter gene (LacZ) expression in the ortholog of each of the above structures in transient transgenic mouse embryos (Supplementary Fig. 2). Importantly, at 48 hpf we detected EGFP expression in structures consistent with Schwann cells along motor nerves (arrows in Fig. 4E and 4F) and lateral line nerves (arrowheads in Fig. 4E and 4F). Reporter expression was concordant in these structures among 3 independent F1 lines, co-staining with an anti-MBP antibody at 72 hpf revealed overlap with EGFP signal (Supplementary Fig. 3), and detailed analyses did not reveal EGFP expression in oligodendrocytes.
Fig. 4.
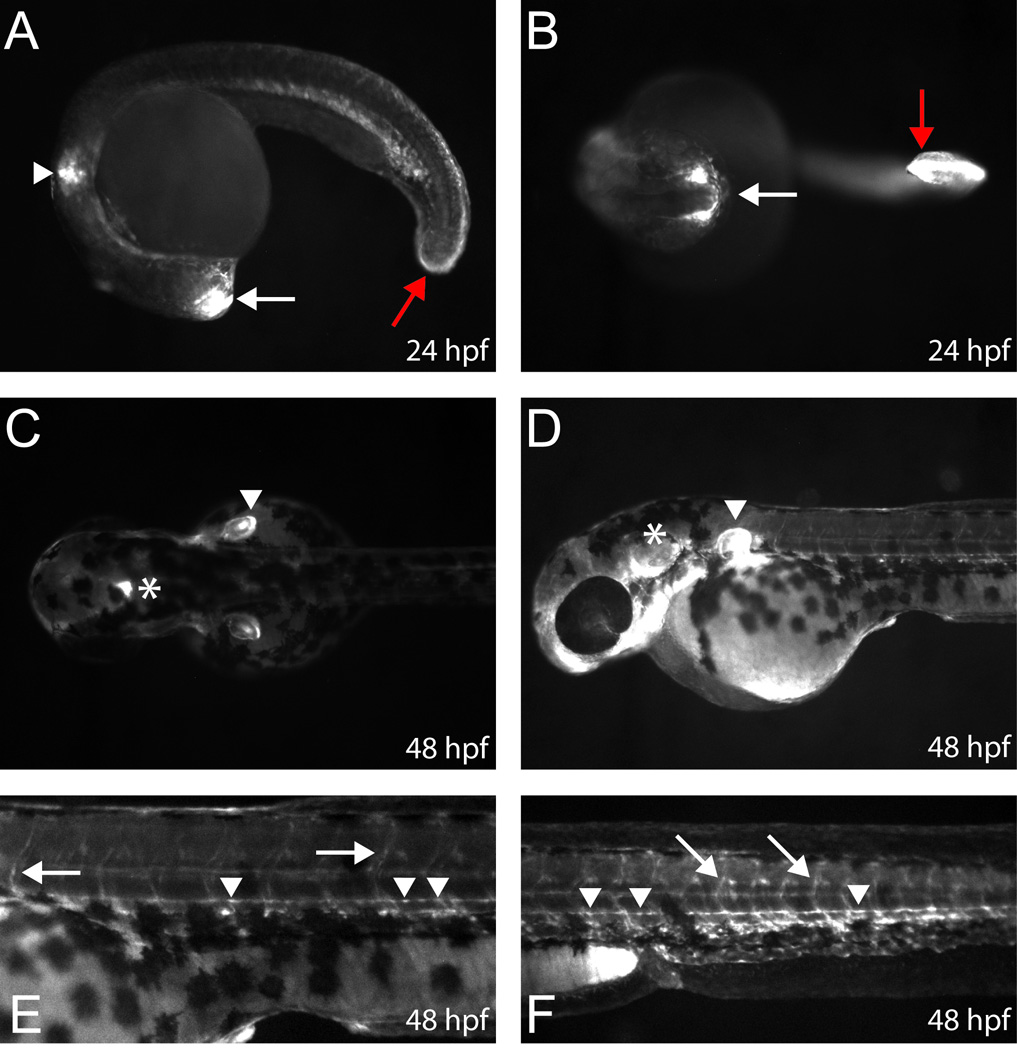
Sh3kbp1 P3 directs EGFP expression in Schwann cells in vivo. Zebrafish embryos were injected with constructs harboring mouse Sh3kbp1 P3 directing enhanced green fluorescent protein (EGFP) expression and analyzed (A and B). Sh3kbp1 P3 directs reporter gene expression in the forebrain (white arrows), fin bud (white arrowheads), and caudal region (red arrows) at 24 hpf. (C–F) By 48 hpf, signal is detected in the hindbrain (C and D; asterisks), fin bud (C and D; white arrowheads), and along motor nerves (E and F; white arrows) and lateral line nerves (E and F; white arrowheads) consistent with Schwann cell expression. The above expression patterns were confirmed in three independent F1 zebrafish strains (see text).
The SOX10 binding site is necessary for Sh3kbp1 P3 activity in S16 cells
To determine the necessity of the SOX10 and POU3F2 binding sites for Sh3kbp1 P3 activity, we deleted each binding site and performed luciferase assays in cultured Schwann (S16) cells. Deletion of the SOX10 binding site alone caused a ~97% reduction in luciferase activity compared to wild-type P3 (Fig. 5A). Deletion of the POU3F2 binding site caused a ~66% reduction in luciferase activity (Fig. 5A). Interestingly, deletion of both the SOX10 and POU3F2 binding sites only caused a ~71% reduction in luciferase activity compared to wild-type P3 (Fig. 5A). To determine if our deletion assay results were an artifact of removing nucleotides from Sh3kbp1 P3, we mutated the SOX10 and POU3F2 binding sites by complementing (but not reversing) each sequence. This approach retains the number of nucleotides as well as the GC content of the sequence, but should ablate transcription factor specificity. Mutation of the SOX10 binding site resulted in a ~90% decrease in luciferase activity compared to wild-type P3 (Fig. 5B). Mutation of the POU3F2 binding site resulted in a ~50% decrease in luciferase activity (Fig. 5B). Finally, mutation of both the SOX10 and POU3F2 binding sites resulted in a ~96% decrease in luciferase activity compared to wild-type P3 (Fig. 5B). Combined, these data indicate that the SOX10 and POU3F2 binding sites are necessary for the promoter activity of Sh3kbp1 P3 in S16 cells.
Fig. 5.
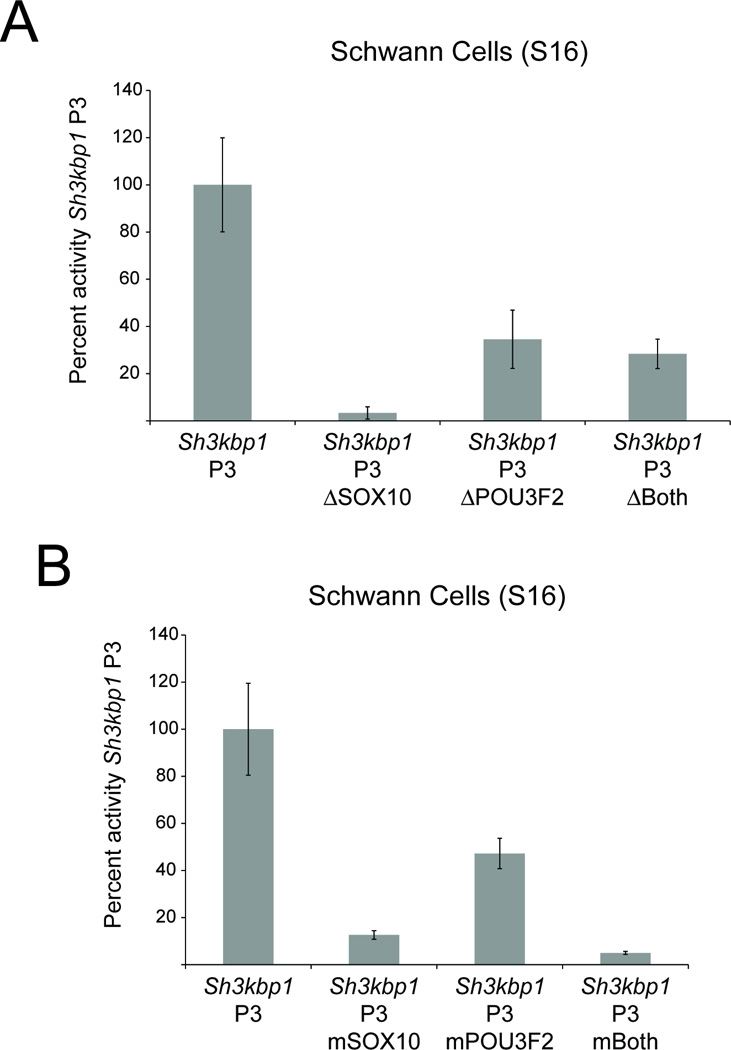
SOX10 binding sites are required for Sh3kbp1 P3 activity. Site-directed mutagenesis was used to delete (A) and mutate (B) either the SOX10 or POU3F2 consensus sequences (or ‘Both’) within Sh3kbp1 P3. Mutagenized constructs were then tested for the ability to direct luciferase expression in cultured Schwann (S16) cells, and compared against wild-type Sh3kbp1 P3. Error bars indicate standard deviations.
Expression of SOX10 in SOX10-negative cells activates Sh3kbp1 P3
Sox10 and Pou3f2 are expressed in S16 cells, while neither is expressed in MN-1 cells (Fig. 2A). Furthermore, Sh3kbp1 P3 is not active in MN-1 cells (Fig. 3B). Thus, MN-1 cells provide a unique system to test for the effect of SOX10 and POU3F2 on the activity of P3. We therefore co-transfected the construct harboring Sh3kbp1 P3 directing a luciferase reporter gene with expression constructs for SOX10 and POU3F2 directed by a strong promoter (pCMV) into MN-1 cells. The addition of a SOX10 expression construct resulted in ~50-fold induction of P3 in MN-1 cells while the addition of a POU3F2 expression construct resulted in ~2-fold induction of P3 in these cells (Fig. 6A). Co-transfection of a SOX10 and POU3F2 expression construct in this system resulted in a slight increase in the mean fold-induction, however this increase was not significantly different from SOX10 transfection alone (Fig. 6A). Co-transfection with an expression vector without a transcription factor cDNA (pCMV only) into MN-1 cells did not have an effect on P3 activity (‘Empty’ in Fig. 6A). These data are consistent with our deletion and mutation studies (Fig. 5A and 5B) and indicate that SOX10 is important for the observed promoter activity in Schwann (S16) cells (Fig. 3A).
Fig. 6.
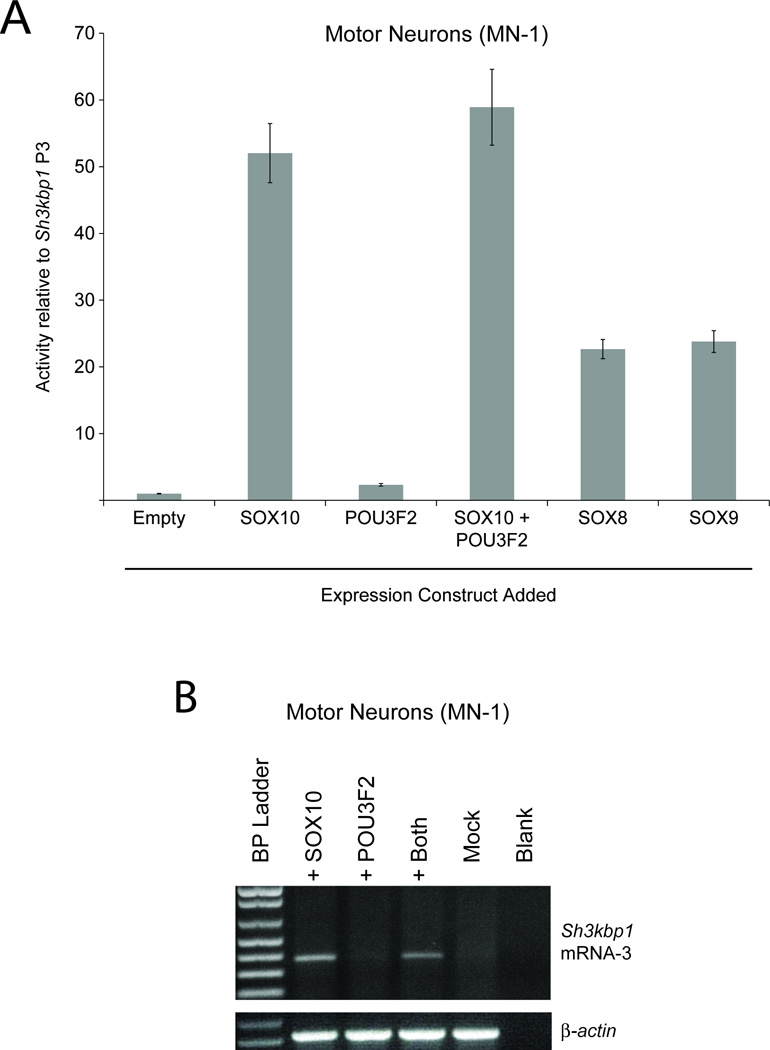
SOX10 is sufficient for Sh3kbp1 P3 activity in SOX10-negative MN1 cells. (A) A construct harboring Sh3kbp1 P3 directing luciferase expression was co-transfected into cultured motor neurons (MN-1) with constructs to express POU3F2, SOX10, SOX8, or SOX9. Luciferase assays were employed to test the activity of Sh3kbp1 P3 in the presence of each construct, relative to the activity in the presence of the control (‘Empty’) vector. Error bars indicate standard deviations. (B) cDNA was generated from cultured motor neurons (MN-1) transfected with constructs to express SOX10 or POU3F2, both proteins, or neither (‘Mock’). RT-PCR was then performed to test for the presence of Sh3kbp1 Exon 1C (mRNA-3). Note that the primers used were the same ones employed in Figure 2B. A negative control without cDNA (‘Blank’) was included and primers for β-actin mRNA were used as a positive control. A base pair marker is provided on the far left of the gel.
SOX10 is a member of the SOXE transcription factor family that also includes SOX8 and SOX9. All three SOXE transcription factors bind to a highly-similar consensus sequence (Mollaaghababa and Pavan, 2003; Peirano and Wegner, 2000; Stolt and Wegner, 2010; Wegner and Stolt, 2005). To test the specificity of the SOX10 binding site in Sh3kbp1 P3, we co-transfected the construct harboring P3 directing a luciferase reporter gene with expression constructs for SOX8 or SOX9 directed by pCMV. SOX10 increased P3 activity in a manner ~2-fold higher than SOX8 and SOX9 (Fig. 6A), suggesting that SOX10 is more specific than SOX8 and SOX9 at Sh3kbp1 P3.
Expression of SOX10 in SOX10-negative cells induces the expression of endogenous Sh3kbp1 mRNA-3
Based on the rationale outlined above, MN-1 cells also provide a system to test for the effect of SOX10 and POU3F2 on the activity of endogenous Sh3kbp1 P3. We transfected expression constructs for SOX10 or POU3F2 directed by a strong promoter (pCMV) into MN-1 cells, isolated RNA after 48 hours, and tested for the presence of Sh3kbp1 mRNAs containing exon 1C. While the addition of a POU3F2 expression construct did not result in detectable mRNA, the addition of a SOX10 expression construct resulted in an RT-PCR harboring exon 1C (Fig. 6B)—PCR products were subsequently gel-isolated and sequenced to confirm the specificity of Sh3kbp1 exon 1C (data not shown). Mock transfection, and transfection of an expression vector without a transcription factor cDNA (pCMV only) did not have an effect on endogenous Sh3kbp1 expression (Fig. 6B and data not shown). Thus, SOX10 is sufficient for directing endogenous Sh3kbp1 mRNA-3 expression from P3 in SOX10-negative MN-1 cells.
SOX10 binds to Sh3kbp1 Promoter 3 in vitro and in vivo
The data presented above support the notion that SOX10 directly regulates Sh3kbp1 P3. To determine if SOX10 binds to this promoter, we employed a previously-described chromatin immunoprecipitation (ChIP) assay (Jang et al., 2006; Jang and Svaren, 2009) using formaldehyde-crosslinked samples from cultured S16 cells. Quantitative PCR analysis of an immunoprecipitation of sonicated chromatin from S16 cells revealed strong binding of SOX10 to Sh3kbp1 P3 compared to the nonspecific IgG control immunoprecipitation (Fig. 7A). In addition, siRNA-mediated knock down of Sox10 in S16 cells dramatically reduced the binding of SOX10 to Sh3kbp1 P3 (Fig. 7A). To determine if this binding occurs in vivo, similar ChIP assays were performed using P15 rat sciatic nerve. Consistent with the S16 ChIP assays, our results indicate that SOX10 occupies Sk3kbp1 P3 in vivo (Fig. 7B).
Fig. 7.
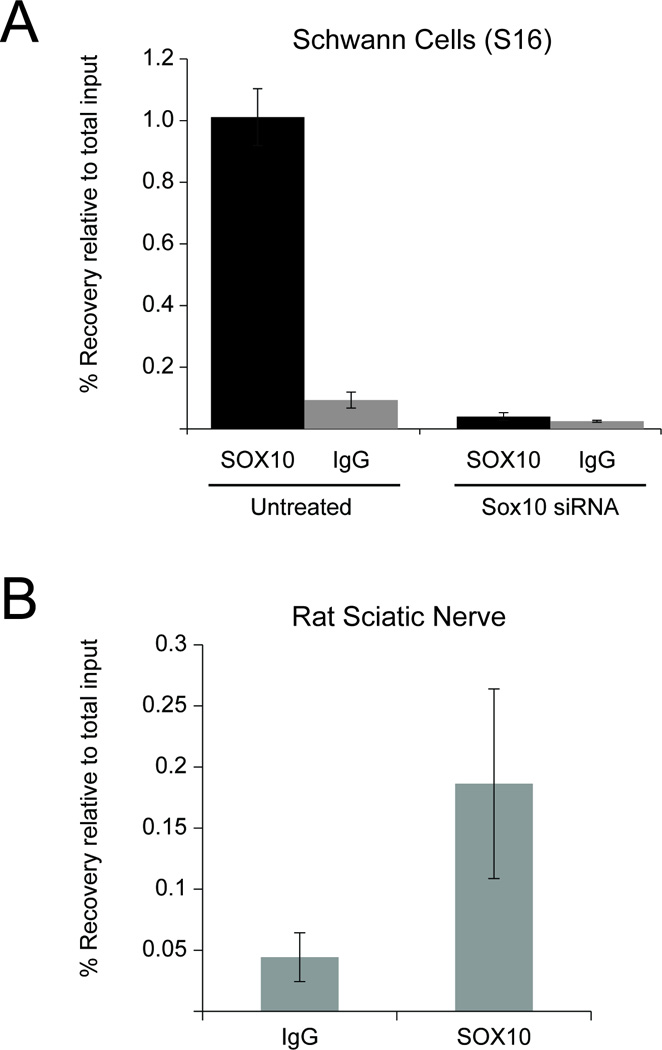
SOX10 binds to Sh3kbp1 P3 in Schwann cells. (A) SOX10 ChIP assays were performed in cultured Schwann (S16) cells treated with either control or Sox10 siRNA. Samples were analyzed using quantitative PCR and percent recovery is calculated relative to input. Error bars represent the standard deviation of at least two technical replicates. (B) In vivo SOX10 ChIP was performed using pooled sciatic nerve from P15 mice alongside an IgG control, and quantitative PCR was performed using the same primer sets as in (A). Error bars represent the standard deviation of three technical replicates.
Reducing Sox10 expression in Schwann cells reduces Sh3kbp1 expression
As an independent test of Sh3kbp1 regulation by SOX10, siRNA directed towards Sox10 was transfected into cultured Schwann (S16) cells to reduce Sox10 levels. Previous analysis of these samples showed that Sox10 expression is reduced at both the mRNA and protein levels (Jones et al., 2011). RNA was purified from the cells at 48 hours post-transfection, and quantitative RT-PCR showed that Sox10 mRNA levels were reduced ~95%, and that total Sh3kbp1 mRNA levels were reduced ~45% compared to a non-targeting siRNA control (Fig. 8A). Importantly, these data are consistent with array-based expression profiling of Sh3kbp1 in a Schwannoma cell line (RT4D6) treated with siRNA against Sox10 (GEO DataSet accession number GSE12007) (Lee et al., 2008).
Fig. 8.
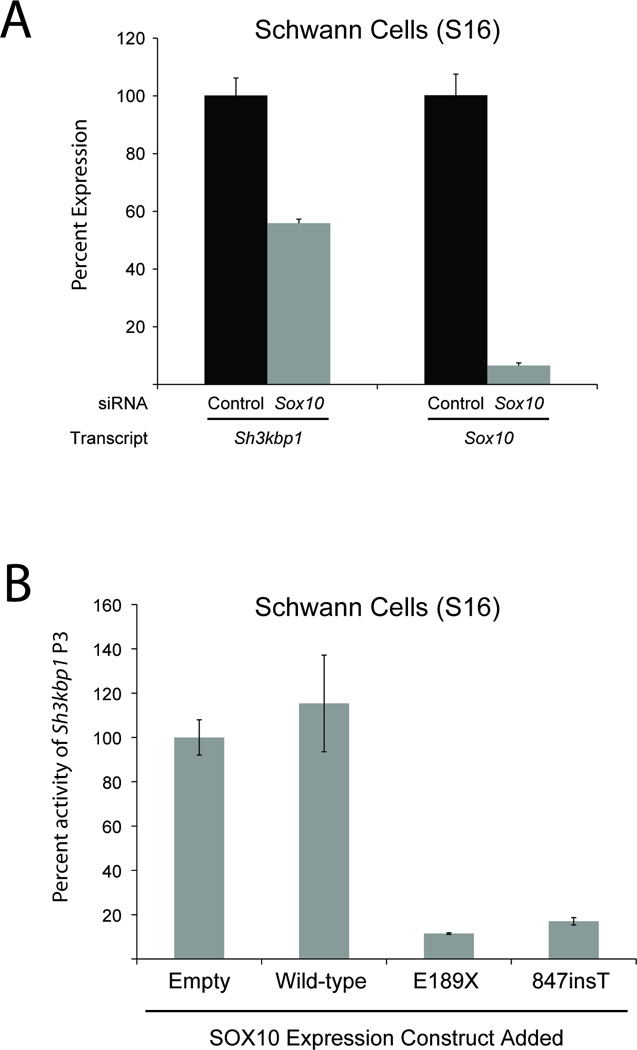
Disrupting SOX10 function reduces Sh3kbp1 expression and P3 activity. (A) Cultured Schwann (S16) cells were transfected with siRNA targeted to Sox10 and quantitative RT-PCR was used to determine the effect on Sh3kbp1 (left) and Sox10 (right) mRNA expression. Levels were normalized to the negative controls and error bars represent standard deviations (n=6). (B) A construct harboring wild-type Sh3kbp1 P3 directing luciferase expression was co-transfected into cultured Schwann (S16) cells with constructs to express wild-type or dominant-negative (E189X and 847insT) SOX10 proteins, or an insert-free construct (‘Empty’). Luciferase assays were then employed to test the activity of Sh3kbp1 P3, relative to the activity in the presence of the control (‘Empty’) vector. Error bars indicate standard deviations.
Dominant-negative SOX10 mutant proteins reduce Sh3kbp1 P3 activity in Schwann cells
Mutations in the human SOX10 gene cause autosomal dominant Waardenburg syndrome type 4A (WS4A) or autosomal dominant peripheral demyelinating neuropathy, central dysmyelination, Waardenburg syndrome, and Hirschsprung disease (PCWH) (Inoue et al., 2004). It was previously shown that mutations associated with WS4 represent null alleles, while those associated with PCWH represent dominant-negative alleles. Importantly, dominant-negative proteins interfere with the function of wild-type SOX10 in activating gene transcription in vitro (Inoue et al., 2004). These findings provide a system to disrupt the function of endogenous SOX10 protein by over-expressing dominant-negative forms of SOX10, and then assessing for an effect on target promoter activity. We co-transfected the construct harboring Sh3kbp1 P3 directing a luciferase reporter gene with expression constructs for wild-type or dominant-negative (E189X and 847insT) SOX10 into S16 cells. The effect on P3 activity was then determined via comparison to similar co-transfection experiments using a control (‘Empty’ in Fig. 8B). Transfection with a wild-type SOX10 expression construct resulted in a modest, but significant, increase (~20%; p=2.84×10−2) in luciferase activity in S16 cells (Fig. 8B). In contrast, transfection with E189X and 847insT SOX10 resulted in a ~90% and a ~80% reduction in luciferase activity in S16 cells, respectively. These data confirm the specificity of SOX10 in regulating Sh3kbp1 P3.
Identification and characterization of the zebrafish sh3kbp1 P3
The conservation of function of the mammalian Sh3kbp1 P3 in zebrafish (Figure 4) suggests that a functional ortholog of this promoter exists in the zebrafish genome. However, the zebrafish sh3kbp1 locus is poorly characterized (UCSC Zebrafish Genome Browser). To address this, we performed comparative sequence analysis between human and zebrafish to identify zebrafish genomic segments that should flank sh3kbp1 exon 1C (data not shown). Subsequently, 5’ RACE was employed to amplify and sequence sh3kbp1 exon 1C (Fig. 9A). Interestingly, analysis of the genomic sequence upstream of sh3kbp1 exon 1C (which we termed sh3kbp1 P3) revealed nine SOX10 and two POU3F2 consensus sequences (Fig. 9A)—all of these have a probability score of 0.9 or higher. We next tested sh3kbp1 P3 for the ability to direct luciferase expression in cultured Schwann (S16) cells. Zebrafish sh3kbp1 P3 directs luciferase expression in a manner ~4-fold higher than the control vector in mammalian Schwann cells. In contrast, sh3kbp1 P3 only directed luciferase expression in a manner ~1.5-fold higher than the control vector in SOX10-negative MN1 cells. Finally, we tested sh3kbp1 P3 for activity in zebrafish Schwann cells in vivo. Indeed, sh3kbp1 P3 directed EGFP expression along motor nerves (consistent with Schwann cell expression) in 34 out of 44 (77%) EGFP-positive transient transgenic zebrafish embryos at 48 hpf (Fig. 9C and 9D). Thus, we have identified the zebrafish ortholog of Sh3kbp1 P3 and have shown that it directs reporter gene expression in both mammalian and zebrafish Schwann cells.
Fig. 9.
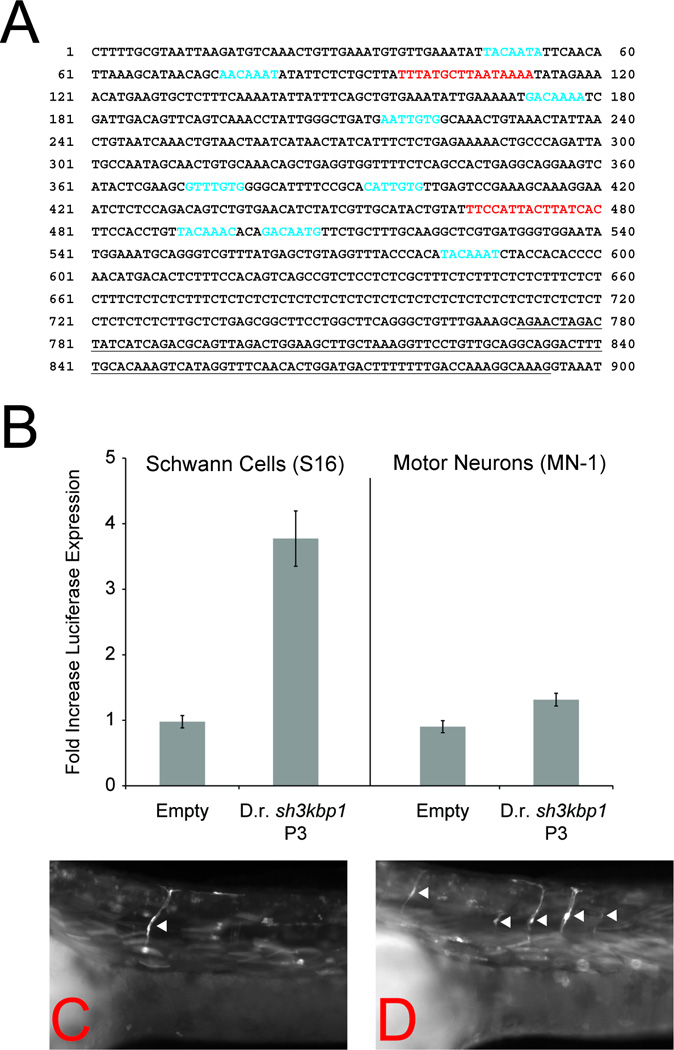
Characterization of the zebrafish sh3kbp1 P3. (A) 5’ RACE, RT-PCR, and DNA sequencing analysis was performed on RNA isolated from 48 hpf zebrafish embryos. The sequence of sh3kbp1 exon 1C is indicated by underlined text. SOX10 (blue text) and POU3F2 (red text) consensus sequences were identified using TRANSFAC and the MATCH algorithm. (B) Zebrafish (D.r.) sh3kbp1 P3 was cloned upstream of a luciferase reporter gene and assayed for enhancer activity in S16 (left panel) and MN-1 (right panel) cells relative to a control vector (‘Empty’). The error bars indicate standard deviations. (C and D) Zebrafish embryos were injected with a construct harboring zebrafish sh3kbp1 P3 directing enhanced green fluorescent protein (EGFP) expression and analyzed at 48 hpf. Zebrafish sh3kbp1 P3 directs reporter gene expression along motor nerves (arrowheads) consistent with expression in Schwann cells. Two representative transient transgenic embryos are shown.
Discussion
The transcription factor SOX10 is essential for the proper development and function of myriad neural crest-derived cell populations (Britsch et al., 2001; Southard-Smith et al., 1998; Stolt and Wegner, 2010) including Schwann cells—specialized glial cells responsible for myelinating axons in the peripheral nervous system. To date, only a handful of validated SOX10 targets have been characterized in these cells (Svaren and Meijer, 2008). The identification of additional SOX10 targets will be critical for a better understanding of Schwann cell biology and for exploring novel candidates for related diseases. Here, we provide computational and functional evidence that SOX10 directly regulates a tissue-specific promoter at the Sh3kbp1 locus. These data add to our knowledge of SOX10 target genes and raise important questions regarding the role of SH3KBP1 in Schwann cells.
Our computational-based approach for identifying SOX10 target genes begins with genome-wide identification of highly-conserved SOX10 binding sites (e.g., Fig. 1F). These analyses have revealed myriad potential transcriptional regulatory elements relevant for downstream characterization in Schwann cells (unpublished data). One such element was identified at an alternative promoter (P3) of the Sh3kbp1 locus (Fig. 1A). Expression analyses revealed the presence of mRNAs directed from Sh3kbp1 promoter 1 (P1) and promoter 3 (P3) in Schwann cells in vitro and in vivo—as well as the absence of mRNAs expressed from promoter 2 (P2). This is the first report of Sh3kbp1 expression in Schwann cells. Our data, combined with previous studies showing that Sh3kbp1 P1 is active in a near-ubiquitous manner (Buchman et al., 2002; Mayevska et al., 2006), suggest that transcripts produced from P3 have a specific function in Schwann cells.
The promoter activities of Sh3kbp1 P1, P2, and P3 in SOX10-positive and SOX10-negative cells (Fig. 3) was consistent with our expression studies and supportive of the hypothesis that SOX10 regulates P3. However, two somewhat unexpected observations were encountered. First, Sh3kbp1 P1 displays a relatively small amount of activity (~2-fold higher than the control vector) in both S16 and MN-1 cells; however, the associated exon (1A) is expressed in both cell lines. One possible explanation is that Sh3kbp1 P1 acts in concert with other, unidentified regulatory elements to direct the expression of mRNAs containing exon 1A. While we did not identify any conserved elements outside of the three promoter regions, a less stringent comparative sequence analysis may reveal additional candidate regulatory elements. Second, Sh3kbp1 P3 displays a low level of promoter activity (~3.5-fold higher than the control vector) in MN-1 cells, even though the associated exon (1C) is not expressed in these cells. One possible explanation is that other factors are expressed in MN-1 cells that act upon P3, but that SOX10 is required for a higher level of promoter activity (see below). Scrutinizing the remaining 132 base pairs of Sh3kbp1 P3 that are fully-conserved between mammals and chicken may provide insight into this possibility and facilitate the identification of other transcription factors that bind to P3.
Analysis of the promoter activity of mouse Sh3kbp1 P3 in zebrafish revealed a tissue-specific expression pattern conserved between diverse vertebrate species (Fig. 4 and Supplementary Figure 2). For example, strong reporter gene expression was observed in the fin bud and limb bud of developing zebrafish and mice, respectively. These data raise important questions regarding the transcriptional regulation of P3 and the function of the associated protein in this structure. However, since SOX10 expression has not been detected in the developing limb, there are likely other transcription factors that mediate P3 activity in this tissue. Importantly, we observed strong Sh3kbp1 P3 promoter activity in structures consistent with zebrafish Schwann cells at 48 and 72 hours post fertilization (hpf). Reporter gene expression patterns (e.g., EGFP) have been well-characterized in the Schwann cells of developing zebrafish (Antonellis et al., 2008; Jung et al., 2010). Specifically, at 48 hpf, a characteristic expression pattern is detected along the lateral line nerves and motor nerves (e.g., see references above in comparison to Fig. 4E and 4F). Therefore, our analyses strongly suggest that Sh3kbp1 P3 is active in Schwann cells in vivo. Furthermore, the activity of mouse Sh3kbp1 P3 in zebrafish cells indicates a conservation of function between these two species, and may underscore a critical role for Sh3kbp1 P3 in vertebrate organisms. These observations highlight an emerging body of evidence showing that zebrafish are a relevant and tractable system for evaluating the tissue-specific activities of highly-conserved mammalian transcriptional regulatory elements (Antonellis et al., 2010; Antonellis et al., 2008; Fisher et al., 2006a).
Further characterization of Sh3kbp1 P3 showed that: (1) SOX10 binds to this region in vitro and in vivo; (2) The identified SOX10 binding site is essential for the observed promoter activity in vitro; and (3) Expressing SOX10 in SOX10-negative cells activates P3 in luciferase assays. Additionally, disrupting endogenous SOX10 function decreases the activity of P3 activity in luciferase assays. All of these data strongly support the hypothesis that SOX10 directly regulates Sh3kbp1 P3. Depleting Sox10 expression with siRNA in cultured Schwann (S16) cells reduces the expression of endogenous Sh3kbp1 by ~45% (Fig. 8A). This observation is consistent with reduced transcription from P3 and maintained expression from P1—both of the associated exons (1A and 1C) are expressed in S16 cells (Fig. 2B). However, our attempts to show a specific decrease in exon 1C levels via qPCR were unsuccessful (data not shown). While there are a number of technical reasons that might explain this discrepancy (e.g., probe specificity), an interesting possibility is that Sh3kbp1 P3 acts as an intronic enhancer that regulates another promoter at Sh3kbp1. However, expressing SOX10 in SOX10-negative cells induces the expression of endogenous Sh3kbp1 exon 1C, indicating that SOX10 is sufficient for Sh3kbp1 P3 activity in these cells.
Our transcription factor binding site predictions revealed the presence of SOX10 and POU3F2 motifs within Sh3kbp1 P3. Since both of these factors have important roles in Schwann cells (Svaren and Meijer, 2008), we considered each a relevant candidate for regulating P3. While our functional data largely implicate SOX10 in this process, similar observations were not made for POU3F2. Specifically: (1) The POU3F2 motif is not as well-conserved as the SOX10 binding site; (2) Mutating the POU3F2 binding site had a smaller effect on P3 activity compared to mutating the SOX10 binding site; (3) Expressing POU3F2 in MN-1 cells did not increase P3 activity in luciferase assays; and (4) Expressing POU3F2 in MN-1 cells did not induce expression of endogenous Sh3kbp1 mRNA-3. However, since mutating both the SOX10 and POU3F2 binding sites had an additive effect on Sh3kbp1 P3 promoter activity (Fig. 5B) and since expressing both SOX10 and POU3F2 had an additive effect on luciferase expression directed by Sh3kbp1 P3 (Fig. 6A), the POU3F2 binding site may play a role in Sh3kbp1 expression. One possibility is that POU3F1, which binds to similar sequences as POU3F2 (Jaegle et al., 2003), may act on this promoter—SOX10 and POU3F1 are known to act synergistically at other Schwann cell loci (Ghislain and Charnay, 2006; Kuhlbrodt et al., 1998). Sh3kbp1 is a widely-expressed gene that spans ~350 kb on mouse XqF4. SH3KBP1 proteins contain a C-terminal coiled-coil domain, a proline-rich region, and one, two, or three src homology 3 (SH3) domains on the N-terminus (Take et al., 2000). The inclusion of either three, two, or one SH3 domains is dictated by the alternative use of P1, P2, and P3 (Fig. 1A), respectively (Buchman et al., 2002). Previous expression assays have shown that at least one of the three major isoforms is expressed in every tissue type (Buchman et al., 2002; Mayevska et al., 2006). However, to date, mRNA-3 (Fig. 1A) has only been detected in Schwann cells (Fig. 2B), heart, skin, and kidney (Buchman et al., 2002).
Combined, our data suggest that the protein encoded from Sh3kbp1 mRNA-3 has a specific role in Schwann cells. Interestingly, the two variants of SH3KBP1 protein predicted to be expressed in Schwann cells (Fig. 2) have opposing functions that may be relevant for this cell population. Specifically, the full-length protein isoform (RukL) is a potent inhibitor of signaling by receptor tyrosine kinsases (Soubeyran et al., 2002), while the shorter protein isoform translated from mRNA-3 (RukM) inhibits the function of RukL (Gout et al., 2000). Therefore, the specific expression of RukM in Schwann cells may underscore the importance of tightly regulated RTK signaling [e.g., ERBB2 and ERBB3 (Newbern and Birchmeier, 2010)] from Schwann cell development through terminal differentiation. A number of future studies may provide further insight into the role of SH3KBP1 in Schwann cells. These include: (i) Determining the effect of over-expressing RukL and RukM on RTK internalization and degradation; (ii) Identifying proteins that differentially bind to RukL and RukM; and (iii) Knocking down mRNA-3 expression in a relevant model system and testing for a Schwann cell phenotype. Indeed, the identification of sh3kbp1 P3 and exon 1C (Fig. 9) will facilitate these latter studies in zebrafish. These lines of experimentation will provide key information on the function of SH3KBP1, and insight into any specific requirement for expression from promoter 3 in Schwann cells.
Experimental Methods
Multiple-species comparative sequence analysis
The following sequences were collected at orthologous Sh3kbp1 loci using the University of California at Santa Cruz (UCSC) Genome Browser (Kent et al., 2002): mouse (chrX:156031013-156426365, mm9), rat (chrX:55956157-56565330, rn4), human (chrX:19433168-20034598, hg19), dog (chrX:15324708-15892001, canFam2), cow (chrX:75469265-76034103, bosTau4), and chicken (chr1:123303813-123714817, galGal3). Repetitive DNA sequences were masked using the UCSC Genome Browser ‘Sequence Formatting Options’, and each genomic sequence was submitted to the MultiPipMaker alignment algorithm (http://pipmaker.bx.psu.edu/cgi-bin/multipipmaker) (Elnitski et al., 2003). Mouse genomic sequence was used as the reference, and the ‘Search both strands’ and ‘Chaining’ options were employed. Subsequently, the MultiPipMaker ‘acgt’ alignment file was submitted to the ExactPlus (EP) algorithm (http://research.nhgri.nih.gov/exactplus/) (Antonellis et al., 2006) using the following settings: Minimum length of exact match to seed = 6; Minimum number of species to seed = 6; and Minimum number of species to extend an alignment = 6. Data were viewed on the UCSC Genome Browser using the EP custom track output file, and a cluster of EP hits was defined as 3 or more EP hits within a 500 base pair window.
Transcription factor binding site predictions
Transcription factor binding site predictions were performed in two phases. First, clustering EP hits (bold entries in Table 1) were analyzed using the MATCH algorithm (Matys et al., 2003) to identify transcription factor bindings sites from vertebrate species with the “minimize false negatives” setting. Second, mouse genomic sequences upstream of the second and third Sh3kbp1 transcription start sites were obtained from the UCSC Genome Browser (P2= chrX:156146410-156146532, mm9; P3= chrX:156278277-156278399, mm9) and specifically assessed for factors involved in Schwann cell development. This was accomplished by using the MATCH algorithm with the “minimize the sum of both error rates” setting, and the following matrices: V$BRN2_01, V$EGR2_01, V$POU3F2_01, V$POU3F2_02, V$SOX10_Q6, and V$TST1_02. The entire zebrafish sh3kbp1 P3 (chr24:26675176-26675949, danRer7) region was evaluated using the above matrices.
RNA isolation and cDNA synthesis
Total RNA was isolated using the Nucleospin RNA L kit (Clontech, Mountain View, CA) according to manufacturer’s instructions. RNA was eluted from columns with 50 uL of RNase-free water and stored at −80°C. RNA concentration and purity (A260 and A280 values) were obtained using a Nanodrop 1000 (Thermo Fischer Scientific, Waltham, MA). cDNA libraries were generated using 1 µg of total RNA from either MN-1 cells, S16 cells, mouse sciatic nerve, or cardiac myocytes, and the High-Capacity cDNA Reverse Transcription Kit (Ambion, Austin, TX) with the provided solution of random reverse-transcription primers. Commercially available cDNA from human oligodendrocytes was obtained from ScienCell Research Laboratories (Carlsbad, CA).
RT-PCR analysis
Isolated cDNA (see above) was subjected to RT-PCR analysis using gene-specific primers (sequences available upon request). For each reaction, 22 µL of PCR Supermix (Invitrogen) was combined with 1 µL each of 20 µM primer solutions and 1 µL of the indicated cDNA sample. Standard PCR conditions were employed for a total of 35 cycles. A cDNA-negative control reaction was included for each primer pair. After standard agarose gel electrophoresis, reactions were scrutinized for the expected band sizes for Sox10 (952 bp), Pou3f2 (592 bp), Sh3kbp1 exon 1A (997 bp), Sh3kbp1 exon 1B (847 bp), Sh3kbp1 exon 1C (380 bp), human SH3KBP1 exon 1C (237 bp), and a human (445 bp) or rodent (520 bp) β-actin amplification control. All PCR products were subsequently resolved on 1.5% low-melt agarose gels in 1X TAE, purified, and subjected to DNA sequencing analysis to confirm specificity.
Western blot analysis
Cultured S16 and MN-1 cells, and mouse sciatic nerve tissue were harvested and resuspended in 37.5 uL of lysis solution containing 10 mM Tris-HCl/1 mM EDTA/50 mM NaCl (Invitrogen) and 12.5 uL of loading buffer containing SDS (Invitrogen). After incubation at 95°C for 5 min, the total protein lysate was electrophoresed on 4–12% Bis-Tris gels (Invitrogen) at 125 V for 100 min in running buffer containing SDS (Invitrogen). Samples were then electroblotted onto nitrocellulose membranes at 25 V for 100 min in NuPAGE transfer buffer (Invitrogen). Membranes were blocked in 1X PBS/0.05% Tween 20/5% nonfat dry milk for 18 h at 4°C and then incubated with a 1:1000 dilution of anti-SOX10 (Santa Cruz Biotechnology) or 1 ug/uL anti-actin (Sigma) antibody in blocking solution for 1 h at room temperature. Membranes were washed three times in 1X PBS plus 0.05% Tween 20, incubated with a secondary antibody conjugated to horseradish peroxidase for 1 h at room temperature, washed three times as described above, and finally incubated with DAB substrate (Roche Diagnostics), according to the manufacturer’s instructions.
Expression and reporter gene construction
Primers were designed to amplify each Sh3kbp1 promoter region using the following UCSC Mouse Genome Browser (mm9) coordinates: P1 (chrX:156064945-156065677), P2 (chrX:156145737-156146679), and P3 (chrX:156278255-156278503). Primers were also designed to amplify the zebrafish sh3kbp1 promoter 3 region using the following UCSC Zebrafish Genome Browser (danRer7) coordinates: chr24:26675176-26675949. PCR products were amplified using Gateway Cloning compatible primers (Invitrogen), so that each PCR product was flanked by attB recombination sites. PCR products were cloned into the pDONR221 entry vector using BP Clonase (Invitrogen) and each resulting construct underwent digestion with BsrGI and DNA sequencing to assure insert integrity. Subsequently, each promoter construct was recombined into expression vectors [directing luciferase expression for enhancer assays or EGFP expression for zebrafish studies] (Antonellis et al., 2006; Fisher et al., 2006b) using LR Clonase (Invitrogen). Mutagenic primers were designed and mutagenesis reactions performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). All primers sequences are available upon request. The wild-type, E189X, and 847insT SOX10 expression constructs (cloned downstream of a pCMV promoter element)(Inoue et al., 2004; Pingault et al., 1998) were a kind gift of Ken Inoue, Jim Lupski, and Michael Wegner. The pCMV-SOX8 and pCMV-SOX9 expression constructs were a kind gift of Stacie Loftus and Bill Pavan. The pCMV-POU3F2 expression construct was obtained from Origene (Rockville, MD).
Cell culture and transfection assays
Immortalized rat Schwann cells (S16) (Toda et al., 1994) and mouse spinal motor neurons (MN-1) (Salazar-Grueso et al., 1991) were grown under standard conditions in DMEM supplemented with 10% fetal bovine serum and 1X glutamine, and were cultured to > 80% confluency before harvesting. For RNA isolation, ~2×107 cells were collected, transferred to a 15 mL conical tube and centrifuged for 20 min at 2000 RPM. For luciferase assays, ~1×104 cells were plated into each well of a 96-well plate and cultured for 24 hours prior to transfection assays.
MN-1 or S16 cells were plated in 6-well or 96-well culture dishes. Lipofectamine 2000 (Invitrogen) was diluted 1:100 in OptiMEM I minimal growth medium (Invitrogen) and incubated at room temperature for 10 minutes. Separately, DNA samples were diluted to a concentration of 8 ng/uL in OptiMEM I. An equivalent mass of each DNA sample was employed for co-transfection experiments. For luciferase assays, DNA from an internal control renilla expression construct was diluted to a concentration of 8 pg/uL in the above-mentioned solution. Subsequently, equal volumes of Lipofectamine and DNA solutions were combined and incubated for 20 minutes at room temperature. Cells were then incubated with transfection solution for 4 hours under standard conditions, after which growth media was added.
Luciferase assays
MN-1 or S16 cells were plated in 96-well plates and transfection assays performed as described above. After 48 hours in culture medium, cells were washed with 1X PBS and lysed for 1 hour at ~25°C using 1X Passive Lysis Buffer (Promega, Madison, WI). A total of 10 µL of the resulting cell lysate was transferred to a white polystyrene 96-well assay plate (Corning Inc., Corning, NY). Luciferase and renilla activities were determined using the Dual Luciferase Reporter 1000 Assay System (Promega) and a Glomax Multi-Detection System (Promega). Each experiment was performed at least 24 times, and the ratio of luciferase to renilla activity, as well as the fold increase in this ratio over that observed for a control luciferase expression vector with no insert, were calculated. The mean (bar height in figures) and standard deviation (error bars in figures) were determined using standard calculations.
Zebrafish husbandry and transgenesis
Zebrafish were raised and bred under standard conditions (Kimmel et al., 1995; Westerfield, 2000). Embryos were maintained at 28°C and staged in accordance with standard methods (Kimmel et al., 1995; Westerfield, 2000). Two-celled zebrafish embryos were dechorionated with Pronase in 1X Holtfrieter’s solution for ~7 minutes. Expression constructs (see above) were injected into embryos (n > 100) at the two cell stage as described (Fisher et al., 2006b). Transient transgenic, EGFP-positive embryos (n=44) were analyzed at 48 hours post fertilization (hpf; see below). To generate F1, transgene-stable progeny, transgene-positive zebrafish were allowed to develop to sexual maturity and crossed with wild-type (AB) zebrafish. Immunohistochemistry analyses were carried out following standard protocols and using a 1:50 dilution of a rabbit anti-MBP antibody [kind gift of Dr. William Talbot (Lyons et al., 2005)], and a 1:50 dilution of a mouse anti-EGFP antibody (Invitrogen). Secondary antibodies (Invitrogen) included a 1:1,000 dilution of goat anti-rabbit IgG conjugated to Alexa Fluor 555 (MBP), and a 1:1,000 dilution of goat anti-mouse IgG conjugated to Alexa Fluor 488 (EGFP). Subsequently, embryos were evaluated for reporter gene expression at 24, 48, and 72 hpf using a Carl Zeiss Lumar V12 Stereo microscope with AxioVision version 4.5 software.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays on pooled male and female rat sciatic nerve at postnatal day 15 (P15) and cultured Schwann (S16) cells were performed as previously described (Jang et al., 2006; Jang and Svaren, 2009), except that S16 cells were crosslinked in phosphate-buffered saline (PBS) containing 1% formaldehyde. In addition, the herring sperm DNA in the blocking procedure was omitted in the in vivo ChIP assays. The antibodies used in this study include anti-SOX10 (Santa Cruz Biotechnology sc-17342X), and control anti-IgG (normal goat IgG: Santa Cruz Biotechnology sc-2028). Following ChIP, quantitative PCR was performed in duplicate to calculate the percent recovery of Sh3kbp1 P3 relative to the total DNA input, using the Comparative Ct method (Livak and Schmittgen, 2001).
siRNA treatment
Either siRNA directed towards Sox10 (Ambion, 4390771) or a negative siRNA control (Negative control #2, Ambion, AM4613) were transiently transfected into cultured Schwann (S16) cells with the Amaxa system (Lonza) using the Rat Neuron Nucleofection Solution. The transfected cells were incubated for 48 hours before harvesting RNA using Tri Reagent (Ambion), and quantitative PCR was performed in duplicate to calculate the fold recovery, using the Comparative Ct method (Livak and Schmittgen, 2001). The following primer set was used to detect Sh3kbp1 expression: Sh3kbp1_F, 5’-GAAGTCTGGCCTGCTCTTTGG-3’; and Sh3kbp1_R, 5’-GCAGGAGAAACGGAAAAGTCAC-3’.
Mouse transient transgenesis
Linearized plasmid DNA was isolated using gel electrophoresis followed by electro-elution into 1X TAE buffer. Microinjections into eggs produced from C57BL/6xSJL crosses were performed as previously described (Hogan et al., 1986). Subsequently, embryos were dissected at embryonic day 11.5 (E11.5) and fixed at 4°C in 1X PBS, 8% formaldehyde, 5mM EGTA, and 2mM MgCl2 for 30 minutes, followed by three 15-minute washes (1X PBS, 2 mM MgCl2, .01% deoxycholate, and 0.02% NP40). Embryos were then stained at 37°C in wash buffer with 12.5 mM K-Ferricyanide, 12.5 mM K-Ferrocyanide, and 2% 50 mg/ml 5-bromo-4-chloro-3-indolyl-b-D-galacto-pyranoside in N, N-dimethyl formamide for 4 hours (embryos with no indication of stain at that time were stained overnight). Subsequently, embryos were washed twice in wash buffer for 30 minutes at room temperature, fixed overnight in 8% formaldehyde in PBS at 4°C, and transferred into 70% ethanol in PBS. Of 200 embryos, 37 showed positive LacZ expression. Of the positively-stained embryos, overlapping expression was seen in the caudal spine (9), forebrain (2), midbrain (4), cerebral cortex midline (10), limb bud (14), eye (9), endolymphatic diverticular appendage of the otic vesicle (19), and the upper and/or lower rhombic lip (11).
5’ RACE
First-strand cDNA was generated from RNA isolated from whole, 48 hour post fertilization, wild-type zebrafish embryos, and a single oligonucleotide primer designed within a predicted sh3kbp1 exon down-stream of exon 1C (5’-AGAGATTCAGGCCGCTCTGG-3’). The cDNA sample was subsequently TdT-tailed according to the manufacturer’s instructions (Invitrogen) and sequential PCR reactions were performed using nested primers (available upon request) and standard PCR conditions. Following separation on 1% agarose gels, PCR products were excised and purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA), and subjected to DNA sequence analysis. All sequencing data was analyzed using Sequencher software (Gene Codes, Ann Arbor, MI).
Supplementary Material
(A) SOX10 protein is expressed in S16 cells and sciatic nerve. Western blot analyses were performed with total protein lysates from S16 and MN-1 cells, and mouse sciatic nerve (mSN) using an anti-SOX10 or anti-actin antibody, as indicated. (B) SH3KBP1 mRNA-3 is not expressed in oligodendrocytes. RT-PCR was performed on cDNA generated from human cultured oligodendrocytes (Oligo) and human cardiac myocytes (Heart) to test for the presence of SH3KBP1 transcripts harboring exon 1C. Cardiac myocytes were analyzed as a positive control for human SH3KBP1 exon 1C. Negative controls without cDNA (‘Blank’) were included for each reaction and primers for β-ACTIN mRNA were used as a positive control. Base pair (b.p.) sizes of markers are provided on the left.
Sh3kbp1 P3 directs tissue-specific expression in mouse embryos. Mouse pronuclear injections were performed using a construct harboring Sh3kbp1 P3 directing a LacZ reporter gene, and embryos were analyzed at embryonic day 11.5 (E11.5; A–D). LacZ expression was observed in the limb buds (A; black arrow, n=14), caudal region (A; inset, black arrowhead, n=9), forebrain (B, n=2), hindbrain (C, n=4), rhombic lip (D; black arrow, n=11), and endolymphatic diverticular appendage of the otic vesicle (D; black arrowhead, n=19).
Co-localization of EGFP and MBP along the motor nerves of Sh3kbp1 P3-EGFP zebrafish embryos. Sh3kbp1 P3-EGFP stable transgenic zebrafish embryos (72 hpf) were stained with antibodies against EGFP (A) and MBP (B). Arrows indicate motor nerves, and the merged image (C) indicates co-localization of the EGFP and MBP signals.
Acknowledgements
The authors gratefully acknowledge Julie Jones for mouse sciatic nerve RNA; Shannon Davis for mouse embryo analysis; Sally Camper for microscopy; Richard Quarles for S16 cells; Kurt Fischbeck for MN-1 cells; Len Pennachio for the gateway hsp68-LacZ vector; Stacie Loftus and Bill Pavan for SOX8 and SOX9 expression constructs and for assistance interpreting LacZ expression in mouse embryos; Jim Lupski, Ken Inoue, and Michael Wegner for SOX10 expression constructs; William Talbot for the anti-MPB antibody; Sundeep Kalantry for mouse dissection, microscopy assistance, and critical evaluation of the manuscript; Thom Saunders, Maggie Van Keuren, and the University of Michigan Transgenic Animal Model Core for assistance with mouse transient transgenesis; Bob Lyons and the University of Michigan DNA Sequencing Core for sequencing assistance; and Miriam Meisler and her laboratory for thoughtful discussions on the experiments and manuscript. This work was supported by grants HD41590 from NIH/NICHD (JS), GM071648 from the NIGMS (ASM), NS062972 from the NINDS (ASM), and NS073748 from the NIH/NINDS (AA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum Mol Genet. 2006;15:259–271. doi: 10.1093/hmg/ddi442. [DOI] [PubMed] [Google Scholar]
- Antonellis A, Dennis MY, Burzynski G, Huynh J, Maduro V, Hodonsky CJ, Khajavi M, Szigeti K, Mukkamala S, Bessling SL, Pavan WJ, McCallion AS, Lupski JR, Green ED. A rare myelin protein zero (MPZ) variant alters enhancer activity in vitro and in vivo. PLoS One. 2010;5:e14346. doi: 10.1371/journal.pone.0014346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Huynh JL, Lee-Lin SQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, Pavan WJ. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N, Girard M, Pingault V, Lemort N, Dubourg O, Goossens M. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum Mol Genet. 2001;10:2783–2795. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Buchman VL, Luke C, Borthwick EB, Gout I, Ninkina N. Organization of the mouse Ruk locus and expression of isoforms in mouse tissues. Gene. 2002;295:13–17. doi: 10.1016/s0378-1119(02)00821-1. [DOI] [PubMed] [Google Scholar]
- Elnitski L, Riemer C, Schwartz S, Hardison R, Miller W. PipMaker: a World Wide Web server for genomic sequence alignments. Curr Protoc Bioinformatics. 2003;Chapter 10(Unit 10):12. doi: 10.1002/0471250953.bi1002s00. [DOI] [PubMed] [Google Scholar]
- Finzsch M, Schreiner S, Kichko T, Reeh P, Tamm ER, Bosl MR, Meijer D, Wegner M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J Cell Biol. 2010;189:701–712. doi: 10.1083/jcb.200912142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006a;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006b;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Ghislain J, Charnay P. Control of myelination in Schwann cells: a Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006;7:52–58. doi: 10.1038/sj.embor.7400573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout I, Middleton G, Adu J, Ninkina NN, Drobot LB, Filonenko V, Matsuka G, Davies AM, Waterfield M, Buchman VL. Negative regulation of PI 3-kinase by Ruk, a novel adaptor protein. EMBO J. 2000;19:4015–4025. doi: 10.1093/emboj/19.15.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Constantini FEL. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1986. [Google Scholar]
- Inoue K, Khajavi M, Ohyama T, Hirabayashi S, Wilson J, Reggin JD, Mancias P, Butler IJ, Wilkinson MF, Wegner M, Lupski JR. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- Inoue K, Tanabe Y, Lupski JR. Myelin deficiencies in both the central and the peripheral nervous systems associated with a SOX10 mutation. Ann Neurol. 1999;46:313–318. doi: 10.1002/1531-8249(199909)46:3<313::aid-ana6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Jaegle M, Ghazvini M, Mandemakers W, Piirsoo M, Driegen S, Levavasseur F, Raghoenath S, Grosveld F, Meijer D. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 2003;17:1380–1391. doi: 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SW, LeBlanc SE, Roopra A, Wrabetz L, Svaren J. In vivo detection of Egr2 binding to target genes during peripheral nerve myelination. J Neurochem. 2006;98:1678–1687. doi: 10.1111/j.1471-4159.2006.04069.x. [DOI] [PubMed] [Google Scholar]
- Jang SW, Svaren J. Induction of myelin protein zero by early growth response 2 through upstream and intragenic elements. J Biol Chem. 2009;284:20111–20120. doi: 10.1074/jbc.M109.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EA, Jang SW, Mager GM, Chang LW, Srinivasan R, Gokey NG, Ward RM, Nagarajan R, Svaren J. Interactions of Sox10 and Egr2 in myelin gene regulation. Neuron Glia Biol. 2007;3:377–387. doi: 10.1017/S1740925X08000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EA, Lopez-Anido C, Srinivasan R, Krueger C, Chang LW, Nagarajan R, Svaren J. Regulation of the PMP22 Gene through an Intronic Enhancer. J Neurosci. 2011;31:4242–4250. doi: 10.1523/JNEUROSCI.5893-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Kim S, Chung AY, Kim HT, So JH, Ryu J, Park HC, Kim CH. Visualization of myelination in GFP-transgenic zebrafish. Dev Dyn. 2010;239:592–597. doi: 10.1002/dvdy.22166. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc SE, Jang SW, Ward RM, Wrabetz L, Svaren J. Direct regulation of myelin protein zero expression by the Egr2 transactivator. J Biol Chem. 2006;281:5453–5460. doi: 10.1074/jbc.M512159200. [DOI] [PubMed] [Google Scholar]
- Lee KE, Nam S, Cho EA, Seong I, Limb JK, Lee S, Kim J. Identification of direct regulatory targets of the transcription factor Sox10 based on function and conservation. BMC Genomics. 2008;9:408. doi: 10.1186/1471-2164-9-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, Nix R, Arana N, Jacobs J, Talbot WS. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15:513–524. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayevska O, Shuvayeva H, Igumentseva N, Havrylov S, Basaraba O, Bobak Y, Barska M, Volod'ko N, Baranska J, Buchman V, Drobot L. Expression of adaptor protein Ruk/CIN85 isoforms in cell lines of various tissue origins and human melanoma. Exp Oncol. 2006;28:275–281. [PubMed] [Google Scholar]
- Mollaaghababa R, Pavan WJ. The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene. 2003;22:3024–3034. doi: 10.1038/sj.onc.1206442. [DOI] [PubMed] [Google Scholar]
- Newbern J, Birchmeier C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Semin Cell Dev Biol. 2010;21:922–928. doi: 10.1016/j.semcdb.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero gene expression is regulated by the glial transcription factor Sox10. Mol Cell Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano RI, Wegner M. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res. 2000;28:3047–3055. doi: 10.1093/nar/28.16.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prehu MO, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith JC, Read AP, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- Reiprich S, Kriesch J, Schreiner S, Wegner M. Activation of Krox20 gene expression by Sox10 in myelinating Schwann cells. J Neurochem. 2010;112:744–754. doi: 10.1111/j.1471-4159.2009.06498.x. [DOI] [PubMed] [Google Scholar]
- Salazar-Grueso EF, Kim S, Kim H. Embryonic mouse spinal cord motor neuron hybrid cells. Neuroreport. 1991;2:505–508. doi: 10.1097/00001756-199109000-00002. [DOI] [PubMed] [Google Scholar]
- Senderek J, Bergmann C, Stendel C, Kirfel J, Verpoorten N, De Jonghe P, Timmerman V, Chrast R, Verheijen MH, Lemke G, Battaloglu E, Parman Y, Erdem S, Tan E, Topaloglu H, Hahn A, Muller-Felber W, Rizzuto N, Fabrizi GM, Stuhrmann M, Rudnik-Schoneborn S, Zuchner S, Michael Schroder J, Buchheim E, Straub V, Klepper J, Huehne K, Rautenstrauss B, Buttner R, Nelis E, Zerres K. Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot-Marie-Tooth type 4C neuropathy. Am J Hum Genet. 2003;73:1106–1119. doi: 10.1086/379525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Wegner M. SoxE function in vertebrate nervous system development. Int J Biochem Cell Biol. 2010;42:437–440. doi: 10.1016/j.biocel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56:1541–1551. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Take H, Watanabe S, Takeda K, Yu ZX, Iwata N, Kajigaya S. Cloning and characterization of a novel adaptor protein, CIN85, that interacts with c-Cbl. Biochem Biophys Res Commun. 2000;268:321–328. doi: 10.1006/bbrc.2000.2147. [DOI] [PubMed] [Google Scholar]
- Toda K, Small JA, Goda S, Quarles RH. Biochemical and cellular properties of three immortalized Schwann cell lines expressing different levels of the myelin-associated glycoprotein. J Neurochem. 1994;63:1646–1657. doi: 10.1046/j.1471-4159.1994.63051646.x. [DOI] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Werner T, Hammer A, Wahlbuhl M, Bosl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) SOX10 protein is expressed in S16 cells and sciatic nerve. Western blot analyses were performed with total protein lysates from S16 and MN-1 cells, and mouse sciatic nerve (mSN) using an anti-SOX10 or anti-actin antibody, as indicated. (B) SH3KBP1 mRNA-3 is not expressed in oligodendrocytes. RT-PCR was performed on cDNA generated from human cultured oligodendrocytes (Oligo) and human cardiac myocytes (Heart) to test for the presence of SH3KBP1 transcripts harboring exon 1C. Cardiac myocytes were analyzed as a positive control for human SH3KBP1 exon 1C. Negative controls without cDNA (‘Blank’) were included for each reaction and primers for β-ACTIN mRNA were used as a positive control. Base pair (b.p.) sizes of markers are provided on the left.
Sh3kbp1 P3 directs tissue-specific expression in mouse embryos. Mouse pronuclear injections were performed using a construct harboring Sh3kbp1 P3 directing a LacZ reporter gene, and embryos were analyzed at embryonic day 11.5 (E11.5; A–D). LacZ expression was observed in the limb buds (A; black arrow, n=14), caudal region (A; inset, black arrowhead, n=9), forebrain (B, n=2), hindbrain (C, n=4), rhombic lip (D; black arrow, n=11), and endolymphatic diverticular appendage of the otic vesicle (D; black arrowhead, n=19).
Co-localization of EGFP and MBP along the motor nerves of Sh3kbp1 P3-EGFP zebrafish embryos. Sh3kbp1 P3-EGFP stable transgenic zebrafish embryos (72 hpf) were stained with antibodies against EGFP (A) and MBP (B). Arrows indicate motor nerves, and the merged image (C) indicates co-localization of the EGFP and MBP signals.


