Abstract
Many anticancer agents damage DNA and arrest cell cycle progression primarily in S or G2 phase of the cell cycle. Previous studies with the topoisomerase I inhibitor SN38 have demonstrated the efficacy of the Chk1 inhibitor UCN-01 to overcome this arrest and induce mitotic catastrophe. UCN-01 was limited in clinical trials by unfavorable pharmacokinetics. SCH900776 is a novel and more selective Chk1 inhibitor that potently inhibits Chk1 and abrogates cell cycle arrest induced by SN38. Like UCN-01, abrogation of SN38-induced arrest enhances the rate of cell death but does not increase overall cell death. In contrast, SCH900776 reduced the growth-inhibitory concentration of hydroxyurea by 20-70 fold. A similar magnitude of sensitization was observed with cytarabine, a 5-10 fold sensitization occurred with gemcitabine, but no sensitization occurred with cisplatin, 5-fluorouracil or 6-thioguanine. Sensitization occurred at hydroxyurea concentrations that marginally slowed DNA replication without apparent activation of Chk1, but this led to dependence on Chk1 that increased with time. For example, when added 18 h after hydroxyurea, SCH900776 induced DNA double-strand breaks consistent with rapid collapse of replication forks. In addition, some cell lines were highly sensitive to SCH900776 alone, and these cells required lower concentrations of SCH900776 to sensitize them to hydroxyurea. We conclude that some tumors may be very sensitive to the combination of SCH900776 and hydroxyurea. Delayed administration of SCH900776 may be more effective than concurrent treatment. SCH900776 is currently in Phase I clinical trials, and these results provide the rationale and schedule for future clinical trials.
Introduction
Many anticancer drugs target DNA resulting in activation of cell cycle checkpoints, arrest of proliferation, and repair, the unfortunate consequence of which is recovery and cell survival. Current efforts to enhance tumor cell killing include combining anticancer agents with inhibitors of DNA checkpoints. Chk1 has been identified as a critical kinase for cell cycle arrest and many inhibitors are currently in preclinical and clinical development (1). The first Chk1 inhibitor to enter clinical trials was 7-hydroxystaurosporine (UCN-01) (2). We initially discovered that UCN-01 was a potent inhibitor of S and G2 arrest induced by cisplatin (3), and subsequently, that it abrogated arrest induced by the topoisomerase I inhibitor SN38 (the active metabolite of irinotecan) (4). The abrogation of arrest occurred preferentially in p53-defective cells suggesting that the enhanced cell killing might be selective for tumors (5,6). Clinical trials with UCN-01 were disappointing because UCN-01 binds avidly to alpha-1 acid glycoprotein in patient plasma which made it difficult to control the concentration of bioavailable inhibitor (7,8). As UCN-01 also inhibits many other kinases, this made it difficult to achieve only the low bioavailable concentration that was relatively selective for Chk1. SCH900776 was developed as a much more selective inhibitor of Chk1 (9). Here, we compare the activity of UCN-01 and SCH900776 in combination with a variety of DNA damaging agents (structures are available in Supplementary Figure 1).
Anticancer agents induce a variety of DNA lesions which elicit cell cycle arrest. γ-Radiation induces DNA double-strand breaks at all phases of the cells cycle whereas topoisomerase I inhibitors form double-strand breaks only in S phase when the replication complex collides with an inhibited topoisomerase (10). Cisplatin causes DNA inter- and intra-strand crosslinks that primarily block replication fork progression (11,12). Many antimetabolites such as cytarabine and gemcitabine inhibit synthesis of DNA by inhibiting either DNA polymerase or ribonucleotide reductase, respectively, but they are also incorporated into DNA where they terminate strand synthesis (13). Hydroxyurea also inhibits ribonucleotide reductase but is not incorporated into DNA. It functions solely by limiting synthesis of deoxyribonucleotides such that replication slows or stops. The stalled replication forks are stabilized by Chk1 such that inhibition of Chk1 leads to collapse of the replication fork and DNA double-strand breaks (14). Furthermore, Chk1 is essential for survival of cells incubated with hydroxyurea (15). For most DNA damaging agents, cell cycle arrest occurs rapidly as a consequence of activation of Chk1. However, hydroxyurea differs in that cell cycle progression is inhibited directly by the lack of DNA precursors and checkpoint activation is not required for the arrest.
Here, we show dramatic sensitization when SCH900776 is combined with concentrations of hydroxyurea that alone cause only slight slowing of DNA synthesis and little if any activation of Chk1. We also demonstrate that some cell lines are highly sensitive to SCH900776 alone. The results suggest that some tumors may be highly and selectively sensitive to the combination of hydroxyurea and SCH900776.
Materials and Methods
Drugs were obtained from the following sources: SN38 (7-ethyl-10-hydroxycampothecin), Pfizer, Kalamazoo, MI; cisplatin, Bristol Myers Squibb, NJ; gemcitabine, Eli Lilly, Indianapolis, IN; hydroxyurea, 5-fluorouracil and cytarabine, Sigma Chemical Co. St Louis, MO; UCN-01, National Cancer Institute, Bethesda, MD; KU55933, Tocris Biosciences, Ellisville, MO. SCH900776 was provided by Merck, Kenilworth, NJ. The 2-arylbenzimadazole “2h” is a selective Chk2 inhibitor and was synthesized according to the published method (16). The concentration of SN38 used in most experiments was 10 ng/ml which is equivalent to 25.5 nmol/L.
The origin and maintenance of MDA-MB-231, MCF10A and U2OS cells and their derivatives, MDA-MB-231ΔChk1 cells and MCF10AΔp53 have been described previously (6,17). The latter two cell lines were derived by stable expression of an appropriate shRNA. All other cell lines were obtained from the Developmental Therapeutics Program, National Cancer Institute, Bethesda and maintained in RPMI1640 medium plus serum and antibiotics (18). Cells were harvested and analyzed by immunoblotting as previously detailed (6) with the following additional antibodies: phosphoserine-1981-ATM (Epitomics), phosphoserine-2056-DNA-PK (Abcam) and γH2AX (Cell Signaling).
Cell cycle analysis was performed by flow cytometry as described previously (19). DNA synthesis was assessed by incubation of cells with 10 μmol/L 5-ethynyl-2′-deoxyuridine (EdU) for 30 min and staining according to the manufacturer’s recommendation (Invitrogen) although the recipe was modified to use 20% of the recommended reactants.
For cell growth assays, cells were seeded at low density (500-1000 cells) in 96-well plates and then incubated with drug for 24 h (8 wells per concentration). Following treatment, cells were washed and grown in fresh media for 5-7 days at 37°C. Prior to attaining confluence, cells were washed, lysed, and stained with Hoechst 33258, as previously described (20). Fluorescence was read on a microplate spectrofluorometer (Spectramax M2). Results are expressed as mean and standard error for the concentration of drug that inhibited growth by 50% (IC50) .
Results
The combination of UCN-01 and SN38
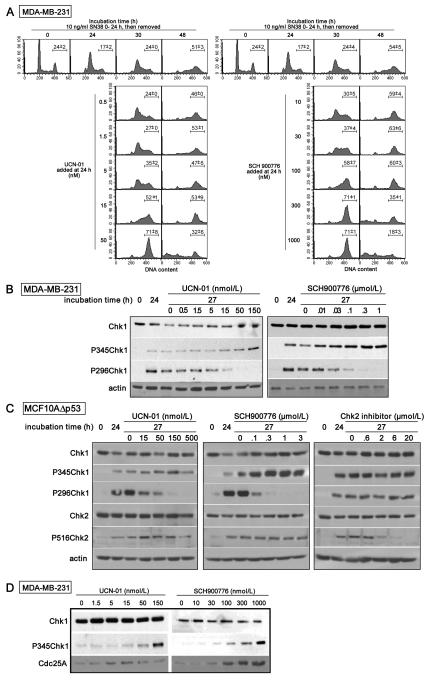
In many cell lines, the topoisomerase I inhibitor SN38 induces a G2 arrest at low concentrations, but late, middle and early S phase at higher concentrations (4,21). Concentrations that inhibit cell cycle progression induce phosphorylation of Chk1. Subsequent incubation with UCN-01 inhibits Chk1, thereby abrogating both S and G2 arrest and driving the cells through a lethal mitosis. An example of these results is shown in Fig. 1A in which MDA-MB-231 cells were incubated with SN38 at a concentration that arrests the cells in early S phase after 24 h. Subsequent incubation with 50 nmol/L UCN-01 results in complete abrogation of S phase arrest within the following 6 h, and abrogation of G2 arrest in many of the cells within 24 h. At these later times, the amount of DNA in G2 appears to increase (values above 400) but this has previously been attributed to decondensation of damaged chromatin which binds more propidium iodide and is not due to further DNA synthesis (4,22). While flow cytometry can not resolve G2 phase from M phase, we previously demonstrated that the cells in G2/M were still positive for geminin, a marker of S and G2, and hence were not arrested in mitosis (17).
Fig. 1.
Comparative efficacy of UCN-01 and SCH900776 at inhibiting Chk1 and abrogating SN38-induced cell cycle arrest. A. MDA-MB-231 cells were incubated with 10 ng/mL SN38 for 24 h. The drug was then replaced with either 0-50 nmol/L UCN-01 (left) or 0-1000 nmol/L SCH900776 (right) for an additional 6 or 24 h. Cells were harvested at the indicated times and analyzed for DNA content by flow cytometry. The experiment with UCN-01 is a replicate of previous publications (4,21), while results with SCH900776 are representative of at least three experiments. The bar reflects the mean and range (n=2) for the percentage of cells in the G2/M phase. B. MDA-MB-231 cells were incubated with 10 ng/mL SN38 for 24 h. The drug was then replaced with either UCN-01 (left) or SCH900776 (right) and the cells harvested after an additional 3 h. Cell lysates were analyzed for Chk1 phosphorylation. C. MCF10AΔp53 cells were incubated as in B, and additionally with the Chk2 inhibitor “2h.” Cell lysates were analyzed by western blotting for the indicated proteins. D. MDA-MB-231 cells were incubated with UCN-01 or SCH900776 for 3 h, then harvested and analyzed for Chk1 phosphorylation and Cdc25A .
Chk1 is phosphorylated at ser345 by ATR, while phosphorylation at ser296 is a consequence of autophosphorylation such that incubation of damaged cells with UCN-01 inhibits only the latter phosphorylation (23). SN38-damaged MDA-MB-231 cells were incubated for 3 h with 0-150 nmol/L UCN-01 to determine the concentration that inhibits Chk1. Significant reduction in ser296 phosphorylation was observed at and above 15 nmol/L while phosphorylation at ser345 actually increased (Fig. 1B).
Chk2 is also activated as a consequence of DNA damage and phospho-ser516 reflects autophosphorylation. Activation of Chk2 is difficult to study in MDA-MB-231 cells because of a low level of this protein. Accordingly, we performed parallel studies in MCF10AΔp53 cells [the shRNA-mediated suppression of p53 prevents damage-mediated downregulation of Chk1 (6)]. Phosphorylation of Chk1 on ser296 is again inhibited by 15 nmol/L UCN-01, but there is no inhibition of Chk2 phosphorylation on ser516 demonstrating that UCN-01 does not inhibit Chk2 at concentrations as high as 500 nmol/L (Fig. 1C). As a control, we also incubated cells with the Chk2 inhibitor “2h” which shows almost complete inhibition of ser516-Chk2 at 6 μmol/L but has no impact on phospho-Chk1.
Comparison of UCN-01 and SCH900776 in combination with SN38
The above experiments provide a prelude for comparison to the novel Chk1 inhibitor SCH900776. In parallel experiments, 300 nmol/L SCH900776 is as effective as 50 nmol/L UCN-01 in abrogating both S and G2 arrest (Fig. 1A). These concentrations were also effective at inhibiting phosphorylation at ser296-Chk1 (Fig. 1B,C). SCH900776 did not inhibit phosphorylation of ser516-Chk2 consistent with its selectivity for Chk1 over Chk2 (9). It is also worth noting that SCH900776 alone had no impact on cell cycle progression as assessed by flow cytomtery at the concentrations studied here (data not shown).
Incubation with either UCN-01 or SCH900776 at the higher concentrations enhanced the SN38-induced phosphorylation of ser345-Chk1. We therefore compared the effect of each inhibitor in the absence of exogenous DNA damage. A 3-h incubation resulted in strong phosphorylation of Chk1 by 100 nmol/L SCH900776 and 150 nmol/L UCN-01 (Fig. 1D). There appears to be no genotoxicity associated with this Chk1 phosphorylation as, unlike with DNA damage, this phosphorylation is reversed within 1 h upon removal of UCN-01 or SCH900776 (data not shown). This is consistent with the report that Chk1 induces feed-back dephosphorylation and inactivation of ATR by protein phosphatase 2A; when Chk1 is inhibited, ATR becomes active and phosphorylates ser345-Chk1 (24). We also investigated the impact of Chk1 inhibition on one of its immediate downstream targets, CDC25A, which is targeted for degradation by Chk1 (25). An increase in CDC25A was observed with both Chk1 inhibitors, although higher concentrations of UCN-01 appeared to be inhibitory, whereas a far more marked increase occurred with SCH9000776 that correlated closely with the phosphorylation of ser345-Chk1 (Fig. 1D).
Comparison of UCN-01 and SCH900776 in cells suppressed for Chk1
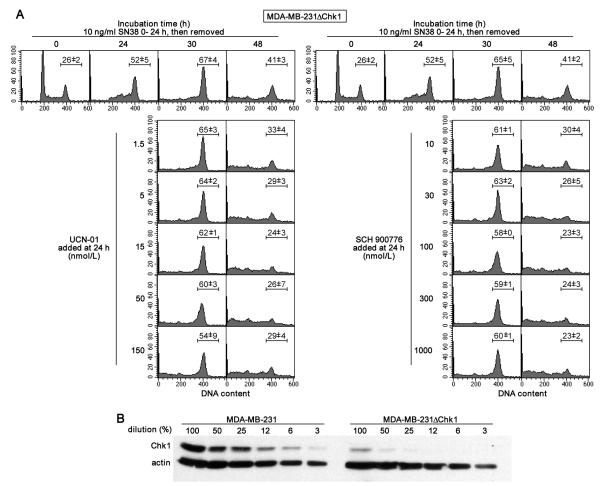
We previously introduced shRNA into MDA-MB-231 cells to suppress Chk1; these cells are termed MDA-MB-231ΔChk1 (17). These cells showed a high constitutive level of CDC25A and little arrest in S phase compared to their parent cell line when incubated with SN38 confirming the critical role of Chk1 in DNA damage-induced S phase arrest. However, the cells arrested in G2 rather than S phase, a phenomenon that was also observed when Chk1-competent parent cells were incubated concurrently with SN38 plus UCN-01; that the cells were arrested in G2 rather than mitosis was confirmed using geminin (17). At that time, we proposed that the G2 arrest might be due to an alternate checkpoint pathway. Other authors have proposed that MAPKAPK2 may be involved in checkpoint regulation although we previously found no evidence to support this claim (17,26). To determine whether UCN-01, but not SCH900776, might be impacting another undefined checkpoint pathway, we compared their efficacy in the MDA-MB-231ΔChk1 cells. After a 24 h incubation, SN38 alone induced arrest primarily in G2, with the remaining cells accumulating in G2 within the following 6 h (Fig. 2). Neither UCN-01 nor SCH900776 had any further impact during this 6 h period. During the 24 h period after removal of SN38, many of the MDA-MB-231ΔChk1 cells appeared to overcome the G2 arrest as reflected in cells with either G1 or sub-G1 DNA content; these cells also exhibited multiple micronuclei commonly associated with mitotic catastrophe (21). Most of the residual G2 arrest was abrogated by both Chk1 inhibitors suggesting that the presence of residual Chk1 rather than another kinase is responsible for the persistent G2 arrest.
Fig. 2.
Comparative efficacy of UCN-01 and SCH900776 at abrogating SN38-induced cell cycle arrest in cells suppressed for Chk1. A. MDA-MB-231ΔChk1 cells were incubated with 10 ng/mL SN38 for 24 h. The drug was then replaced with either 0-150 nmol/L UCN-01 (left) or 0-1000 nmol/L SCH900776 (right) for an additional 6 or 24 h. Cells were harvested at the indicated times and analyzed for DNA content by flow cytometry. The experiment with UCN-01 is a replicate from a previous publication (17), while results with SCH900776 are representative of at least three experiments. The bar reflects the mean and range (n=2) for the percentage of cells in the G2/M phase. B. The level of Chk1 in the ΔChk1 cells was assessed by loading 15 μg of protein in the first lane (100%) followed by serial 2-fold dilutions. Comparison was made to the levels of Chk1 in the parent cell line.
On further analysis of the Chk1 expression levels, we found that there was indeed a very low level of Chk1 present. By comparing to a dilution series of the parent cells, we conclude that the MDA-MB-231ΔChk1 cells have 5-10% of the parent level (Fig. 2B). Importantly, such a low level of Chk1 appears unable to induce an S phase arrest in damaged cells but can still contribute to G2 arrest. The low level of Chk1 in these cells will be important in some of the experiments discussed below.
Impact of UCN-01 and SCH900776 on SN38-mediated cytotoxicity
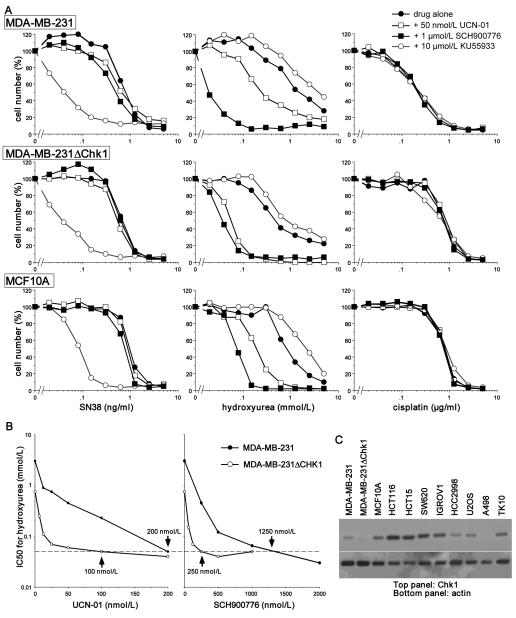
We have consistently observed that UCN-01 can enhance cell death when added 24 h after cisplatin or SN38, but most of the previous experiments were performed over the subsequent 24 h similar to those shown in Figs. 1 and 2 (4,27). However, when growth was followed for 5-7 days after removal of drug there was no sensitization by UCN-01 as reflected in the IC50 (27). We conclude that incubation of arrested cells with UCN-01 accelerates the rate of cell death but does not significantly enhance the overall extent of death. We have extended those studies by investigating the impact of concurrent incubation with SN38 and Chk1 inhibitors. We anticipated that the combination would show enhanced toxicity because cells would be unable to arrest proliferation, thereby continuing to replicate their DNA in the presence of SN38. Surprisingly, neither Chk1 inhibitor enhanced growth inhibition following a 24 h concurrent incubation with SN38 as measured over the following 6 days (Fig. 3, left). In contrast, concurrent incubation with the ATM inhibitor KU55933 caused a dramatic sensitization to SN38 with a decrease in the IC50 of about 20 fold. This result shows that it is possible to enhance the sensitivity of cells to SN38 by inhibiting an alternate checkpoint protein, but the underlying mechanism is still under investigation. Neither the Chk1 nor ATM inhibitors sensitized cells to concurrent incubation with cisplatin (Fig. 3 right). Similar results were seen in the MDA-MB-231 cells, their ΔChk1 derivative and MCF10A cells.
Fig. 3.
The impact of checkpoint inhibitors on sensitivity of cells to SN38, hydroxyurea and cisplatin. A. MDA-MB-231 (top), MDA-MB-231ΔChk1 (middle) and MCF10A cells (bottom) were incubated with SN38 (left), hydroxyurea (middle) or cisplatin (right) for 24 h either alone or in combination with 50 nmol/L UCN-01, 1 μmol/L SCH900776 or 10 μmol/L KU55933. The drugs were replaced with fresh media and analyzed after an additional 5-7 days for cell growth. B. MDA-MB-231 and MDA-MB-231ΔChk1 cells were incubated with hydroxyurea for 24 h in combination with a range of concentrations of UCN-01 (left) or SCH900776 (right). Cell growth was assessed after 6 days and are recorded as the 50% inhibitory concentration (IC50) of hydroxyurea at each concentration of the Chk1 inhibitors. Values are highlighted for the concentration of each Chk1 inhibitor that reduces the IC50 for hydroxyurea to 50 μmol/L (the dotted line). C. Lysates were made from the indicated cell lines and analyzed for expression of Chk1.
Comparison of UCN-01 and SCH900776 in combination with hydroxyurea
Recent experiments have established that Chk1 is essential for viability of cells when their replication fork is arrested by incubation with the ribonucleotide reductase inhibitor hydroxyurea (15). We therefore compared the impact of UCN-01 and SCH900776 when used in combination with hydroxyurea. For these experiments, MDA-MB-231 cells were incubated concurrently with the Chk1 inhibitors and hydroxyurea for 24 h, then drug-free media was added for another 6 days. SCH900776 (1 μmol/L) caused a 30-fold decrease in the IC50 for hydroxyurea. UCN-01 (50 nmol/L) also sensitized the cells but only by about 5 fold, albeit as noted below, this was a sub-optimal concentration of UCN-01. A similar degree of sensitization was observed in the MCF10A cells.
To further define the relative efficacy of the two inhibitors, additional growth curves were performed in MDA-MB-231 cells to determine the IC50 for hydroxyurea when combined with a range of concentrations of each Chk1 inhibitor. Both Chk1 inhibitors reduced the IC50 for hydroxyurea by about 60 fold (from 3 mmol/L to 50 μmol/L); this required 200 nmol/L UCN-01 or 1.25 μmol/L SCH900776 (Fig. 3B). The MDA-MB-231ΔChk1 cells were about 6-fold more sensitive to hydroxyurea alone, while concurrent addition of the Chk1 inhibitors produced another 12-fold sensitization thereby resulting in the same overall sensitivity as seen in the parent cell lines. The concentrations of each Chk1 inhibitor required to attain maximum sensitization in the ΔChk1 cells were 2.5 - 5-fold less than in the parent cells; 100 nmol/L UCN-01 and 250 nmol/L SCH900776. This result is consistent with their being very little residual Chk1 to inhibit, but also suggests that >95% inhibition of Chk1 is required to obtain maximum sensitization.
We analyzed a panel of cell lines, focusing on the sensitization achieved by SCH900776. Based on the results in Fig. 3B, we assessed the IC50 for hydroxyurea when combined with either 200 nmol/L or 2 μmol/L SCH900776. While the various cell lines showed different initial sensitivity, the IC50 was reduced to 70 μmol/L or lower in all but one case following incubation with 2 μmol/L SCH900776 (Table 1). In some cell lines, this extent of sensitization was achieved with 200 nmol/L SCH900776. We questioned whether this difference in sensitivity related to the level of Chk1 in the cells as might be predicted from the data in Fig. 3B. There was a large range of expression of Chk1 across the cell lines but both sensitive and resistant cells were seen to have relatively low Chk1 (Fig. 3C). A second possible explanation for the difference in response of the cells could be that Chk2 provides a backup checkpoint for cells with low or inhibited Chk1. However, HCT15 cells are defective for Chk2 yet were relatively resistant. Hence it seems unlikely that the basal level of either Chk1 or Chk2 predicts response of the cells to combination of hydroxyurea plus SCH900776. We suggest this sensitivity depends on either differences in events downstream of Chk1 or to defects in a complementary pathway (see Discussion).
Table 1.
Sensitivity of cell lines to hydroxyurea and Chk1 inhibitors alone or in combination
| CELL LINE | IC50 HYDROXYUREA (mmol/L) | IC50 CHK1 INHIBITOR | |||
|---|---|---|---|---|---|
| control | +200 nmol/L 900776 |
+2 μmol/L 900776 |
900776 (μmol/L) |
UCN-01 (μmol/L) |
|
| MDA-MB-231 | 1.6 ± 0.42 | 0.4 ± 0.06 | 0.045 ± 0.006 | >10 ± 0 | 0.4 ± 0.13 |
| 231ΔChk1 | 0.36 ± 0.05 | 0.03 ± 0.01 | 0.033 ± 0.003 | 8.8 ± 1.3 | >2 ± 0 |
| MCF10A | 0.8 ± 0.31 | 0.18 ± 0.11 | 0.045 ± 0.018 | >10 ± 0 | 1.3 ± 0.75 |
| HCT116 | 2.1 ± 0.3 | 1.8 ± 0.73 | 0.067 ± 0.012 | >10 ± 0 | 1.4 ± 0.13 |
| HCT15 | 4.3 ± 0.33 | 1.8 ± 0.44 | 0.117 ± 0.044 | >10 ± 0 | 1.8 ± 0.25 |
| SW620 | 3.1 ± 1.1 | 0.7 ± 0.25 | 0.070 ± 0.02 | >10 ± 0 | 1.3 ± 0.75 |
| IGROV1 | 2.2 ± 1.3 | 0.08 ± 0.012 | 0.03 ± 0 | 6 ± 4 | 0.55 ± 0.05 |
| HCC2998 | 1.1 ± 0.32 | 0.1 ± 0.003 | 0.043 ± 0.019 | 0.5 ± 0 | 0.63 ± 0.26 |
| U2OS | 2.1 ± 0.32 | 0.06 ± 0.01 | DEAD | 0.55 ± 0.05 | 0.1 ± 0 |
| A498 | 4.3 ± 0.7 | 0.09 ± 0.007 | DEAD | 0.5 ± 0 | 0.1 ± 0 |
| TK-10 | 0.5 ± 0 | 0.04 ± 0.007 | DEAD | 0.23 ± 0.03 | 0.1 ± 0.01 |
DEAD = few residual cells on SCH900776 alone so no cumulative cytotoxicity calculable Values represent mean ± standard error of the mean (columns 2-4, n≥3; columns 5, 6 n=2)
Sensitivity of cells to UCN-01 and SCH900776 alone
Some cell lines were very sensitive to SCH900776 alone. Most of the cell lines tolerated incubation with 10 μmol/L SCH900776 alone for 24 h, but four of the lines were inhibited by incubation in 0.5 μmol/L for 24 h (Table 1). These four cell lines (A498, U2OS, TK-10 and HCC2998) were also highly sensitive to hydroxyurea when combined with low concentrations of SCH900776. Accordingly, we anticipate that the mechanism of sensitivity to SCH900776 alone also impacts the sensitivity of cells to the combination.
Concurrently, we compared the sensitivity of these cell lines to UCN-01 alone (Table 1). While the cell lines sensitive to SCH900776 were generally more sensitive to UCN-01, there were exceptions, and the degree of difference between cell lines was much less striking. This is attributed to the fact that UCN-01 inhibits many additional kinases whose inhibition likely contributes to the observed cytotoxicity. These results are consistent with the much greater selectivity of SCH900776 for Chk1 (9).
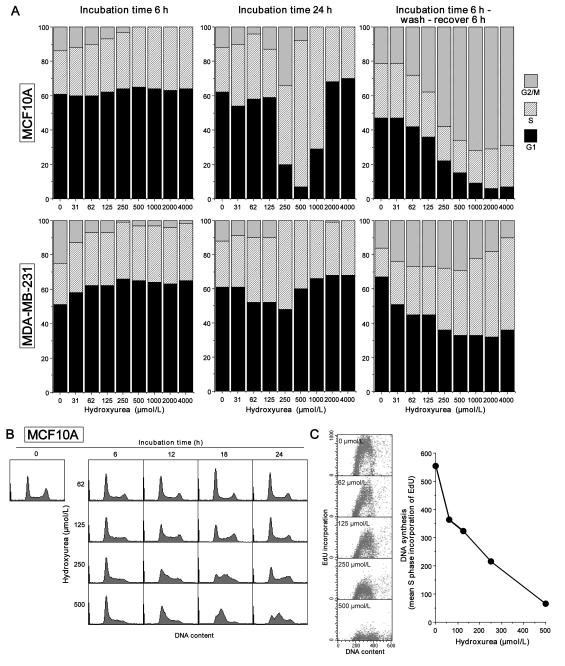
Cell cycle perturbation induced by hydroxyurea
It is intriguing that Chk1 inhibition can enhance sensitivity of cells to concentrations of hydroxyurea that alone did not inhibit growth. However, the data in Fig. 3 and Table 1 were obtained 6-7 days after a 24 h incubation with hydroxyurea. We therefore investigated the cell cycle perturbation and checkpoint activation during the initial 24 h incubation. Incubation of MCF10A cells with 2 mmol/L hydroxyurea results in an almost immediate arrest of DNA synthesis; after both 6 and 24 h, the only change in cell cycle distribution is the loss of the G2/M population which will not be perturbed by hydroxyurea (Fig. 4A). At 0.5–1 mmol/L hydroxyurea, loss of the G2/M population is also observed by 6 h consistent with inhibition of DNA synthesis. However, by 24 h, there is a marked increase in the percentage of cells in S (>80%) and between 12 and 24 h, a peak of cells can be seen slowly progressing through S (Fig. 4B). At the lowest concentrations of hydroxyurea (62–125 μmol/L) that are sensitized by SCH900776, there is a slight decrease in the G2/M population at 6 h, but there is no dramatic change in the cell cycle by 24 h.
Fig. 4.
Analysis of cell cycle perturbation induced by hydroxyurea. A. MCF10A and MDA-MB-231 cells were incubated with 0 – 4 mmol/L hydroxyurea for 6 h (left), 24 h (center), or were allowed to recover for 6 h after the initial 6-h incubation with hydroxyurea (right). Cells were analyzed by flow cytometry and the results analyzed by ModFit for the proportion in G1, S or G2/M phase. B. Representative examples of MCF10A cells incubated with 62-500 μmol/L hydroxyurea for 0-24 h. C. MCF10A cells were incubated for 24 h with 0 – 500 μmol/L hydroxyurea; 10 μmol/L EdU was added for the final 30 min. Cells were harvested and analyzed by 2-dimensional flow cytometry for DNA content and incorporation of EdU. The data were gated to calculate the mean incoporation of EdU into S phase cells.
To further visualize the impact of hydroxyurea during the first 6 h, we removed the drug and allowed the cells to recover for an additional 6 h (Fig. 4A). There was a dose-dependent decrease in G1 cells with an increase in the proportion of cells in G2/M above 62 μmol/L demonstrating that the brief incubation with even low concentrations of hydroxyurea slowed the rate of DNA synthesis and partially synchronized the cells.
Similar conclusions can be drawn from experiments in MDA-MB-231 cells (Fig. 4A). A decrease in G2/M was observed at both 6 and 24 h, although the increase in S phase after 24 h at the intermediate concentrations was much less dramatic than in the MCF10A cells. Release from hydroxyurea after 6 h resulted in a decrease in G1 cells over the following 6 h, although many of the cells were still in S phase at the time of analysis. The conclusion from both cell lines is that low concentrations of hydroxyurea can partially synchronize the cells and that the arrest is rapidly reversible upon removal of the drug.
These experiments were complemented by analysis of the incorporation of the nucleoside analog EdU (Fig. 4C). After a 24 h incubation of MCF10A cells with hydroxyurea, DNA synthesis was inhibited by about 30% at 62 μmol/L and >90% at 500 μmol/L. These results further demonstrate that at low concentrations of hydroxyurea DNA replication is slowed, and this stress is presumably sufficient to make the cells sensitive to SCH900776.
Activation of Chk1 by hydroxyurea
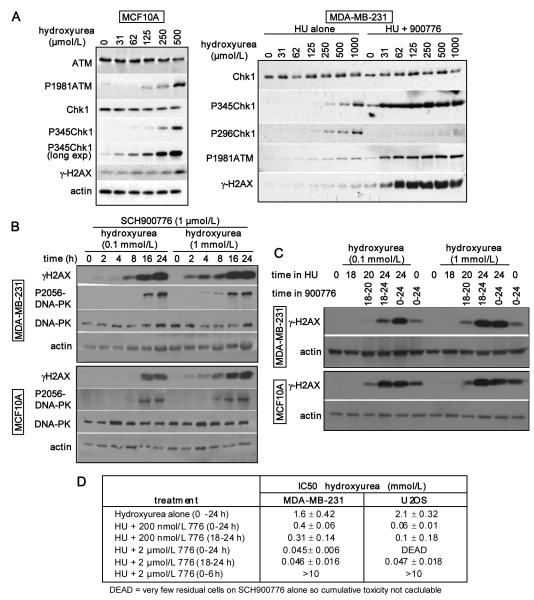
One question raised by these observations is whether Chk1 is activated at the low concentrations at which replication is only slowed slightly. Cell lysates were analyzed after a 24 h incubation of MCF10A cells with 0-500 μmol/L hydroxyurea (Fig. 5A). Phosphoserine-345-Chk1 was readily observed at 250 and 500 μmol/L hydroxyurea, a concentration at which the majority of cells were S phase arrested, but a longer exposure of the film was required to see the marginal phosphorylation at the lower concentrations. Phospho-ATM was also observed only at the higher concentrations of hydroxyurea while minimal γ-H2AX was observed.
Fig. 5.
Impact of concentration and schedule of hydroxyurea and SCH900776 on DNA damage and cytotoxicity A. MCF10A (left) and MDA-MB-231 cells (right) were incubated with hydroxyurea for 24 h and lysates analyzed for the indicated proteins. MDA-MB-231 cells were also coincubated with 1 μmol/L SCH900776. B. MDA-MB-231 and MCF10A cells were incubated with 0.1 or 1 mmol/L hydroxyurea in combination with 1 μmol/L SCH900776 for 0-24 h. C. MDA-MB-231 and MCF10A cells were incubated with 0.1 or 1 mmol/L hydroxyurea for 0 – 24 h. 1 μmol/L SCH900776 was added as indicated at time 0 or after 18 h. In the latter case, the cells were harvested after an additional 2 h (20 h) or 6 h (24 h). D. MDA-MB-231 and U2OS cells were incubated with hydroxyurea and SCH900776 at the indicated conditions and schedules. Drugs were removed and growth inhibition (IC50) assessed over the following 6 days.
A similar experiment in MDA-MB-231 cells showed little activation of Chk1 or ATM at the lower concentrations of hydroxuyrea, and almost no detectable γ-H2AX (Fig. 5A). However, when hydroxyurea was co-incubated with SCH900776, there was a dramatic increase in phosphorylation of ser345-Chk1, ATM and H2AX even at low hydroxyurea concentrations. Phosphorylation of H2AX is usually a reflection of DNA double-strand breaks (28) and therefore consistent with the collapse of stalled replication forks. The increase in phosphoserine-345-Chk1 is much greater than in cells incubated with SCH900776 alone and is presumably a consequence of this enhanced DNA damage. The absence of phosphoserine-296-Chk1 reiterates that SCH900776 is inhibiting Chk1.
Schedule dependence of sensitization to hydroxyurea by SCH900776
Considering that SCH900776 dramatically increased the DNA damage induced by hydroxyurea, we assessed the kinetics of appearance of γ-H2AX. In cells incubated with SCH900776 and either 0.1 or 1 mmol/L hydroxyurea, a very low level of γ-H2AX was observed at 2 – 8 h but a dramatic increase occurred at 16 h (Fig. 5B). Concurrently, we observed the induction of phospho-DNA-PK, another indicator of double-strand breaks. As the appearance of double-strand breaks reflects collapse of stalled replication forks, this suggests that the arrested forks are not immediately dependent on Chk1 for stability. To further investigate this phenomenon, we delayed the addition of SCH900776 until 18 h after addition of hydroxyurea; γ-H2AX rapidly increased and within 6 h was generally as high as if SCH900776 had been present for the entire 24 h (Fig. 5C). Similar results were obtained in both MDA-MB-231 and MCF10A cells.
Finally, we compared the sensitivity of cells to hydroxyurea when SCH900776 was added concurrently for 24 h, or only for the last 6 h. In MDA-MB-231 cells, the delayed addition of 2 μmol/L SCH900776 induced a 50-fold decrease in IC50 which was only slightly less than if the two drugs had been added concurrently (Fig. 5D). However, if the two drugs were administered concurrently for only 6 h, no cytotoxicity was observed. In U2OS cells which are sensitive to SCH900776 alone, we again found that the delayed addition was almost as effective as concurrent treatment, although this dramatic decrease in IC50 was achieved with only 200 nmol/L SCH900776. A 6 h concurrent treatment again failed to kill cells. These results suggest that cells accumulate stress over the time frame that replication is slowed by hydroxyurea and these replication forks become more dependent on Chk1 for stability with increasing time.
Combination of SCH900776 with other DNA damaging drugs
Our results show that SCH900776 potently sensitizes cells to hydroxyurea but has no impact on cells incubated with SN38 or cisplatin (Fig. 3). We therefore assessed the impact of SCH900776 in combination with other antimetabolites. A 3-10 fold sensitization was observed with gemcitabine. Specifically, in the MDA-MB-231 cells, the IC50 for gemcitabine was reduced from 10 (±0) to 4 (±1) or 0.9 (±0.38) nmol/L when combined with 200 or 2 μmol/L SCH900776 respectively; in MCF10A cells the comparable values were 7.3 (±1.3) to 5.9 (±2.2) or 3 (±1) nmol/L. These cell lines were also assessed for sensitivity to cytarabine: in MCF10A cells, the IC50 for cytarabine was reduced from 78 (±11.6) to 8.2 (±0.9) or 2.3 (±0.3) nmol/L upon addition of 200 or 2 μmol/L SCH900776, respectively; in MDA-MB-231 the equivalent values were 56 (±6.25) to 7.25 (±0.25) or 3.7 (±0.2) nmol/L, respectively. These results show 15 – 35-fold sensitization by SCH900776 which is comparable to the sensitization observed with hydroxyurea. In contrast, at concentrations of 5-fluorouracil or 6-thioguanine that reduced cell growth, no further decrease in IC50 was observed in these cells when SCH900776 was added (n=3).
Discussion
Many Chk1 inhibitors are at various stages of preclinical and clinical development in combination with a variety of anticancer DNA damaging agents (1). Chk1 is required for cell cycle arrest, recombination repair and replication fork stability in damaged cells, hence it is believed that inhibiting Chk1 will enhance cell killing induced by DNA damage. Whether this is selective for the tumor remains to be established although it has been suggested that either p53 or p38mapk may selectively protect normal cells (5,29,30). In addition, prior experiments demonstrated that human diploid fibroblasts were not sensitized to gemcitabine by SCH900776, nor were myelosuppressive effects of gemcitabine enhanced by SCH900776 in mouse models (9). The experiments reported here used both a p53-wildtype (MCF10A) and p53-defective cell line (MDA-MB-231), both of which were sensitive to antimetabolites when Chk1 was inhibited. We confirm that both UCN-01 and SCH900776 inhibit Chk1 but not Chk2 in cells. Previous experiments have variously suggested that UCN-01 does or does not inhibit Chk2 but those experiments were performed with purified protein (31-33). The current experiments took advantage of the autophosphorylation sites on each protein that permit assessment of the kinase inhibitory action in intact cells.
The results presented here are particularly significant because they show that inhibition of Chk1 causes dramatic sensitization of cells to hydroxyurea with up to 70-fold decrease in the concentration required to inhibit growth. While the assay used reflects an inability of the cells to grow over 6 days following a 24 h incubation with drugs, the few cells remaining after 6 days were usually dead as assessed by loss of refractive index. The lack of sensitization to SN38 and cisplatin might appear to contradict the expectation that Chk1 protects cells from DNA damage. Our previous studies (27) and those presented here (Fig. 1) show that UCN-01 and SCH900776 enhance cell killing induced by SN38, although termination of those experiments at 48 h could not establish whether the surviving cells would eventually die. Our current results show that most of the cells that are growth inhibited by SN38 alone but still surviving at 48 h eventually die. The arrest induced by SN38 results when the replication fork collides with an inhibited topoisomerase I which is thought to result in DNA double-strand breaks and activation of Chk1; if Chk1 is then inhibited, it is these arrested cells that are killed. There is presumably no decrease in the IC50 concentration as Chk1 is not activated at the lower concentrations of SN38. In contrast, an inhibitor of ATM did sensitize cells to low concentrations of SN38 even though ATM was not activated, perhaps reflecting inhibition of some unrecognized role for non-activated ATM.
The results with hydroxyurea are in stark contrast to what was observed with SN38. At low concentrations of hydroxyurea, replication forks slowed with little if any activation of Chk1. However, when Chk1 was inhibited, there was strong phosphorylation of ATM, DNA-PK and H2AX, all suggesting the collapse of replication forks. These observations are consistent with a well-established mechanism whereby Chk1 is required to stabilize the replication fork (14,34). A related and important observation is that the stalled replication forks do not appear to rely initially on Chk1 for stability. Removal of hydroxyurea after 6 h results in rapid recovery of DNA synthesis and no cytotoxicity. Even concurrent incubation with SCH900776 and hydroxyurea is reversible for at least 6 h. The combination did not induce γ-H2AX until 16 h, but when SCH900776 was added after 18 h, the replication forks appeared to collapse very quickly. Another recent study has also suggested that stalled replication forks evolve with time, yet the results of that study are intriguingly different from ours as they observed γ-H2AX within 2 h of addition of hydroxyurea (35). We observed almost no γ-H2AX upon incubation with hydroxyurea alone, yet the cells still evolved with time to become Chk1 dependent. While we used MDA-MB-231 and MCF10A cells, the previous study used U2OS cells which we show here are much more sensitive to SCH900776. Hence the difference between these studies may shed light on the difference in sensitivity to SCH900776.
These observations reflect two different Chk1-dependent processes. In the case of SN38, the cells arrest because of DNA damage-induced activation of Chk1. In the case of hydroxyurea, the cells arrest due to inhibition of DNA synthesis which does not directly damage DNA and does not require Chk1 activation. However, in this replication arrest, Chk1 is required to stabilize the replication fork such that inhibition of Chk1 creates new DNA damage and induces a dramatic enhancement in response to the antimetabolite. The nature of the accumulating stress that results from stalled replication forks remains to be established.
It is interesting that SCH900776 did not induce the same extent of sensitization to several other antimetabolites. Hydroxyurea solely inhibits ribonucleotide reductase thereby limiting the availability of DNA precursors. Gemcitabine also inhibits ribonucleotide reductase but is also incorporated into DNA. In contrast, cells were not sensitized to 5 fluorouracil. To assess the importance of ribonucleotide reductase, we investigated the impact of cytarabine which does not inhibit ribonucleotide reductase but inhibits DNA synthesis through incorporation into DNA. We found that SCH900776 dramatically enhanced sensitivity to cytarabine suggesting inhibition of ribonucleotide reductase is not essential. It remains to be determined why SCH900776 did not sensitize cells to 5-fluorouracil despite using concentrations that inhibited cell cycle progression (data not shown). This observation contrasts several reports that suppression of Chk1 can sensitize cells to 5-fluorouracil (36,37), but this may reflect a different experimental design. Whereas the previous authors constitutively suppressed Chk1 and incubated cells continuously in 5-fluorouracil, our experiments only involved co-incubation for 24 h followed by 5-7 day recovery. This shorter exposure time may be insufficient to kill the cells such that they were still able to recover.
One other aspect of the sensitization to hydroxyurea is worth noting. The MDA-MB-231ΔChk1 cells have about 5% of the level of Chk1 as the parent cells, yet they are only about 5-fold more sensitive to hydroxyurea. Incubating the parental cell line with 200 nmol/L SCH900776 also results in 5-fold sensitization suggesting that this concentration inhibits Chk1 by about 95%. Hence, in both cases the remaining Chk1 still provides significant protection. In the ΔChk1 derivative, 200 nmol/L SCH900776 elicits another 10-fold sensitization to hydroxyurea, while this level of sensitization is achieved in the parent cell line at 2 μmol/L. Consequently, to obtain the maximum degree of sensitization, it is necessary to inhibit far more than 95% of Chk1. We suggest there may be two different pools of Chk1 in the cell. One pool is responsible for cell cycle arrest in the face of DNA damage as reflected in the lack of S phase arrest in SN38-treated ΔChk1 cells. Alternatively, it has been shown that Chk1 can bind DNA polymerase alpha and this may represent a second pool that is more tightly linked to replication fork stability (38).
While screening a panel of cell lines, we identified four that were much more sensitive to SCH900776 alone. While the same cell lines were also more sensitive to UCN-01, this effect was more pronounced with SCH900776 because of its much greater selectivity. At 0.5 μM SCH900776, the only other kinase significantly inhibited was Pim-1 (9), yet its action counteracts Chk1 by, for example, activating rather than inhibiting Cdc25A (39). Accordingly, inhibition of Pim-1 appears contradictory to the cell cycle abrogation observed here, so it seems unlikely that Pim-1 contributes to the observed cytotoxicity induced by SCH900776. The sensitivity to SCH900776 also did not correlate with the level of Chk1. Sensitivity to SCH900776 could arise because of high endogenous damage in some cells, or because of differences in downstream consequences of Chk1 inhibition. Alternately, sensitivity could arise through a defect in a complementary protein or pathway required for replication fork stabilization. For example, an siRNA screen identified defects in the Fanconi anemia pathway as eliciting sensitivity to Chk1 inhibition (40). Whatever the reason, this sensitivity may provide a chemical synthetic lethal interaction that would be selective for the tumor. In addition, the fact that these cells are extremely sensitive to the combination of low concentrations of both SCH900776 and hydroxyurea could have significant clinical impact.
Hydroxyurea has been used for many years as an anticancer drug, primarily in leukemia, but it is also approved as a radiosensitizer in head and neck carcinomas. The regularly administered doses and schedules are unlikely to cause sustained suppression of DNA synthesis rather transient suppression may occur which, based on our cell culture data, the cells are likely to tolerate. Repeated administration of hydroxyurea to patients every 4 h can maintain the plasma concentration in excess of 400 μmol/L for 72 h (41). As hydroxyurea is freely diffusible, adequate concentrations can likely be achieved in tumors that would slow replication such that cells would become dependent on Chk1. Our results also suggest that it is not necessary to inhibit Chk1 until 18 h making it possible to administer a single bolus of SCH900776 at that time and potentially enhance cell killing. Whether such a schedule would provide an increased therapeutic window or whether it would equally enhance toxicity to normal tissues remains to be determined. However, the observation that some tumor cell lines are highly sensitive to SCH900776 alone and in combination with hydroxyurea suggests that an enhanced therapeutic window should be feasible for these tumors. Ongoing experiments will assess the potential efficacy of these drug combinations in further preclinical and clinical studies.
Supplementary Material
Acknowledgments
Grant Support NIH grant CA117874 (A. Eastman), Cancer Center Support grant CA23108 to the Norris Cotton Cancer Center, and a NIH training grant T32 09658 (K.M. Garner).
Footnotes
Disclosure of Potential Conflicts of Interest D.A. Parry was an employee of Merck who developed SCH900776. Other authors have no potential conflicts.
References
- 1.Bucher N, Britten CD. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer. 2008;98:523–528. doi: 10.1038/sj.bjc.6604208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sausville EA, Arbuck SG, Messmann R, Headlee D, Bauer KS, Lush RM, et al. Phase I trial of 72-hour continuous infusion UCN-01 in patients with refractory neoplasms. J Clin Oncol. 2001;19:2319–2333. doi: 10.1200/JCO.2001.19.8.2319. [DOI] [PubMed] [Google Scholar]
- 3.Bunch RT, Eastman A. Enhancement of cisplatin-induced cytotoxicity by 7-hydroxystaurosporine (UCN-01), a new G2-checkpoint inhibitor. Clin Cancer Res. 1996;2:791–797. [PubMed] [Google Scholar]
- 4.Kohn EA, Ruth ND, Brown MK, Livingstone M, Eastman A. Abrogation of the S phase DNA damage checkpoint results in S phase progression or premature mitosis depending on the concentration of UCN-01 and the kinetics of Cdc25C activation. J Biol Chem. 2002;277:26553–26564. doi: 10.1074/jbc.M202040200. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Fan S, Eastman A, Worland PJ, Sausville EA, O’Connor PM. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Nat Cancer Inst. 1996;88:956–965. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 6.Levesque AA, Kohn EA, Bresnick E, Eastman A. Distinct roles for p53 transactivation and repression in preventing UCN-01-mediated abrogation of DNA damage-induced S and G2 cell cycle checkpoints. Oncogene. 2005;24:3786–3796. doi: 10.1038/sj.onc.1208451. [DOI] [PubMed] [Google Scholar]
- 7.Fuse E, Tanii H, Kurata N, Kobayashi H, Shimada Y, Tamura T, et al. Unpredicted clinical pharmacology of UCN-01 caused by specific binding to human α1-acid glycoprotein. Cancer Res. 1998;58:3248–3253. [PubMed] [Google Scholar]
- 8.Perez RP, Lewis LD, Beelen AP, Olszanski AJ, Johnston N, Rhodes CH, et al. Modulation of cell cycle progression in human tumors: a pharmacokinetic and tumor molecular pharmacodynamic study of cisplatin plus the Chk1 inhibitor UCN-01 (NSC 638850) Clin Cancer Res. 2006;12:7079–7085. doi: 10.1158/1078-0432.CCR-06-0197. [DOI] [PubMed] [Google Scholar]
- 9.Parry D, Shanahan F, Davis N, Wiswell D, Seqhezzi W, Pierce R, et al. Targeting the replication checkpoint with a potent and selective CHK1 inhibitor. Proc Am Assoc Cancer Res. 2009;50 abstract-2490. [Google Scholar]
- 10.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nature Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 11.Eastman A. Reevaluation of the interaction of cis-dichloro(ethylenediamine)platinum(II) with DNA. Biochemistry. 1986;25:3912–3915. doi: 10.1021/bi00361a026. [DOI] [PubMed] [Google Scholar]
- 12.Sorenson CM, Eastman A. Mechanism of cis-diamminedichloroplatinum(II)-induced cytotoxicity: role of G2 arrest and DNA double-strand breaks. Cancer Res. 1988;48:4484–4488. [PubMed] [Google Scholar]
- 13.Iwasaki H, Huang P, Keating MJ, Plunkett W. Differential incorporation of ara-C, gemcitabine and fludarabine into replicating and repairing DNA in proliferating human leukemia cells. Blood. 1997;90:270–278. [PubMed] [Google Scholar]
- 14.Zegerman P, Diffley JFX. DNA replication as a target of the DNA damage checkpoint. DNA Repair. 2009;8:1077–1088. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 15.Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle. 2005;4:131–139. doi: 10.4161/cc.4.1.1299. [DOI] [PubMed] [Google Scholar]
- 16.Arienti KL, Brunmark A, Axe FU, McClure K, Lee A, Blevitt J, et al. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimadoles. J Med Chem. 2005;48:1873–1885. doi: 10.1021/jm0495935. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W-H, Poh A, Fanous AA, Eastman A. DNA damage-induced S phase arrest in human breast cancer depends on CHK1, but G2 arrest can occur independently of Chk1, Chk2 or MAPKAPK2. Cell Cycle. 2008;7:1668–1677. doi: 10.4161/cc.7.11.5982. [DOI] [PubMed] [Google Scholar]
- 18.Garner KM, Eastman A. Variations in Mre11/Rad50/Nbs1 status and DNA damage-induced S-phase arrest in cell lines of the NCI60 panel. BMC Cancer. 2011;11:206. doi: 10.1186/1471-2407-11-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demarcq C, Bunch RT, Creswell D, Eastman A. The role of cell cycle progression in cisplatin-induced apoptosis in Chinese hamster ovary cells. Cell Growth Different. 1994;5:983–993. [PubMed] [Google Scholar]
- 20.Rao J, Otto WR. Fluorometric DNA assay for cell growth estimation. Anal Biochem. 1992;207:186–192. doi: 10.1016/0003-2697(92)90521-8. [DOI] [PubMed] [Google Scholar]
- 21.Levesque AA, Fanous AA, Poh A, Eastman A. Defective p53 signaling in p53 wildtype tumors attenuates p21waf1 induction and cyclin B repression rendering them sensitive to Chk1 inhibitors that abrogate S and G2 arrest. Mol Cancer Therap. 2008;7:252–262. doi: 10.1158/1535-7163.MCT-07-2066. [DOI] [PubMed] [Google Scholar]
- 22.Darzynkiewicz Z, Traganos F, Kapuscinski J, Staiano-Coico L, Melamed MR. Accessibility of DNA in situ to various fluorochromes: relationship to chromatin changes during erythroid differentiation of Friend leukemia cells. Cytometry. 1984;5:355–363. doi: 10.1002/cyto.990050411. [DOI] [PubMed] [Google Scholar]
- 23.Clarke AAL, Clarke PR. DNA-dependent phosphorylation of Chk1 and claspin in a human cell-free system. Biochem J. 2005;388:705–712. doi: 10.1042/BJ20041966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung-Pineda V, Ryan CE, Piwnica-Worms H. Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol Cell Biol. 2006;26:7529–7538. doi: 10.1128/MCB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci USA. 2002;99:14795–14800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-Deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eastman A, Kohn EA, Brown MK, Rathman J, Livingstone M, Blank DH, et al. A novel indolocarbazole, ICP-1, abrogates DNA damage-induced cell cycle arrest and enhances cytotoxicity. Similarities and differences to the cell cycle checkpoint abrogator UCN-01. Mol Cancer Therap. 2002;1:1067–1078. [PubMed] [Google Scholar]
- 28.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–505. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 29.Levesque AA, Eastman A. p53-based cancer therapies: is defective p53 the Achilles heel of the tumor? Carcinogenesis. 2007;28:13–20. doi: 10.1093/carcin/bgl214. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Bravo V, Guaita-Esteruelas S, Salvador N, Bachs O, Agell N. Different S/M checkpoint responses of tumor and non-tumor cell lines to DNA replication inhibition. Cancer Res. 2007;67:11648–11656. doi: 10.1158/0008-5472.CAN-07-3100. [DOI] [PubMed] [Google Scholar]
- 31.Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, et al. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 32.Busby EC, Leistritz DF, Abraham RT, Karnitz LM, Sarkaria JN. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 2000;60:2108–2112. [PubMed] [Google Scholar]
- 33.Yu Q, Rose JL, Zhang H, Takemura H, Kohn KW, Pommier Y. UCN-01 inhibits p53 up-regulation and abrogates γ-radiation-induced G2-M checkpoint independently of p53 by targeting both of the checkpoint kinases, Chk2 and Chk1. Cancer Res. 2002;62:5743–5748. [PubMed] [Google Scholar]
- 34.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Molecular Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao Z, Xue J, Sowin TJ, Rosenberg SH, Zhang H. A novel mechanism of checkpoint abrogation conferred by Chk1 downregulation. Oncogene. 2005;24:1403–1411. doi: 10.1038/sj.onc.1208309. [DOI] [PubMed] [Google Scholar]
- 37.Robinson HM, Jones R, Walker M, Zachos G, Brown R, Cassidy J, et al. Chk1-dependent slowing of S-phase progression protects DT40 B-lymphoma cells against killing by the nucleoside analogue 5-fluorouracil. Oncogene. 2006;25:5359–5369. doi: 10.1038/sj.onc.1209532. [DOI] [PubMed] [Google Scholar]
- 38.Taricani L, Shanahan F, Parry D. Replication stress activates DNA polymerase alpha-associated Chk1. Cell Cycle. 2009;8:482–489. doi: 10.4161/cc.8.3.7661. [DOI] [PubMed] [Google Scholar]
- 39.Shah N, Pang B, Yeoh K-G, Thorn S, Chen CS, Lilly MB, et al. Potential roles for the PIM1 kinase in human cancer - a molecular and therapeutic appraisal. Eur J Cancer. 2008;44:2144–2151. doi: 10.1016/j.ejca.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 40.Chen CC, Kennedy RD, Sidi S, Look AT, D’Andrea A. Chk1 inhibition as a strategy for targeting fanconi anemia (FA) DNA repair pathway deficient tumors. Mol Cancer. 2009;8:24. doi: 10.1186/1476-4598-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belt RJ, Haas CD, Kennedy J, Taylor S. Studies of hydroxyurea administed by continous infusion: toxicity, pharmacokinetics, and cell synchronization. Cancer. 1980;46:455–462. doi: 10.1002/1097-0142(19800801)46:3<455::aid-cncr2820460306>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.