Summary
The multi-protein kinetochore complex must assemble at a specific site on each chromosome to achieve accurate chromosome segregation. Defining the nature of the DNA-protein interactions that specify the position of the kinetochore and provide a scaffold for kinetochore formation remain key goals. Here, we demonstrate that the centromeric histone-fold containing CENP-T-W and CENP-S-X complexes co-assemble to form a stable CENP-T-W-S-X heterotetramer. High-resolution structural analysis of the individual complexes and the heterotetramer reveals similarity to other histone fold-containing complexes including canonical histones within a nucleosome. The CENP-T-W-S-X heterotetramer binds to and supercoils DNA. Mutants designed to compromise heterotetramerization or the DNA-protein contacts around the heterotetramer strongly reduce the DNA binding and supercoiling activities in vitro and compromise kinetochore assembly in vivo. These data suggest that the CENP-T-W-S-X complex forms a unique nucleosome-like structure to generate contacts with DNA, extending the “histone code” beyond canonical nucleosome proteins.
Keywords: Kinetochore, CENP-T-W, CENP-S-X, The X-ray Structure, DNA binding
Introduction
To facilitate faithful chromosome segregation during mitosis, the kinetochore is assembled on centromere DNA to form a dynamic interface with microtubules from the mitotic spindle (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). To establish a functional kinetochore structure, a subset of kinetochore proteins must make strong and specific contacts with centromeric DNA. Defining the molecular mechanisms by which kinetochore proteins specify the position on the chromosome and generate stable contacts with DNA to drive kinetochore assembly remain key goals. Nucleosomes containing the centromere-specific histone H3 variant CENP-A provide an important mark to establish a centromere-specific chromatin structure (Black and Cleveland, 2011). However, although CENP-A deposition is necessary for kinetochore specification, it is not strictly sufficient for the formation of functional kinetochores in vertebrate cells (Van Hooser et al., 2001; Gascoigne et al., 2011). This suggests that there are additional proteins that are required to generate centromere-specific chromatin and direct sequence-independent kinetochore assembly.
To form a scaffold for kinetochore assembly, a subset of kinetochore proteins must associate with centromeric DNA. We have previously identified the CENP-T-W complex, which contains histone-fold domains required to associate with DNA and target to kinetochores (Hori et al., 2008). In addition to a histone-fold domain, the CENP-T-W complex provides an important platform for kinetochore assembly through a long N-terminal flexible region that binds directly to outer kinetochore proteins (Gascoigne et al., 2011; Suzuki et al., 2011). Ectopic localization of CENP-T induces kinetochore formation and can at least partially bypass the requirement for CENP-A nucleosomes (Gascoigne et al., 2011). However, little is known about how the CENP-T-W complex is targeted to endogenous centromeres. Although the CENP-T-W complex requires CENP-A for its recruitment to centromeres, CENP-T does not bind directly to CENP-A nucleosomes (Hori et al., 2008). Thus, the binding of CENP-T to DNA may provide an important scaffold to direct kinetochore formation.
To define the role of CENP-T in kinetochore assembly, we analyzed the structural, biochemical, and functional basis for its association with DNA. We demonstrate that the CENP-T-W complex associates with a second group of histone-fold containing proteins, the CENP-S-X complex (Amano et al., 2009). CENP-S and X are conserved kinetochore-localized proteins (Perpelescu and Fukagawa, 2011), but have also been identified as Fanconia Anemia M (FANCM) associated proteins where they were named MHF1 and MHF2 (Yan et al., 2010; Singh et al, 2010) suggesting a possible dual function. Association of the CENP-T-W and CENP-S-X complexes generates a stable CENP-T-W-S-X heterotetramer with structural similarity to canonical nucleosomes. The CENP-T-W-S-X heterotetramer supercoils DNA suggesting that this complex bends DNA to form a nucleosome-like structure. Mutants predicted to compromise heterotetramerization or the DNA contacts around the heterotetramer show a dramatic reduction in DNA binding and supercoiling activities and fail to form functional kinetochores. In total, our results suggest that a CENP-T-W-S-X containing nucleosome-like structure functions as a scaffold for kinetochore formation.
Results
The CENP-T-W complex forms a dimer of histone fold domains
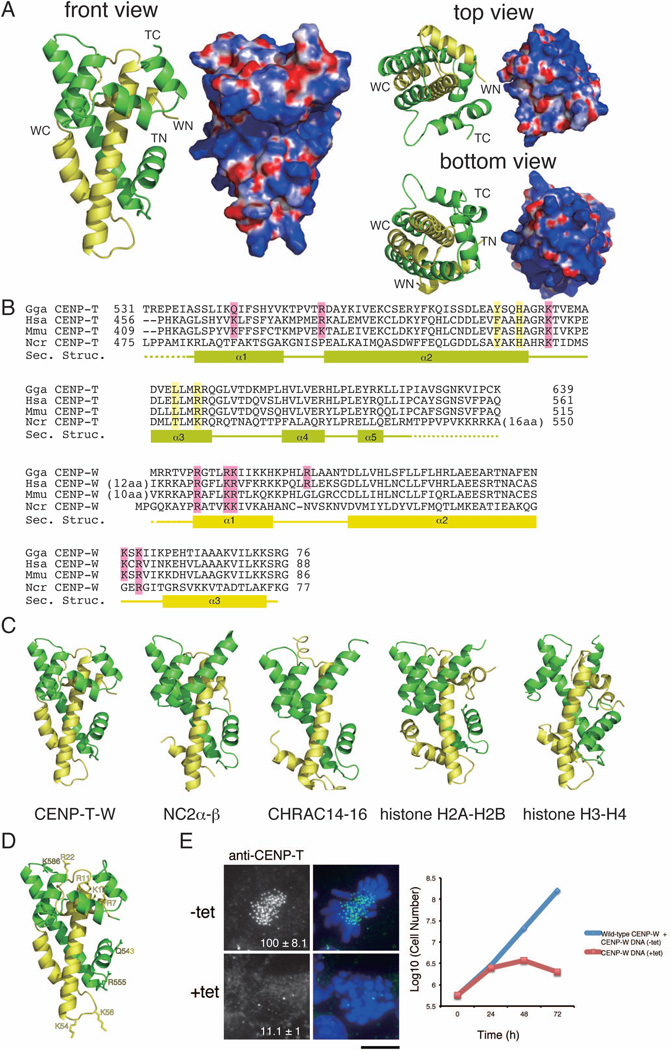
To define the mechanisms by which the CENP-T-W complex associates with DNA, we began by conducting a structural analysis of the C-terminal histone-fold regions of chicken CENP-T (531–639 aa) and CENP-W. Following co-expression of the recombinant complex and crystallization, we determined the structure by multiple wavelength anomalous dispersion (MAD) from selenomethionine derivatives (Figure 1A and Table S1). Two crystal forms of the CENP-T-W complex were obtained that were both refined to 2.2 Å resolution with refinement statistics of Rwork = 0.220 (Rfree = 0.269) and Rwork = 0.203 (Rfree = 0.261), respectively.
Figure 1. (refers to Supplemental Figure S1 and Table S1). The CENP-T-W complex is a heterodimer with structural similarity to other histone-fold containing protein complexes.
(A) Ribbon diagram representations of the CENP-T-W dimer (left) and its surface charges (right). Three orthogonal views of the CENP-T-W dimer are shown. CENP-T is colored in green and CENP-W is colored in yellow. The N- and C-terminus of CENP-T and CENP-W are shown as TN, TC, WN, and WC, respectively. Electrostatic surface charges of CENP-T and CENP-W were calculated by APBS and are contoured from −8.0 (red) to 8.0 (blue).
(B) Sequence alignment of CENP-T (top) and CENP-W (bottom) from chicken, human, mouse, and Neurospora. Boxes (α1 – α5), solid lines, and dashed lines indicate α-helices, random coil regions, and disordered residues, respectively. Residues marked by pink are predicted as DNA binding sites by comparison with canonical histones and residues marked by yellow are predicted as the tetramerization interface based on comparison to CENP-S-X.
(C) Structural comparison of the CENP-T-W complex with other histone-fold containing protein complexes including NC2α-β (PDB ID: 1JFI), CHRAC14-16 (PDB ID: 2BYK), and histone H2A–H2B and H3–H4 in the nucleosome (PDB ID: 1KX5). The structural alignment is based on the DALI structure alignment server with the structures viewed from the same angle as in (A).
(D) Prediction of the DNA binding sites for the CENP-T-W dimer based on structural similarities of the DNA binding surface in histone H3–H4.
(E) Analysis of DT40 cells in which endogenous CENP-W is replaced with the CENP-WDNA mutant (+tet). In absence of tetracycline (−tet), cells express both wild-type CENP-W and the CENP-WDNA mutant and in the presence of tetracycline (+tet), cells express only the CENP-WDNA mutant. The signal intensity of CENP-T at each kinetochore detected by immunofluorescence was measured relative to an adjacent background signal (n = more than 10 cells; average intensity +/− standard deviation). Bar, 10 µm. The growth curve was measured using Trypan blue exclusion.
CENP-T and CENP-W contain five and three α helices, respectively and form a heterodimer in the crystal (Figure 1A and B). The composition of these α helices and the entire structure of the CENP-T-W dimer are highly homologous to the structure of the histone H2A–H2B or H3–H4 dimers within the nucleosome (Figure 1C). Additional searches of the structural database using the DALI server (Holm and Rosenström, 2010) indicate that the structure of other histone-fold containing protein complexes, such as NC2α-β and CHRAC14-16, are also similar to that of the CENP-T-W complex (Figure 1C). In total, our structural analysis reveals that the CENP-T-W complex is a dimer of histone-fold domains.
Mutants that disrupt the DNA binding activity of CENP-T-W complex fail to form functional kinetochores
Although we have previously predicted likely DNA binding amino acids in CENP-W (Hori et al., 2008), the structural analysis described above defines clear DNA binding residues for CENP-T (Q543, R555, and K586) and CENP-W (R7, R11, K12, R22, K54, and K56) based on structural similarity to the DNA binding interface of canonical histones (Figure 1B and D). To test the role of these residues, we generated alanine mutants for Q543, R555, K586 in CENP-T and R7, R11, K12, R22, K54 and K56 in CENP-W (termed CENP-TDNA and CENP-WDNA) and performed DNA binding assays with recombinant CENP-T-W proteins. The wild type CENP-T-W complex binds to DNA, but did not show an apparent DNA binding specificity (data not shown). Thus, we used the classical “601” DNA positioning sequence (Vasudevan et al., 2010) for further DNA binding assays. Although the CENP-T-W complex binds to DNA, the majority of the DNA-CENP-T-W complexes do not enter the acrylamide gel (Figure S1A). Under low salt concentrations, some DNA-CENP-T-W complexes can enter the gel (Figure S1A and B). Importantly, this DNA binding activity was reduced in the CENP-TDNA-WDNA mutant (Figure S1C). However, we still observed a large assembly of DNA with the CENP-TDNA-WDNA mutant complex. This suggests that there may be multiple DNA binding sites in the CENP-T-W complex in addition to sites predicted based on similarity to canonical histones (Figure 1B), including possibly some non-specific binding interactions.
To test significance of the predicted DNA binding sites in vivo, we analyzed chicken DT40 cells in which endogenous CENP-W is replaced with the CENP-WDNA mutant and found that these cells died following strong mitotic defects (Figure 1E). We also found that CENP-T is degraded when the CENP-T-W complex is unable to localize to kinetochores (Figure 1E and S1D). In addition, we analyzed human CENP-WDNA mutants in HeLa cells in which endogenous CENP-W is disrupted by RNAi and found that both CENP-WDNA and the outer kinetochore protein Ndc80 failed to localize to kinetochore in these cells (Figure S1E). Ndc80 is a key outer kinetochore microtubule binding protein that is directly downstream of CENP-T for kinetochore assembly pathways (Hori et al., 2008; Gascoigne et al., 2011). These data suggest that the DNA binding activity of the CENP-T-W complex is essential for kinetochore formation.
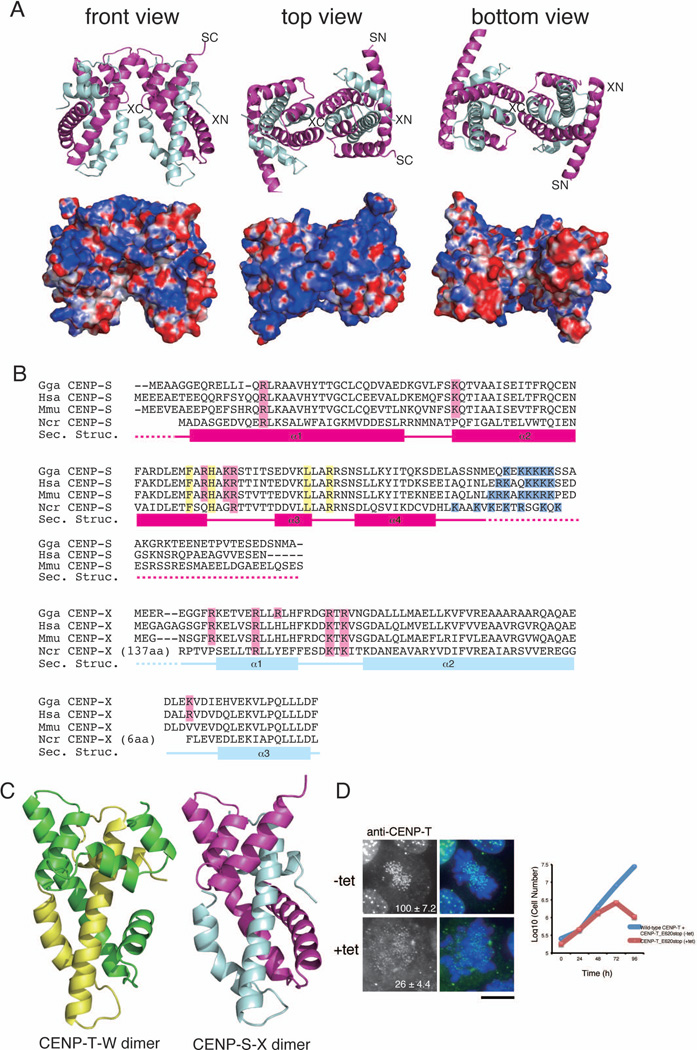
Structural analysis reveals that the CENP-S-X complex forms a (CENP-S-X)2 tetramer
In addition to the CENP-T-W complex, we have previously identified a second kinetochore-localized histone fold-containing complex – the CENP-S-X complex (Amano et al., 2009). CENP-T was detected as a CENP-S-associated protein in immunoprecipitations, suggesting a possible interaction between these complexes. To test the relationship between the CENP-T-W and CENP-S-X complexes, we next conducted a high-resolution structural analysis of the CENP-S-X complex. For these studies, we co-expressed truncated chicken CENP-S (CENP-SΔC; lacking the C-terminal 33 amino acid unstructured region) with full-length chicken CENP-X. Following crystallization of the recombinant complex, the structure was determined by single wavelength anomalous dispersion (SAD) from mercury derivatives and MAD from selenomethionine derivatives (Table S2). The CENP-S-X crystal structure was refined to 2.15 Å resolution with refinement statistics of Rwork = 0.186 (Rfree = 0.235).
Similar to the CENP-T-W complex, the structure of the CENP-S-X complex is closely related to that of canonical histones (Figure 2A–C). However, the CENP-S-X complex forms a (CENP-S-X)2 tetramer (Figure 2A and S2A), whereas the CENP-T-W complex forms a dimer. CENP-S uses the α2 and α3 helices to form the tetramer interface, similar to the histone (H3–H4)2 interface within the nucleosome (Luger et al., 1997). As occurs for histone H3, the CENP-S-CENP-S interface is comprised of hydrophobic interactions and a critical hydrogen-bond network (Figure S2A). In addition to the hydrophobic interactions, electrostatic interactions are used for the CENP-S-CENP-S interface. To confirm the importance of the α2 and α3 helices of CENP-S for tetramerization, we mutated critical amino acids in these helices (F65E, H68E, L81R, R84E: CENP-Stet protein; Figure 2B and S2B–C). Although CENP-Stet associates with CENP-X and forms a heterodimer in vitro, the estimated size of the CENP-Stet-X complex is half that of the wild-type CENP-S-X complex based on size exclusion chromatography (Figure S2B). Similar results were observed using CENP-StetΔC-X (Figure S2C). These results indicate that the CENP-S-X complex forms a (CENP-S-X)2 tetramer both in crystals and in solution.
Figure 2. (refers to Supplemental Figure S2 and Table S2). The CENP-S-X complex forms a tetramer with structural similarity to the CENP-T-W complex.
(A) Ribbon diagram representations of the (CENP-S-X)2 tetramer (top) and its surface charges (bottom). Three orthogonal views of the (CENP-S-X)2 tetramer are shown. CENP-S is colored in magenta and CENP-X is colored in cyan. The N- and C-terminus of CENP-S and CENP-X are shown as SN, SC, XN, and XC, respectively. Electrostatic surface charges of CENP-S and CENP-X were calculated by APBS and are contoured from −8.0 (red) to 8.0 (blue).
(B) Sequence alignment of CENP-S (top) and CENP-X (bottom) from chicken, human, mouse, and Neurospora. Boxes (α1 – α4), solid lines, and dashed lines indicate α-helices, random coil regions, and disordered residues, respectively. Residues marked by pink are predicted as DNA binding sites by comparison with canonical histones and residues marked by yellow indicate the tetramerization interface. Basic residues marked by blue are additional DNA binding sites of CENP-S.
(C) Structural comparison of the CENP-T-W and CENP-S-X complexes. Both complexes are viewed from the same angle as in (A).
(D) Immunofluorescence analysis of DT40 cells in which endogenous CENP-T is replaced by the CENP-TE620stop mutant with deletion of the α5 helix (+tet). In the absence of tetracycline (−tet), cells express both wild-type CENP-T and the CENP-TE620stop mutant. In the presence of tetracycline (+tet), cells express only CENP-TE620stop mutant. The growth curve was shown. Bar, 10 µm.
DNA binding defective CENP-S-X mutants compromise kinetochore assembly
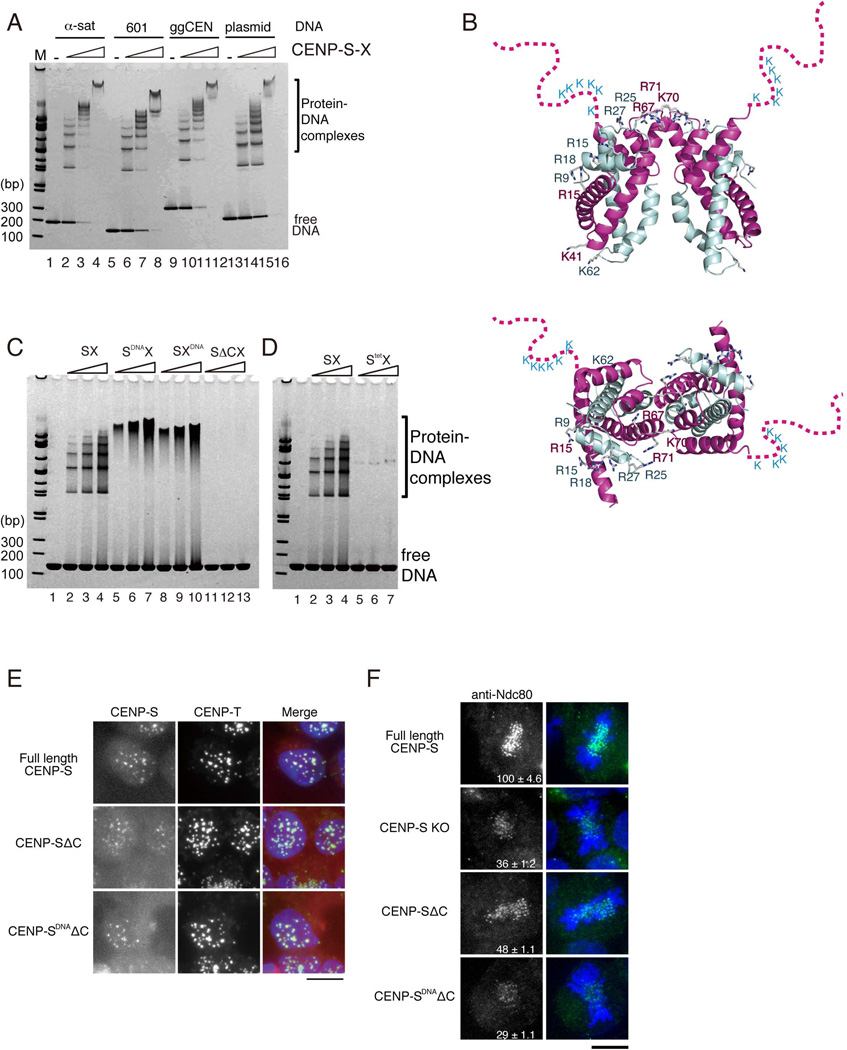
When full length CENP-S and CENP-X were used for the DNA binding assays, we observed a regular ladder of protein-DNA complexes (Figure 3A), suggesting that multiple CENP-S-X complexes bind to a single DNA molecule at regular intervals. Structural comparison of CENP-S-X to histone H3–H4 suggested that R15, K41, K67, K70, and K71 of CENP-S and R9, R15, R18, R25, and R27 of CENP-X represent the DNA binding interface (Figure 2B and 3B). Mutant CENP-S-X complexes (containing CENP-S (R15A, K41A, K70A) or CENP-X (R9A, R27A, K62A); denoted as CENP-SDNA or CENP-XDNA; Figure S3A) were defective in forming a regular ladder in DNA binding assays and instead formed a smear-like pattern (lane 5–7 for CENP-SDNA-X and lane 8–10 for CENP-S-XDNA in Figure 3C) suggestive of DNA binding derived other regions of these proteins. Indeed, we found that the conserved basic, unstructured C-terminal region of CENP-S, which is in close vicinity to the histone-fold DNA binding interface, is required to interact with DNA (CENP-SΔC-X; lane 11–13 in Figure 3C). In addition to the residues predicted to make direct contacts with DNA, tetramerization of the CENP-S-X complex is also required for full DNA binding activity (CENP-Stet-X; Figure 3D).
Figure 3. (refers to Supplemental Figure S3). The CENP-S-X complex binds to DNA.
(A) DNA binding assays of the CENP-S-X complex to α-satellite DNA (α-sat: 186 bp), the 601 DNA sequence (601: 145 bp), a chicken centromere sequence (ggCEN: 217bp), and pBluescript plasmid DNAs (plasmid: 217 bp). Different concentrations of protein complex (2.5 µM, 5 µM, 10 µM) were mixed with 2.5 µM DNA. The mixtures were analyzed by 5–20% gradient native PAGE.
(B) A ribbon diagram of the CENP-S-X tetramer shown in orthogonal views. Basic residues on the positively charged surface are shown as stick representations. The disordered CENP-S C-terminal tail, which contains conserved lysine residues, is schematically depicted.
(C) Different concentrations of the indicated CENP-S-X mutant complexes (2.5 µM, 5 µM, 10 µM) were mixed with 2.5 µM 145 bp dsDNA (601 sequence). The mixtures were analyzed using 5–20% gradient native PAGE.
(D) DNA binding activity of the CENP-Stet-X tetramer mutant analyzed as in (C).
(E) Localization analysis of CENP-SΔC- or CENP-SDNAΔC-mRFP fusion proteins in DT40 cells. Cells expressing these fusion proteins were stained with anti-CENP-T antibodies. Bar, 10 µm.
(F) Immunofluorescence analysis of Ndc80 in CENP-S-deficient cells (CENP-S KO) expressing full-length CENP-S, CENP-SΔC, or CENP-SDNAΔC proteins. The kinetochore signal intensity of Ndc80 was measured relative to an adjacent background signal. Bar, 10 µm.
To determine whether the DNA binding activity of the CENP-S-X complex is required for kinetochore formation in vivo, we tested the localization of mutant CENP-S proteins in DT40 cells. We expressed CENP-SΔC and CENP-SDNAΔC in CENP-S-deficient cells and found that both CENP-SΔC and CENP-SDNAΔC mutant proteins localized to kinetochores (Figure 3E). This demonstrates that DNA binding is not essential for CENP-S kinetochore localization, and suggests that the interaction of CENP-S with the CENP-T-W complex may recruit this to kinetochores. We confirmed that mutant CENP-S proteins associate with CENP-T in cells based on co-immunoprecipitation using CENP-T antibodies (Figure S3B). However, we found that cells in which either CENP-SΔC or CENP-SDNAΔC replaces endogenous CENP-S displayed reduced localization of the outer kinetochore protein Ndc80 (Figure 3F), similar to what is observed in CENP-S-deficient cells (Amano et al., 2009). This suggests that outer kinetochore proteins are not assembled properly in cells expressing DNA binding CENP-S mutants. We conclude that the DNA binding activity of CENP-S-X is essential for function of the complex.
The C-terminal helices of CENP-T are critical for kinetochore localization
Structural comparison of the CENP-T-W and CENP-S-X complexes revealed that the α4 helix of the CENP-S-X complex is different from that of the CENP-T-W complex. The corresponding region in the CENP-T-W complex is composed of two helices (α4 and α5) and forms a helix-turn-helix structure (Figure 1B and 2C). CENP-T mutants lacking the α4 or α5 helices (CENP-TE620stop or CENP-TH610stop) form a complex with CENP-W similar to that of wild-type CENP-T (data not shown). However, DT40 cells in which endogenous CENP-T is replaced with CENP-TE620stop or CENP-TH610stop do not grow and kinetochore localization of these CENP-T mutants is strongly reduced (Figure 2D and S2D). Similarly, human CENP-T deleted for these helices (hCENP-TΔ30) also failed to localize to kinetochores in HeLa cells (Figure S2E). However, CENP-TE620stop-W or CENP-TH610stop-W mutant complexes displayed DNA binding activity in vitro (data not shown), suggesting that the α4/α5 helices of CENP-T are necessary for kinetochore localization independent of DNA binding activity. To test whether these helices of CENP-T are sufficient for kinetochore targeting, we analyzed the localization of chimeric proteins in which the α4/α5 helices from CENP-T were exchanged with corresponding regions of histone H2A, histone H3, NC2, or CHRAC. We also generated constructs in which the α4/α5 helices from CENP-T were added to the C-terminus of these histone related proteins. However, in both cases, we did not observe kinetochore localization of these chimeric proteins with the α4/α5 helices of CENP-T (data not shown), suggesting that additional sites also contribute to CENP-T kinetochore targeting.
The CENP-T-W and CENP-S-X complexes associate to form a stable CENP-T-W-S-X heterotetramer
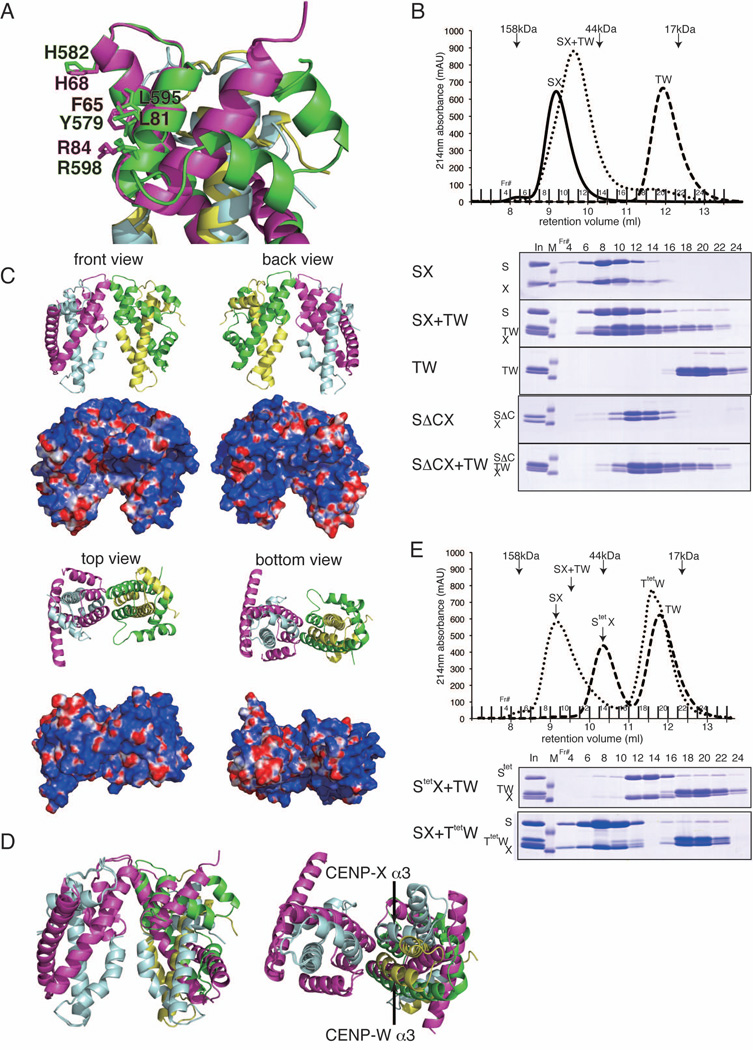
Although we found slight structural differences between the CENP-S-X and CENP-T-W complexes, the overall structure of these complexes is quite similar. Indeed, when the structure of the CENP-S-X complex is superimposed on the CENP-T-W complex, the tetramer interface of the CENP-S-X complex is nearly identical to corresponding sites in the CENP-T-W complex (Figure 4A). Considering this structural similarity together with the functional relationship between these complexes (Amano et al., 2009), we hypothesized that these two histone-fold containing complexes may interact. Indeed, when these complexes are combined in equimolar ratios, CENP-S-X and CENP-T-W co-migrate and form a single stoichiometric complex (Figure 4B and S4A). Based on the observed migration, these data suggest that one CENP-S-X dimer interacts with one CENP-T-W dimer to form a CENP-T-W-S-X heterotetramer. The tail region of CENP-S is not required for this interaction, as the CENP-SΔC-X complex bound similarly to the CENP-T-W complex (Figure 4B). Human CENP-S-X and CENP-T-W also form a single stoichiometric complex as observed for the chicken complexes (Figure S4B). Although the CENP-S-X and CENP-T-W complexes associate strongly with each other, we did not observe clear association of these complexes or the CENP-T-W-S-X heterotetramer with histones under the high salt conditions used to assemble canonical nucleosomes (data now shown).
Figure 4. (refers to Supplemental Figure S4 and Table S3). Formation of a stable CENP-T-W-S-X heterotetramer.
(A) Superimposed image of the CENP-T-W and CENP-S-X structures. Amino acids responsible for tetramer formation are shown (F65, H68, L81, and R84 for CENP-S and Y579, H582, L595, and R598 for CENP-T).
(B) Stoichiometric amounts of CENP-S-X (25 µM) or CENP-ΔS-X (25 µM) and CENP-T-W (25 µM) were mixed and incubated for 15 min at room temperature. The mixture was separated by gel filtration using a Superdex 75 column. Peak fractions were analyzed by SDS-PAGE.
(C) Ribbon diagram representations of the CENP-T-W-S-X heterotetramer (top) and its surface charges (bottom). Four orthogonal views of the CENP-T-W-S-X heterotetramer are shown. CENP-S, CENP-X, CENP-T, and CENP-W are colored in magenta, cyan, green, and yellow, respectively.
(D) Superimposed image of the CENP-T-W-S-X and (CENP-S-X)2 structures.
(E) Stoichiometric amounts of CENP-Stet-X (25 µM) and CENP-T-W (25 µM) or CENP-S-X (25 µM) and CENP-Ttet-W (25 µM) were mixed and incubated. Gel filtration and SDS-PAGE were performed as in (B).
To define the properties of this CENP-T-W-S-X heterotetramer, we next conducted a structural analysis. Following reconstitution of the recombinant complex and crystallization, we determined the structure by molecular replacement methods (Figure 4C and Table S3). Two crystal forms of the CENP-T-W-S-X complex were obtained that were refined to 2.4 Å and 3.4 Å resolutions with refinement statistics of Rwork = 0.188 (Rfree = 0.244) and Rwork = 0.211 (Rfree = 0.273) respectively. For these analyses, we used full-length CENP-S rather than CENP-SΔC, although the C-terminus was disordered in the crystal. The entire structure of the CENP-T-W-S-X heterotetramer is similar to that of (CENP-S-X)2, but there is some tilt in the CENP-S α2 helix within the complex and the α3 helix of CENP-X does not overlap with that of CENP-W (Figure 4D). The area of the tetramer interface for the CENP-T-W-S-X heterotetramer is 559 Å2 and the free energy for this interaction is calculated as −4.3 kcal/mol based on the PDBePISA server (Krissinel and Henrick 2007). In contrast, the area of the tetramer interface for (CENP-S-X)2 is 681 Å2 and the free energy for this interaction is calculated as −0.9 kcal/mol, suggesting that the CENP-T-W-S-X heterotetramer is more stable than (CENP-S-X)2.
The tetramer interface of the CENP-S-X complex (F65, H68, L81, R84 in CENP-S) is nearly identical to the corresponding region of the CENP-T-W complex (Y579, H582, L595, R598 in CENP-T) (Figure 4A). To test the involvement of these residues in tetramerization, we isolated recombinant complexes with CENP-Stet or CENP-Ttet (Y579A, H582E, L595R, R598E) mutants. Although CENP-Stet or CENP-Ttet mutants form dimers with CENP-X or CENP-W, respectively, the CENP-Stet-X or CENP-Ttet-W mutant complexes are unable to assemble the heterotetrameric complex (Figure 4E).
Based on these structural and biochemical analyses, we conclude that the CENP-T-W-S-X heterotetramer is stably formed using unique tetramerization interfaces on the α2 and α3 helices of CENP-S and CENP-T.
Tetramer defective CENP-S and CENP-T mutants fail to assemble functional kinetochores
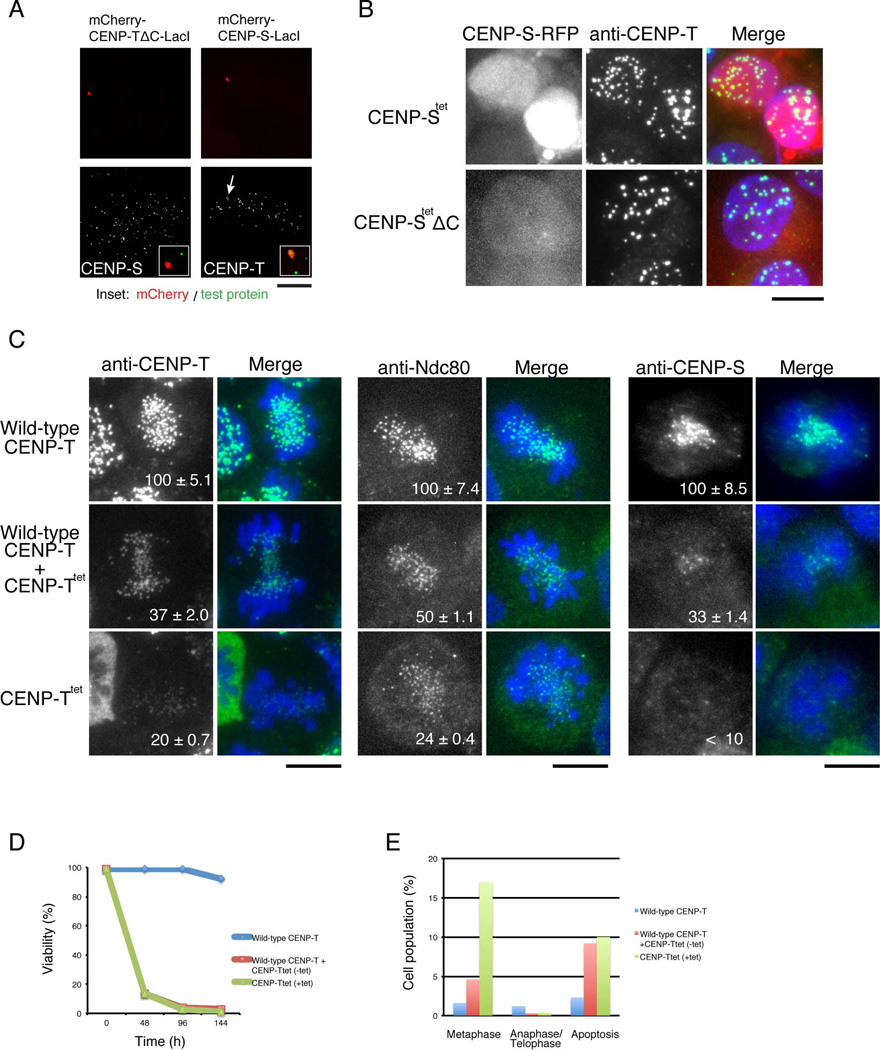
CENP-S and CENP-X proteins are delocalized from kinetochores in CENP-T-deficient cells (Figure S5A). Similarly, although CENP-T still localizes to kinetochores in CENP-S-deficient cells, the length of the region occupied by CENP-T at the kinetochore outer plate based on immuno-EM analysis is reduced (Figure S5B), suggesting close functional relationship between the CENP-T-W and CENP-S-X complexes. To test the association of the histone-fold domains of these complexes in vivo, we ectopically localized CENP-S to non-centromere regions in human cells using a LacI-LacO system (see Gascoigne et al., 2011) and observed co-recruitment of CENP-T to this site (Figure 5A, right). In contrast, CENP-S is not recruited by CENP-TΔC, which lacks the histone-fold domain. These LacI-LacO system experiments suggest that CENP-T-W-S-X forms a heterotetramer using the histone-fold regions in vivo.
Figure 5. (refers to Supplemental Figure S5). The tetramerization interface of the CENP-S-X and CENP-T-W is required for functional kinetochore assembly in vivo.
(A) Ectopic targeting of mCherry-CENP-TΔC-LacI (lacking the histone-fold domain) or CENP-S-LacI. Both fusion proteins are localized on LacO repeats at a non-centromeric region. Bar, 5 µm.
(B) Localization of the CENP-Stet- or CENP-Stet ΔC-mRFP fusion proteins in CENP-S-deficient DT40 cells co-stained with anti-CENP-T antibodies. Bar, 10 µm.
(C) Immunofluorescence analysis of CENP-T, Ndc80, and CENP-S in wild-type CENP-T cells, CENP-T+CENP-Ttet cells (expressing both wild type CENP-T and CENP-Ttet; −tet), and CENP-Ttet cells (expressing only CENP-Ttet; +tet). The signal intensity of each kinetochore was measured. Bars, 10 µm.
(D) Cell viability analysis for wild-type CENP-T cells, CENP-T+CENP-Ttet cells, and CENP-Ttet cells.
(E) Percentage of cells in metaphase, anaphase/telophase, or apoptosis for wild-type CENP-T cells, CENP-T+CENP-Ttet cells, and CENP-Ttet cells (120 h after tetracycline addition). Cells were stained with anti-α-tubulin antibodies. More than 800 cells were analyzed for each condition.
We next sought to test the functional significance of the tetramer interface in vivo. CENP-Stet-mRFP and CENP-StetΔC-mRFP tetramerization defect mutants failed to localize to kinetochores in CENP-S-deficient DT40 cells and instead localized diffusely throughout the nucleus (Figure 5B). We also generated a CENP-T conditional knockout DT40 cell line stably expressing CENP-Ttet (Figure S5C). In the absence of tetracycline, these cells express both wild-type CENP-T and CENP-Ttet (referred as CENP-T+CENP-Ttet), but in the presence of tetracycline, only CENP-Ttet is expressed (referred as CENP-Ttet). A defect in tetramerization compromises CENP-T localization as CENP-Ttet was reduced to ~20% of wild-type CENP-T levels in CENP-Ttet cells (Figure 5C). Consistent with this, CENP-T+CENP-Ttet and CENP-Ttet cells both displayed a strong reduction in viability (Figure 5D). In addition, cells expressing only CENP-Ttet showed reduced levels of Ndc80 and CENP-S (Figure 5C), and a strong mitotic delay (Figure 5E). Similarly, we found that HeLa cells expressing CENP-Ttet and depleted for endogenous CENP-T by RNAi showed reduced levels of CENP-Ttet and Ndc80 at kinetochores and an increase in misaligned chromosomes (Figure S5D). These results indicate that tetramer defective mutants for CENP-S or CENP-T result in a strong reduction of CENP-T-W-S-X levels at kinetochores, which causes a failure to assemble functional kinetochores.
The CENP-T-W-S-X heterotetramer forms a distinct complex with DNA
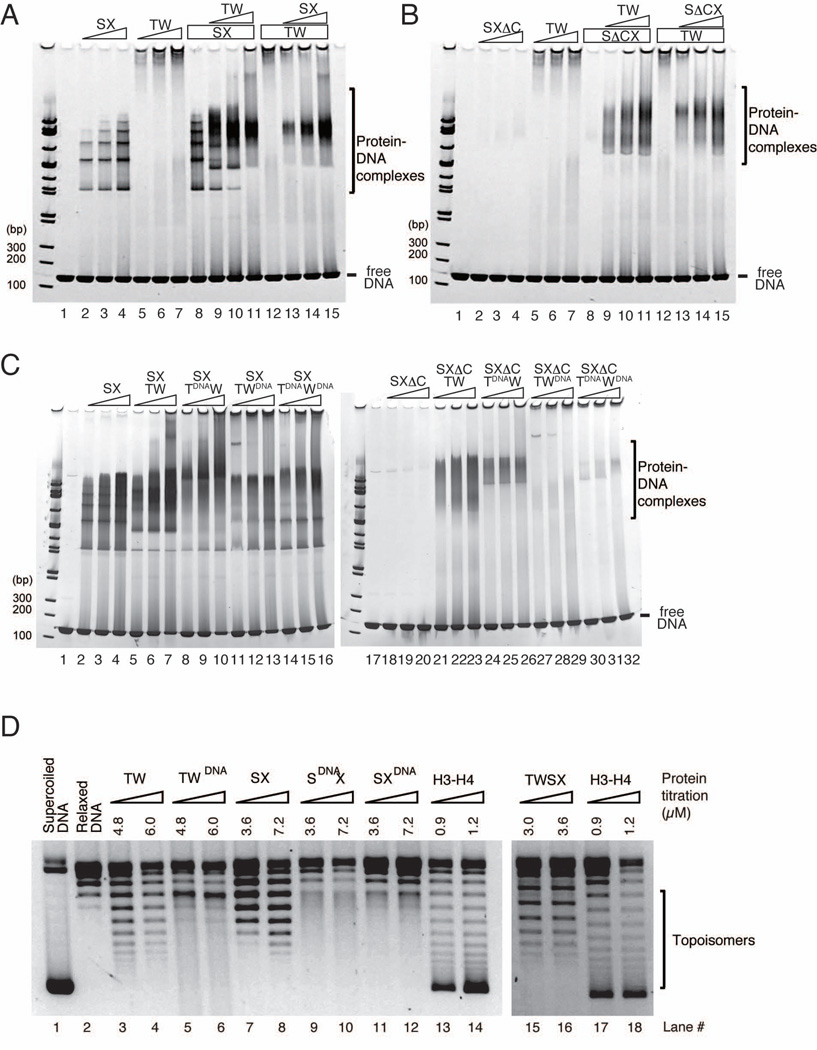
Both the CENP-T-W and CENP-S-X complexes show independent DNA binding activity. However, whereas the CENP-S-X complex forms a regular ladder profile in gel shift assays (lane 2–4 in Figure 6A), the majority of DNA-CENP-T-W complexes do not enter the acrylamide gel (lane 5–7 in Figure 6A). To analyze the properties of the DNA-CENP-T-W complex, we used dynamic light scattering (DLS), which indicated that the CENP-T-W complex forms large assemblies with DNA even in presence of 400 mM NaCl (Figure S6A). Interestingly, when the CENP-S-X and CENP-T-W complexes were combined, the large assembly was dissolved and the DNA-protein complex displayed a band at a distinct position in the gel (lane 8–15 in Figure 6A). Although the CENP-SΔC-X complex does not bind well to DNA on its own (lane 2–4 in Figure 6B), the CENP-SΔC-X-T-W heterotetramer complex binds to DNA similar to the wild-type heterotetrameric complex (lane 8–15 in Figure 6B). However, there is strong reduction in DNA binding activity for the heterotetramerization defective CENP-Stet-X mutants when combined with CENP-T-W (Figure S6B), indicating that heterotetramerization is crucial for DNA binding.
Figure 6. (refers to Supplemental Figure S6). The CENP-T-W-S-X heterotetramer binds to DNA and introduces supercoils into DNA.
(A) Gel shift assays for CENP-S-X (lanes 2–4) and CENP-T-W (lanes 5–7). Different concentrations of CENP-S-X (lanes 2, 13 - 2.5 µM; lanes 3, 14 - 5 µM; lanes 4, 8–11, 15 – 10 µM) and CENP-T-W (lanes 5, 9 - 2.5 µM; lanes 6, 10 - 5 µM; lanes 7, 11, 12–15 - 10 µM) were mixed and incubated. The mixture was further incubated in the presence of 2.5 µM 145 bp dsDNA for 15 min. The mixtures were analyzed by native PAGE.
(B) DNA binding assays for CENP-ΔS-X and CENP-T-W conducted as in (A).
(C) DNA binding assays for the CENP-T-W-S-X or CENP-T-W-SΔC-X complexes with the indicated mutant CENP-T-W complexes (lanes 8–16 or 24–32). Wild type or mutant CENP-T-Ws (lanes 8, 11, 14, 24, 27, 30 - 5 µM; lanes 9, 12, 15, 25, 28, 31 - 10 µM; lanes 10, 13, 16, 26, 29, 32 - 20 µM) were mixed with CENP-S-X (lanes 2, 5, 8, 11, 14 - 5 µM; lanes 3, 6, 9, 12, 15 - 10 µM; lanes 4, 7, 10, 13, 16 – 20 µM) or CENP-SΔC-X (lanes 18, 21, 24, 27, 30 - 5 µM; lanes 19, 22, 25, 28, 31 - 10 µM; lanes 20, 23, 26, 29, 32 - 20 µM).
(D) Plasmid supercoiling assays performed with CENP-T-W, CENP-T-WDNA, CENP-S-X, CENP-SDNA-X, CENP-S-XDNA, and CENP-T-W-S-X complexes. Histone H3–H4 was used a positive control. Topoisomers were observed with CENP-T-W, CENP-S-X, and CENP-T-W-S-X similar to histone H3–H4, but these topoisomers were abolished with CENP-T-WDNA, CENP-SDNA-X, and CENP-S-XDNA mutant complexes.
We also analyzed the CENP-TDNA and CENP-WDNA DNA binding mutants in the context of the heterotetramer. The CENP-TDNA-W-S-X, CENP-T-WDNA-S-X, and CENP-TDNA-WDNA-S-X complexes each displayed a reduction in the formation of the discrete DNA binding complex observed for the wild-type heterotetramer (left gel of Figure 6C). Similarly, we observed a strong reduction of DNA binding for the CENP-TDNA-W, CENP-T-WDNA, and CENP-TDNA-WDNA complexes in combination with the CENP-SΔC-X mutant that does not show DNA binding activity on its own (right gel of Figure 6C). This indicates that mutation of the predicted DNA binding sites for CENP-T and CENP-W reduces the DNA binding activity of the CENP-T-W-S-X heterotetramer.
For our mutational analyses, we predicted the DNA binding sites for the CENP-T-W-S-X heterotetramer from a comparison with the DNA binding surface of canonical histones. As the histone octamer wraps DNA along the surface of the nucleosome, it is possible that DNA is also bent along the surface of the CENP-T-W-S-X heterotetramer to form a nucleosome-like structure. To test this, we used a DNA supercoiling assay for the CENP-T-W, CENP-S-X, and CENP-T-W-S-X complexes. In this assay, if a protein causes the plasmid DNA to be wrapped or bent, topoisomers are introduced that migrate faster based on gel electrophoresis. We detected supercoiling activities for the CENP-T-W, CENP-S-X, and CENP-T-W-S-X complexes similar to, although with weaker activity than, histone H3–H4 tetramers (Figure 6D). Importantly, topoisomers were abolished with the CENP-T-WDNA, CENP-SDNA-X, or CENP-S-XDNA mutant complexes (Figure 6D) despite the fact that these complexes show residual DNA binding activity (Figure 3C). These results suggest that DNA is bent or partially wrapped along the CENP-T-W-S-X complex similar to a nucleosome.
The DNA-CENP-T-W-S-X complex shows a unique DNA-protein structure
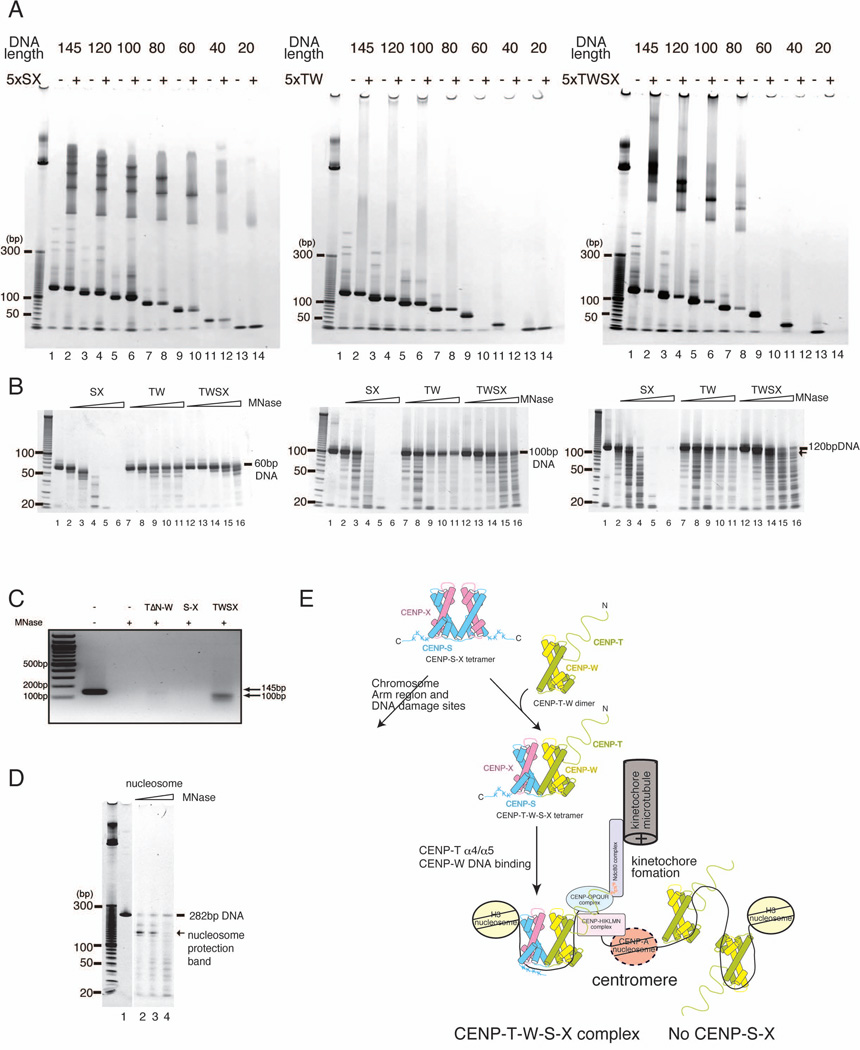
To define the DNA binding properties of the CENP-T-W-S-X heterotetramer, we next tested binding to various sized linear DNA fragments (Figure 7A). The CENP-S-X does not bind to less than 40 bp DNA, but binds to DNA with a regular ladder when more than 60 bp of DNA is present. The number of DNA-protein species increases proportionally as the length of the DNA is increased (Figure 7A). In contrast, the CENP-T-W complex binds to as little as 40 bp based on the disappearance of free DNA from the gel, but does not show a regular DNA binding pattern (middle of Figure 7A; Figure 6A). Although the CENP-T-W-S-X heterotetramer binds to short DNA fragments based on the disappearance of free DNA, this may reflect the formation of a larger assembly similar to the CENP-T-W complex as there is not a discrete band observed in the gel when less than 60 bp DNA is present. Importantly, the CENP-T-W-S-X heterotetramer displays a discrete pattern of DNA binding when more than 80 bp DNA is used (Figure 7A), indicating that the heterotetramer can form a stoichiometric complex with >80 bp DNA.
Figure 7. The CENP-T-W-S-X heterotetramer binds to ~100 bp DNA.
(A) DNA binding assays of the CENP-S-X, CENP-T-W, and CENP-T-W-S-X complexes (5 µM) with various length of DNAs (1 µM). The CENP-S-X binds DNA greater than 60 bp. The number of ladder bands increases corresponding to the length of the DNA (left). The CENP-T-W-DNA binds DNA greater than 40 bp based on reduction of free DNA (middle). A discrete DNA-CENP-T-W-S-X complex is observed when more than 80 bp DNA was used (right). DNA fragments for this assay were generated by PCR. Small DNA bands around 20 bp in all lanes represent primer DNA.
(B) MNase digestion experiments for DNA-protein complexes using either a 60 bp (left), 100 bp (middle), or 120 bp (right) fragment of DNA. For 60 bp DNA (left), both the CENP-T-W and CENP-T-W-S-X complexes protect the entire fragment from MNase digestion. The CENP-S-X complex protects a 50 bp band (lane 3 of left). The CENP-T-W and CENP-T-W-S-X complexes also protect 100 bp DNA (middle). The 120 bp DNA bound by the CENP-T-W-S-X complex produced a ~100 bp MNase-protected DNA fragment (lane 14 of right).
(C) MNase digestion experiments using the human CENP-T-W-S-X complex and 145 bp DNA. The 145 bp DNA bound by the human CENP-T-W-S-X complex produced a ~100 bp MNase-protected DNA fragment.
(D) MNase digestion of canonical nucleosomes produces 146 bp DNA.
(E) Schematic diagram of functional kinetochore formation by CENP-T-W-S-X. A (CENP-S-X)2 tetramer interacts with a CENP-T-W dimer and CENP-S-X alternately co-assembles with the CENP-T-W dimer to form a stable CENP-T-W-S-X heterotetramer. The CENP-T-W-S-X complex binds to ~100bp nucleosome free DNA and forms a nucleosome-like structure. The DNA-CENP-T-W-S-X structure is likely to be associated histone H3 containing nucleosome and recognized by other kinetochore proteins.
To determine the precise size of DNA that binds to these proteins, we next digested the DNA-protein complexes with micrococcal nuclease (MNase) (Figure 7B and C). As a control, we added histone octamers to 282 bp DNA and digested the nucleosome with MNase. As expected, we detected a ~146 bp protected DNA fragment when histone octamers were added - the predicted size that is wrapped by the nucleosome (Figure 7D). In contrast, the DNA-CENP-S-X complex was easily digested even with low concentrations of MNase. For the CENP-T-W complex, the entire region of DNA was protected by the CENP-T-W complex independent of the size of DNA used, which may be caused by a large assembly with DNA (Figure 7B). Importantly, the CENP-T-W-S-X heterotetramer produced a ~100 bp MNase-protected DNA fragment from a 120 bp DNA starting size (lane 14 in right of Figure 7B). We also detected a ~100 bp MNase-protected DNA fragment using the human CENP-T-W-S-X complex and 145 bp DNA (Figure 7C). These data are consistent with the CENP-T-W-S-X complex forming a distinct complex with ~100 bp DNA.
In total, we propose that the CENP-T-W-S-X heterotetramer forms a unique stoichiometric complex that binds to ~100 bp DNA and bends DNA similar to canonical histones to form a nucleosome-like structure (Figure 6D and 7).
Discussion
Formation of a CENP-T-W-S-X heterotetrameric complex
Previous work on kinetochore function has demonstrated that the kinetochore is specified in vertebrates by DNA sequence-independent epigenetic mechanisms. The centromere-specific histone H3-variant CENP-A is an important epigenetic mark for kinetochore specification (Barnhart et al., 2011; Black and Cleveland, 2011; Guse et al., 2011; Tachiwana et al., 2011). Indeed, CENP-T localization requires CENP-A despite the presence of an intrinsic DNA binding activity in CENP-T. Prendergast et al. (2011) recently reported that the CENP-T-W complex is not inherited by daughter cells, unlike CENP-A. This is consistent with a model in which CENP-T functions in kinetochore assembly downstream of kinetochore specification by CENP-A. However, CENP-T does not interact directly with CENP-A nucleosomes (Hori et al., 2008; Ribeiro et al., 2010) and synthetic kinetochores generated by ectopic targeting of CENP-T to non-centromere regions bypass the requirement for CENP-A nucleosomes (Gascoigne et al., 2011). Therefore, it is important to define the nature of the CENP-T-derived scaffold for kinetochore formation. Here, based on the combination of the structural, biochemical, and functional experiments, we propose that a CENP-T-W-S-X complex forms a tetrameric nucleosome-like structure to function as a scaffold for kinetochore formation (Figure 7E).
The CENP-T-W-S-X heterotetramer forms a nucleosome-like structure
Our high-resolution structural analysis demonstrates that both the CENP-S-X and CENP-T-W complexes contain histone-like structures with α helices that are homologous to those of canonical histones. Many proteins possess histone-fold regions, such as TFIID (Xie et al., 1996), NC2α-β (Kamada et al., 2001), and CHRAC 14–16 (Hartlepp et al., 2005). Although there is a debate regarding whether such histone-fold containing complexes bind to and wrap DNA similar to the histone octamer (Luger and Richmond, 1998), several reports have suggested that histone-fold containing proteins use a DNA binding surface similar to that of canonical histones (Oelgeschläger et al., 1996; Burley et al., 1997; Kamada et al., 2001). Our supercoiling experiments suggest that the CENP-T-W-S-X complex bends or partially wraps DNA along its surface and forms a nucleosome-like structure on DNA (Figure 6D). The DNA binding surface of histone H3–H4 faces the minor grove of DNA to bend DNA into a superhelix (Luger and Richmond, 1998). This canonical DNA binding surface is conserved in the CENP-S-X and CENP-T-W complexes and mutations of these residues results in the reduction of the supercoiling activities in vitro and functional kinetochore formation in vivo. We have previously shown that CENP-T-associated DNA isolated from cells is centromeric and the pattern of MNase digestion from CENP-T-associated chromatin is different from that of DNA isolated by CENP-A immunoprecipitations (Hori et al., 2008). This suggests the existence of a unique CENP-T-containing chromatin structure in vivo. Although obtaining definitive proof that this structure exists in vivo is a major future challenge, our mutational analyses strongly suggest that tetramer formation and DNA binding of the CENP-T-W-S-X is critical for kinetochore assembly in vivo.
Identification of non-canonical nucleosome-like structures
In this study, we demonstrated that the CENP-T-W-S-X heterotetramer has a number of structural and functional similarities with canonical nucleosomes. For canonical histone octamers, there are multiple potential combinations of histone variants including the H2A variants H2AZ or macro H2A, and the H3 variants H3.3 or CENP-A. Altering nucleosome composition using these variants can allow for diverse functions. The identification of a nucleosome-like CENP-T-W-S-X complex suggests that the “menu” of potential histones may extend beyond the canonical histone molecules to include additional molecules. The CENP-T-W-S-X heterotetramer plays a key role in centromere function, but similar nucleosome-like structures may also be found at other genomic locations. Indeed, the CENP-S-X complex has been shown to interact with FANCM at DNA damage sites (Yan et al., 2010; Singh et al, 2010). At these locations, the CENP-S-X complex may act as a tetramer to associate with DNA, or may associate with additional histone fold proteins to generate distinct chromatin structures in each region. Importantly, CENP-T, -W, -S, and -X are conserved throughout eukaryotes, suggesting that these tetrameric nucleosome-like structures have broad relevance. In total, these tetrameric nucleosome-like structures extend the “histone code” beyond the canonical nucleosome proteins to provide a new mechanism to form contacts with DNA.
Experimental procedures
Protein preparation
Chicken CENP-T and CENP-W or CENP-S and CENP-X were cloned into the pRSFDuet co-expression vector. 6xHis-TEV-CENP-S and StrepII-TEV-CENP-X were co-expressed in BL21(DE3)Star-pRARE2LysS by the addition of 0.2 mM IPTG for 5 h at 37°C. The CENP-S-X or CENP-T-W complexes were purified using Ni-Sepharose, StrepTactin Sepharose, TEV cleavage, and a Superdex200 column. Mutant CENP-T-W or CENP-S-X complexes were generated by site-directed PCR mutagenesis. Selenomethionine derivatives of the CENP-T-W or CENP-S-X complexes were prepared from bacteria grown in Se-met core medium (Wako) containing selenomethionine.
Crystallization and structural determination of CENP-S-X, CENP-T-W, and CENP-T-W-S-X
The complex of the histone-fold domains of CENP-T and CENP-W were crystallized by mixing equal amounts of the protein solution (20 mg/ml) and 100 mM Citrate-NaOH pH 5.0, 30% PEG 600. Crystals were harvested in a solution containing 30% PEG600 and cryoprotected. X-ray diffraction data were collected at BL38B1 and BL44XU at the SPring8 synchrotron facility. As selenomethionine crystals contained mixtures of both, methionines (M590, M607) in CENP-T were mutated to leucines. We also made cysteine mutants (CENP-T C564A, C638A/CENP-W S56C) to obtain uniform crystals. Selenomethionine multiple wavelength anomalous dispersion data was collected from the selenomethionine mutant and the phase was determined by the Phenix package (Adams et al., 2010). The model was refined using iterative modeling and refinement. The model was used to solve the cysteine mutant crystal by molecular replacement.
The CENP-SΔC-X complex was crystallized by mixing equal amount of protein solution (10 mg/ml) and 100 mM Citrate-BisPropane pH 8.2, 500 mM NaCl, 100 mM MgSO4, 30% PEG600. The selenomethionine derivative was crystallized in similar conditions, but using Citrate-BisPropane pH 9.5. Initially formed small crystals were seeded into a crystallization mixture containing 20% PEG600. Crystals were harvested in solution containing 30% PEG600 and cryoprotected. X-ray diffraction data were collected at BL38B1 and BL44XU. The initial phase of the CENP-SΔC-X complex was determined from a methyl HgCl soaked crystal using single wavelength anomalous dispersion. Selenomethionine multiple wavelength anomalous dispersion data was collected from the selenomethionine derivative to improve the phase. Two mercury atoms and four selenomethionine atoms were identified using the Phenix package. After density modification, clear electron density was found and a CENP-S-X model was built. The final model contains E6 to N104 of CENP-S and E5 to F79 with an extra Val at the C-terminus of CENP-X that was incorporated during the cloning procedure.
The CENP-T-W-S-X complex was prepared by mixing equal volumes of the CENP-T-W histone fold with the CENP-S-X complex. The complex was crystallized either directly (cubic form) or after purification by gel filtration (tetragonal form). The cubic crystal was formed by mixing equal amounts of the CENP-S-X and the CENP-T-W complex in solution at a final concentration of 10 mg/ml and mixing with an equal volume of crystallization solution containing 100 mM Tris-HCl pH 8.5, 8% PEG8000. The crystal was cryoprotected by 20% glycerol. A tetragonal crystal was formed by mixing equal amounts of protein solution (10 mg/ml) and crystallization solution containing 100 mM Tris-HCl pH 8.5, 250mM NaCl, 160 mM MgCl2, 25% PEG 4000. The crystal was cryoprotected by 20% glycerol. X-ray diffraction data were collected at BL38B1 (tetragonal form) and BL44XU (cubic form). Diffraction data were processed by the HKL2000 package. The tetragonal form structure was determined by molecular replacement program using the Phenix package using structures for the CENP-T-W histone fold and the CENP-S-X heterodimer as search models. The cubic form structure was determined by molecular replacement using the refined CENP-T-W-S-X structure as a search model.
All crystal structures shown in the Figures were created using PyMOL (DeLano Scientific LLC).
Immunofluorescence and light microscopy
Chicken DT40 cells were cultured and transfected as described previously (Okada et al., 2006). Immunofluorescent staining of DT40 cells was performed as described previously using anti-CENP-T, anti-CENP-S, or anti-Ndc80 antibodies (Hori et al. 2008; Amano et al., 2009). Immunofluorescence images were collected with a cooled EM CCD camera (QuantEM, Roper Scientific) mounted on an Olympus IX71 inverted microscope with a 100X objective together with a filter wheel and a DSU confocal unit. 15–25 Z-sections were acquired at 0.3 µm steps. Fluorescence intensity measurements were conducted using MetaMorph software (Molecular Devices). Kinetochore fluorescence intensities were determined by measuring the integrated fluorescence intensity within a 6 × 6 pixel square positioned over a single kinetochore and subtracting the background intensity of a 6 × 6 pixel square positioned in a region of cytoplasm lacking kinetochores. Maximal projected images were used for these measurements.
Supercoiling assay
Relaxed plasmid DNA (pBluescript containing ggCEN1 sequence, 4.8 kb) was pre-treated with wheat germ topoisomerase I (Promega) (4 U/1 µg of DNA) at 37°C for 60 min. Purified protein complex and the relaxed-plasmid DNA (0.3 µg) were mixed in buffer A (20 mM Tris-HCl, pH 7.5, 50 or 100 mM NaCl, 0.1 mM EDTA, 5 mM MgCl2) at total volume of 10 µl and incubated at 37°C for 15 min. Topoisomerase I (4 U) was then added and incubated for 150 min at 37°C. After the reaction, proteins were treated with proteinase K and DNA was extracted by phenol-chloroform. Samples were electrophoresized in 1 % agarose gels.
Dynamic Light Scattering
Dynamic light scattering analyses for the CENP-T-W complex or the CENP-S-X complex with DNA were performed using Zetasizer µV (Malvern Inc). A 3 µl sample was loaded onto the microcuvette. Each sample was measured three times with varying integral cycles depending on the amount of scattering and analyzed automatically using the manufacturer’s software.
MNase protection assay
Protein-DNA mixtures used for the gel shift assay were further incubated at 42°C for 1 h. The mixture was then digested by the addition of 1 µl of 1 U/µl MNase (Takara-Bio) and 2 µl of 10× buffer containing 200 mM Tris-HCl, pH 8.0, 50 mM NaCl, 25 mM CaCl2. The MNase mixture was incubated at 25°C for 10 min. After digestion, 80 µl of ProtenaseK mixture containing 20 mM Tris-HCl pH 7.5, 20 mM EDTA, 0.5% SDS was added and incubated at 37°C for 30 min. The mixture was treated with phenol-chloroform and DNA was precipitated with ethanol. The DNA mixture was resuspended in 1× loading buffer, run on 10–20 % native PAGE, and stained with ethidium bromide. Human protein-DNA mixtures were incubated for 5 min at room temperature with 0.1 U MNase (Sigma-Aldrich), then digested with proteinase K as above and run on a 2.5% agarose gel.
Supplementary Material
Acknowledgments
The authors are grateful to M. Takahashi, K. Suzuki, K. Nakaguchi, and K. Kita for technical assistance, C. Davey for the generous gift of the pUC57-601-145 plasmid, and H. Kurumizaka for the advice on histone experiments. The synchrotron radiation experiments were performed at the BL38B1 and BL44XU of SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal No. 2010B1059, 2010B1060, and 2011A1211). This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Cabinet Office, Government of Japan through its "Funding Program for Next Generation World-Leading Researchers" to TF and grants from the Searle Scholars Program and the NIH/National Institute of General Medical Sciences (GM088313) to IMC. TN is supported by The International Human Frontier Science Program Organization grant and by Grants-in-Aid for Scientific Research from MEXT. KEG is supported by an EMBO long-term fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The Protein Data Bank (PDB) ID code of the chicken CENP-S-X complex is 3B0B. PDB IDs for the chicken CENP-T histone fold-CENP-W complex are 3B0C and 3B0D. PDB IDs for the CENP-T-W-S-X heterotetramer are 3VH5 and 3VH6.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echools N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic Centromere Propagation and the Nature of CENP-A Nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Xie X, Clark KL, Shu F. Histone-like transcription factors in eukaryotes. Curr. Opin. Struct. Biol. 1997;7:94–102. doi: 10.1016/s0959-440x(97)80012-7. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nature Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced Ectopic Kinetochore Assembly Bypasses the Requirement for CENP-A Nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucl. Acids Res. 2010;38:545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlepp KF, Fernández-Tornero C, Eberharter A, Grüne T, Müller CW, Becker PB. The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions. Mol. Cell. Biol. 2005;25:9886–9896. doi: 10.1128/MCB.25.22.9886-9896.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang YH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Kamada K, Shu F, Chen H, Malik S, Stelzer G, Roeder RG, Meisterernst M, Burley SK. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell. 2001;106:71–81. doi: 10.1016/s0092-8674(01)00417-2. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luger K, Richmond TJ. DNA binding within the nucleosome core. Curr. Opin. Struct .Biol. 1998;8:33–40. doi: 10.1016/s0959-440x(98)80007-9. [DOI] [PubMed] [Google Scholar]
- Oelgeschläger T, Chiang CM, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, III, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, Hoischen C, Diekmann S, Sullivan KF. Premitotic Assembly of Human CENPs -T and -W Switches Centromeric Chromatin to a Mitotic State. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001082. e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. MHF1–MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, Fukagawa T. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J. Cell Biol. 2011;193:125–140. doi: 10.1083/jcb.201012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Vasudevan D, Chua EY, Davey CA. Crystal structures of nucleosome core particles containing the '601' strong positioning sequence. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- Xie X, Kokubo T, Cohen SL, Mirza UA, Hoffmann A, Chait BT, Roeder RG, Nakatani Y, Burley SK. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.