Abstract
The field of mechanobiology has witnessed an explosive growth over the past several years as interest has greatly increased in understanding how mechanical forces are transduced by cells and how cells migrate, adhere and generate traction. Actin, a highly abundant and anomalously conserved protein, plays a large role in forming the dynamic cytoskeleton that is so essential for cell form, motility and mechanosensitivity. While the actin filament (F-actin) has been viewed as dynamic in terms of polymerization and depolymerization, new results suggest that F-actin itself may function as a highly dynamic tension sensor. This property may help explain the unusual conservation of actin’s sequence, as well as shed further light on actin’s essential role in structures from sarcomeres to stress fibers.
Actin is a central player in many aspects of cell biology, it has been intensively studied for more than 60 years, but surprisingly we continue to realize how little we still understand about this protein. While actin was first studied in muscle, most research on actin today is focused on the crucial roles that actin plays in the cytoskeleton and non-muscle motility. The burgeoning field of mechanobiology [1] addresses questions of how mechanical forces are sensed and generated by cytoskeletal elements, and it has become clear that the transduction of such mechanical signals [2] is as important as the sensing of molecules. The cell has elaborate mechanisms for generating different actin networks in different parts of the cell, each with distinct binding proteins and functions, and our understanding of the mechanisms responsible for such specialization is still unfolding [3]. New areas of study, such as the nucleoskeleton incorporating actin, have recently emerged, while less than 10 years ago the existence of actin within the nucleus was fiercely debated. Advances in electron cryo-microscopy (Fig. 1) have provided unprecedented insights into actin filament structure and dynamics [4,5]
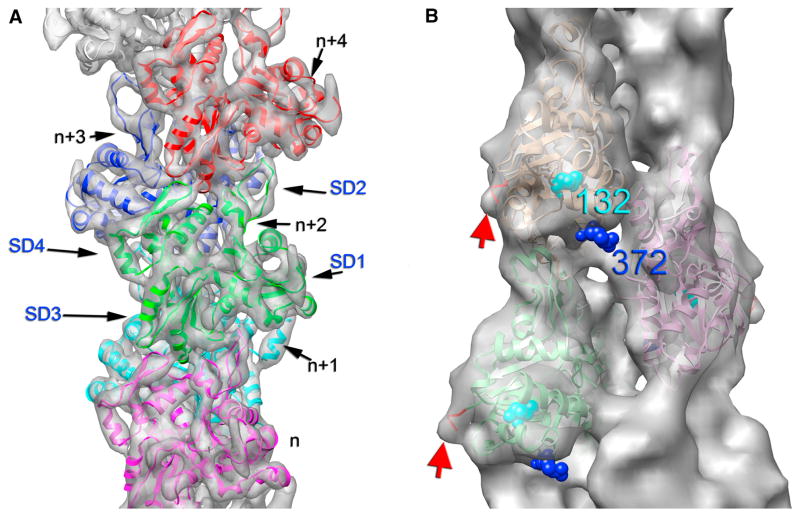
Figure 1. Structure of F-actin.
(a) The highest resolution achieved to date for an F-actin reconstruction comes from Fujii et al. [4]. An atomic model (PDB ID 3MFP) is shown built into the electron density map. Each actin subunit is in a different color in this ribbon representation. Sequential subunits in the filament are labeled from “n” (magenta) to “n+4” (red). The twist of the actin filament involves a rotation of ~ 167° between each successive subunit, and an axial rise of 27.6 Å. Cofilin, which rotates each actin subunit by ~ 5° [36], would rotate subunit “n+4” by ~ 10° away from the position of subunit “n+2”. The four subdomains of the actin subunit are labeled (SD1–SD4) within subunit “n+2”. Subdomains 1 and 2 form one major domain, while subdomains 3 and 4 form the second major domain of actin. The contact between subdomain 2 of one protomer and subdomain 1 within the protomer above it accounts for the highest radius contact in the filament, and thus can modulate the flexural rigidity of the actin filament [40]. This contact involves the DNase I-binding loop of actin in subdomain 2, which forms the top right corner of the green subunit. (b) The N-terminus in actin (red arrow) has been seen by crystallography to be an unstructured region of the protein, but changes in three of these seven amino acids is the difference between life and death for yeast [50]. The H372R mutation (blue spheres), fairly distant from the N-terminus, can rescue lethal changes in the N-terminus through a putative allosteric pathway [14]. Such a pathway could explain why mutations in buried residue 132 (cyan spheres) cause hereditary myopathies [15]. Figure reproduced with permission from [5].
One of the most striking features about actin, in addition to its abundance, has been its exquisite degree of sequence conservation. From chickens to humans, an evolutionary distance of more than 300 million years, every one of the 375 residues in the skeletal muscle isoform has been conserved. If one looks at an evolutionary distance of more than 1 billion years, ~ 90% of the residues are identical between yeast actin and the cytoplasmic isoform of human actin. While suggestions have been made about why almost all actin residues might be under selective pressure, we have no definitive answer at this point.
One possibility for actin’s anomalous sequence conservation is that the interaction of actin with more than 100–200 actin-binding proteins might constrain many residues. But this argument ignores the fact that many actin-binding proteins have significantly diverged over the same evolutionary distances (e.g., from yeast to humans). Further, the residues in actin that are not absolutely conserved [6] are mainly on the surface of the filament where they would directly interact with actin-binding proteins. A quite different argument comes from the observation that highly expressed proteins evolve slowly [7], presumably as a means to prevent protein misfolding. This argument may explain some of the anomalous sequence conservation, since actin is one of the most highly expressed proteins in many cells, but is unlikely to explain why every amino acid appears to be under rather intense selective pressure. We would like to advance a different hypothesis in this Minireview, one supported by a series of recent papers [4,5,8–11], suggesting that cooperative and allosteric properties of the actin filament are essential to cellular function, and that the internal networks within the actin subunit needed to maintain such allosteric linkages [12] have placed every residue under selective pressure.
A Working Hypothesis
Allosteric interactions may explain why buried residues in actin, which cannot interact with actin-binding proteins in muscle such as myosin, tropomyosin, troponin and α-actinin, are responsible for hereditary myopathies [13]. For example, it has been shown that replacing residue 372 in yeast actin with the residue found at this position in vertebrate muscle actin (the H372R mutation) led to severe growth defects [14]. However, substitution of four N-terminal muscle actin residues into the N-terminus of yeast actin restored the viability of cells with the H372R mutation [14]. Since these two regions are widely separated in both G- and F-actin (Fig. 1b), the best explanation for this effect involves an allosteric linkage between these regions [14]. Structural results showing coupled conformational states in F-actin are completely consistent with such an allosteric linkage [5], and this can explain why mutations in residue 132, buried in the subunit but located between the N- and C-terminus (Fig. 1b), can cause hereditary myopathies [15].
In contrast, the bacterial actin homologs have diverged considerably in sequence, so much so that many of them are as different from each other as they are from actin [16]. While it is still an open question as to whether all or even most of these bacterial proteins form filaments, the filaments formed by all bacterial actin-like proteins studied thus far are significantly different from F-actin [17–21]. If our hypothesis is correct, the bacterial actin-like filaments will not display the cooperativity and allostery observed for F-actin [6]. Some specific predictions can therefore be made in this review about how the bacterial actin-like filaments will behave differently from F-actin.
A Finely Tuned Filament
To explore this idea of a highly tuned actin filament that has emerged from extensive evolutionary selection we must start by abandoning the notion that F-actin is merely a passive cable, existing in a single state, to which other proteins can bind. The notion that physical stresses on the actin filament might modulate the interaction with actin-binding proteins has appeared in a number of places. A recent paper on the interaction of formins with actin [22] concluded “Our data have opened up the possibility that actin elongation and remodeling could be regulated by axial torsion in the filament.” It has previously been shown that nucleation of an actin filament by formins can cause long-range conformational changes in the actin filament [23], just as nucleation of actin filaments by gelsolin has been shown earlier to cause such long-range changes [24,25]. These long-range perturbations tell us that the different conformational states accessible by actin must be comparable energetically, so that nucleation by a particular protein is able to bias the distribution of states. Nucleation of actin filaments by different proteins thus provides a means for the cell to differentiate one actin filament from another [3].
A less static and more active picture of the actin filament has arisen from new insights into the dynamic properties of actin filaments. Actin-based structures that were considered to be static, such as the core of the stereocilium (responsible for the mechanotransduction of sound) have now been shown to be dynamic, at least in the sense that there is a continuous flux of actin subunits through these filaments [26]. Consider muscle, where a passive view of actin has been dominant historically. The regulation of myosin heads binding to F-actin, and therefore the generation of force, has been viewed largely due to tropomyosin strands moving across a fixed actin surface. But a number of papers have shown that actin can be modified, either chemically [27–30], by mutation [31] or by proteolysis [32] in a way that inhibits myosin force generation without inhibiting either the binding of myosin to actin or the actin-activated activation of myosin’s ATPase activity. The simplest explanation for these observations is that actin must undergo structural transitions during actomyosin force generation, and these modifications of actin inhibit such structural transitions. Supporting the notion of structural transitions in F-actin, we have shown that naked actin filaments in vitro exist in a multiplicity of discrete structural states [5].
Reconciling Two Different Views
A different picture of F-actin was presented in another recent paper [4], where it was argued that F-actin is quite homogeneous structurally, and that “F-actin is not so flexible” with respect to the large literature showing that the helical twist of F-actin can be quite variable [33–36]. For example, the protein cofilin changes the average twist of F-actin (Fig. 1) by ~ 5° per subunit [36], while in bundles with scruin [35] the twist of actin subunits ranged from 142.5° to 176.5°, deviating widely from the average twist within these filaments of ~ 167°. In the actin angle layered aggregate [37], which is formed in solution prior to specimen preparation for electron microscopy, the angular disorder is locked into the structure, so specimen preparation can be discounted as a source of the variability in twist of actin. These aggregates yielded an rms deviation of ~ 6° per subunit [37].
How can these very different observations, of variable twist and polymorphic filaments, versus relatively fixed twist and a single structure, be reconciled? We think that the answer lies in specimen preparation for cryo-EM, and that understanding the differences between the results obtained is likely to have great biological significance. In preparing a sample for cryo-EM, filaments in buffer are applied to a holey carbon film on an EM grid, and then blotted so that a thin film is formed prior to plunging into a cryostat for vitrification. In the process of creating this film, very large forces can exist on filaments due to both fluid flow and transverse compression. Fujii et al. [4] explicitly stated that the high resolution they achieved was due in part to the use of very thin films which improved the signal to noise ratio, and that blotting conditions were chosen to make “F-actin as straight as possible.” We normally think of the straightness of filaments in solution, in the absence of external forces, as arising from only two physical parameters: the temperature of the solution (T) and the flexural rigidity of the polymer (a). Thus, the persistence length λ for a filament is simply given by λ =a/kT, If blotting conditions are changing the observed flexibility or straightness of these filaments, it is a prima facie argument that forces are being introduced. The use of fluid flow to intentionally stretch and straighten polymers is not novel, and has been used in many experiments involving DNA. But such straightening may also arise in the case of F-actin from the compressive forces perpendicular to the filament axis when the filament experiences the surface tension resulting from a very thin film. Tomographic reconstructions of axonemes in thin ice showed extreme flattening, with the suggestion that the flattening arose from this large surface tension [38]. Most importantly, observations have already been made by Greene et al. [39] about F-actin filaments confined between two mica surfaces (Fig. 2), which appear to be a good analog for the thin films being used for cryo-EM. Surprisingly, these filaments become anomalously stiff under compression, which is consistent with the structural homogeneity and straightening of the actin filaments seen in Fujii et al. [4]. Greene et al. proposed a two state model for actin, and suggested that compression leads to the stress-stiffening of filaments by forcing subunits into a state that is stiffer than the one normally populated. This is consistent with all of the EM results.
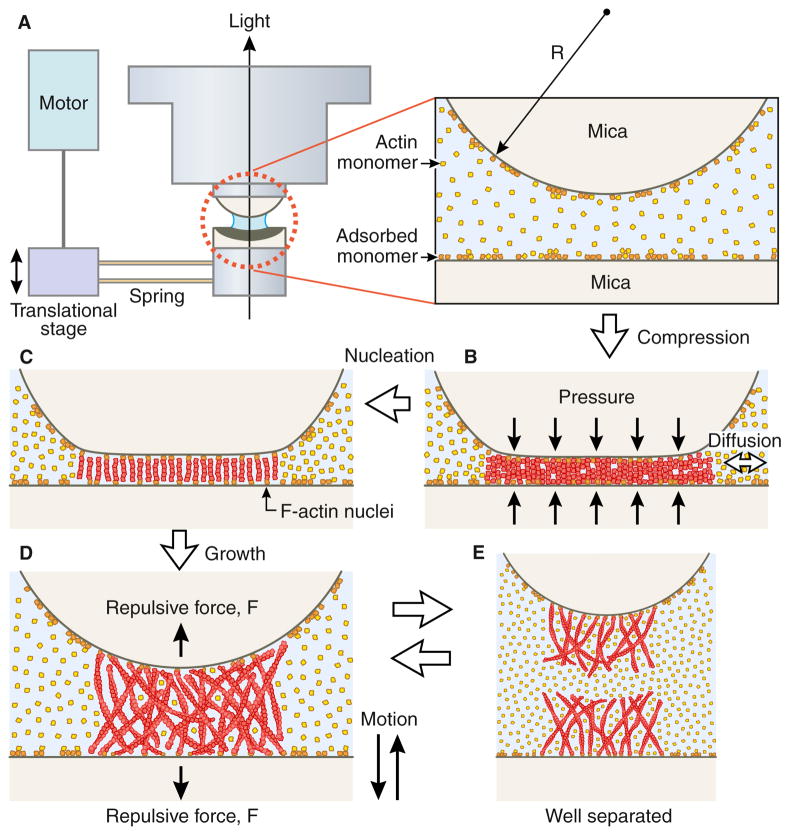
Figure 2. Mechanical Compression of Actin Filaments.
An apparatus was built to mechanically compress actin filaments between two mica surfaces [39]. Surprisingly, the authors found actin filaments stiffened under compression, suggesting that the actin filament, in the absence of any other proteins, might function as a sensing element in cellular systems. Given a persistence length for “normal” F-actin of ~ 10 μ, when the two mica surfaces come close together (to less than 0.1 μ) we would expect that all filaments will be oriented nearly parallel to the mica surfaces, and not perpendicular as drawn. We thus think that this apparatus may be a good model for the filaments confined to a thin film in Fujii et al [4]., and explain why compression leads to a stiffening of actin filaments. We see the stiffening as likely arising from mechanically forcing all actin subunits into one structural state, one in which subdomain 2 is highly ordered. Figure reproduced from [39] with permission.
We may extend the assumption of Greene et al. to suggest that actin filaments may exist in at least three states of macroscopic flexibility. When filaments are under axial tension or transverse compression there is a stiff state, while in the absence of forces there is an ensemble average over a number of states [5] that yields the persistence length for “normal” F-actin. However, it has been shown that under conditions where subdomain 2 of actin becomes disordered, the filaments become anomalously flexible [40], which we can treat as a third macroscopic state. As we have shown [8], cofilin can both substantially displace subdomain 2 of actin as well as cause it to be disordered. One would thus expect that cofilin binding to F-actin might make it more flexible, and the cofilin-induced increase in F-actin flexibility has already been reported [41]. This change in F-actin flexibility makes simple physical sense, since the resistance to bending will scale as the fourth power of the radial mass distribution, and subdomain 2 forms the highest radius contact in the actin filament (Fig. 1a). If one compares ~ 80 crystal structures of actin that now exist, the greatest structural variance is in subdomain 2 and the DNase I-binding loop within subdomain 2. Strikingly, subdomain 2 and the DNase I-binding loop are among the regions of lowest structural variance in the EM reconstruction of Fujii et al. [4], in keeping with our argument that the compressive thin films are inducing this unusual structural homogeneity. The structural homogeneity of subdomain 2 is what is giving these filaments their anomalous rigidity. It is difficult to reconcile such structural homogeneity with spectroscopic observations from filaments in solution suggesting large, and discrete, changes in the conformation of subdomain 2 [42].
What is the molecular mechanism that couples axial tension on a filament (or transverse compression) to a stabilization of subdomain 2? At this point we cannot answer the question, and we expect that this issue will motivate many studies in the future. However, it is tempting to speculate that since the contact between subdomain 4 of one subunit and subdomain 3 of a subunit above it is a relatively invariant interface in F-actin [5], tension along subdomains 3 and 4 within a subunit may be communicated through the hinge region separating the two major domains of actin forcing subdomain 2 into a specific vertical orientation.
Testable Predictions
The hypothesis that we are proposing immediately leads to a number of testable predictions. One is that the elastic stiffening observed for F-actin [39] should not be seen for the filaments formed by bacterial actin-like proteins, such as ParM and AlfA. Another is that tension on an actin filament should be observable either biochemically or spectroscopically, since this tension should change the distribution of structural states. Such a result has already been observed [43], where a change in fluorescence of a probe attached to the C-terminus of actin was observed as a function of tension. The resulting labeled actin filament was therefore described as a “bio-nano strain gauge”. Our explanation for this effect arises from the fact that in the absence of tension the C-terminal region of F-actin, like subdomain 2, can exist in a number of discretely different states [5]. Under conditions where the filament is compressed in a thin film [4,39] we suggest that this region becomes structurally homogeneous.
A third prediction is that proteins which change the twist of F-actin, such as cofilin/ADF [36], should bind much less avidly to actin filaments under tension, since tension should greatly reduce the variability in twist within these filaments. A new paper, entitled “Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament” has observed precisely this expected behavior [9]. We believe that we can explain the molecular basis for this observation, and it also provides insights into the coupling between multiple structural states and variable twist in F-actin.
Just as Fujii et al. have observed structurally homogeneous actin filaments in contrast to the structural heterogeneity that we observe in thicker ice, they observe a distribution of twist that can be parameterized as corresponding to a random angular disorder of ~ 2.5° per subunit, while our filaments have an observed distribution of ~ 6° per subunit (Fig. 3), consistent with earlier estimates from both negatively stained single actin filaments [34] and angle layered aggregates [37]. It has been shown that cofilin actually stabilizes an existing twist of F-actin that is present in vitro in naked actin filaments, rather than imposing a twist that would never be seen in the absence of cofilin [44]. Several papers have suggested that the slow initial binding of cofilin/ADF to F-actin can be explained by the limited number of sites where cofilin can initially attach [45,46], as one would expect in a structurally heterogeneous F-actin (while all actin subunits would be in identical environments in a homogeneous filament). Further, cofilin needs to shift subdomain 2 of actin when it binds to the filament [8]. Since we suggest that tension on a filament or transverse compression in a thin film stabilizes subdomain 2, tension should inhibit cofilin from binding to F-actin. Using both in vivo and in vitro assays, the authors show that when an actin filament is under tension cofilin binding is reduced by a factor of two to three. They used optical tweezers to apply tension to single actin filaments in vitro, and used bundles of actin filaments in vivo which could be stretched by micromanipulation. As a consequence of the reduced binding, they show that the severing of F-actin by cofilin is decreased when a filament is under tension. This has great cell biological significance, since such tension may regulate which actin filaments, whether in stress fibers, filopodia or cleavage furrows, will be severed by cofilin
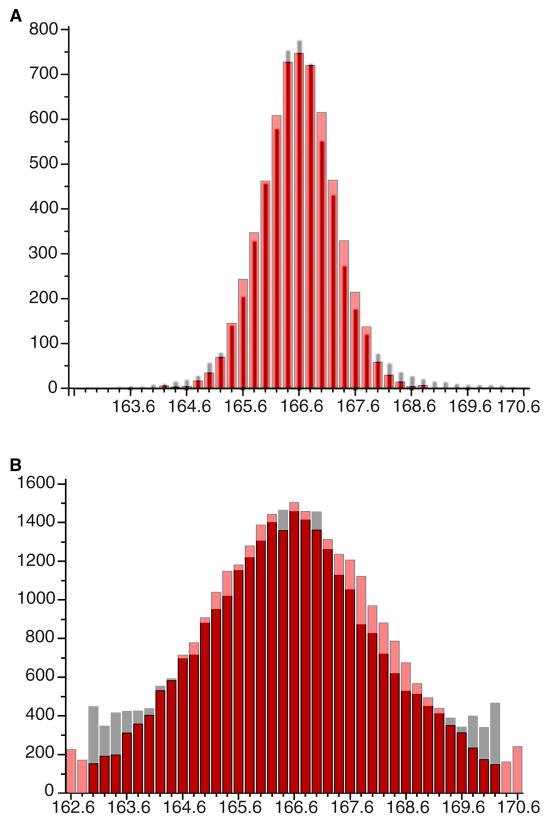
Figure 3. Variability in Twist within F-Actin, As Observed by Cryo-EM.
a, The histogram for twist measured by Fujii et al.[4] is shown in grey, while a simulation involving 2.5° rms cumulative random angular disorder is shown in red. In both cases, segments of F-actin containing ~ 20 subunits are sorted by multi-reference alignment using 41 different atomic models of F-actin, having a twist from 162.6° to 170.6°. In the simulation, actin subunits are added to a previous subunit with a twist of 166.6° + δ, where δ is a random variable with zero mean and a standard deviation of 2.5°. The random disorder is thus cumulative, as deviations from the expected angular position build as the square root of the number of subunits. b, The variability in twist measured for our filaments [5] using the same method described in (a), where the real data are in gray. In contrast to Fujii et al.[4] where the filament segments contained ~ 20 actin subunits, our boxes [5] contained ~ 17 subunits, and this has been used in the simulation. The reduction in box length from 20 to 17 subunits should introduce a broadening by ~ 8% (20½/17½). The simulation (in red) provides the best match when δ has a standard deviation of 6°.
The opposite prediction can also be made, which is that proteins that bind to F-actin and stretch it should show a higher affinity of binding to an actin filament under tension. Remarkably, this has also now been observed [11]. It was shown by x-ray diffraction that the rise per subunit of actin increased by ~ 0.4 % when muscle goes into full tension [47]. It was subsequently shown [48] that the binding of myosin heads in the absence of tension can elongate the actin filament by 0.2%, explaining half of the extension observed. We also know that the conformation of actin in the rigor complex (in the absence of ATP) with myosin is quite similar to the structure of the homogeneous naked actin filament under axial tension/transverse compression [4]. So the prediction is that if an actin filament is under tension, myosin should bind more avidly, and that is exactly what Uyeda and colleagues have now observed in vivo [11]. As with the cofilin result [9], this has enormous cell biological implications, creating another means for the cell to regulate the binding of myosin to F-actin in addition to the regulation provided by a large repertoire of other proteins such as tropomyosin, troponin, calponin, myosin binding protein C, etc. It also has implications for further understanding stretch-activation of muscle [49]. Since actin filaments, in both muscle and non-muscle cells, are associated with a large number of actin-binding proteins, including nucleators, capping proteins and cross-linking proteins, it will be extremely interesting to understand how these other proteins modulate and regulate the response of an actin filament to tension.
Summary
We have proposed a hypothesis that tension on an actin filament can induce structural transitions from a multiplicity of states [5] to largely a single state [4], and that this single state will have a higher affinity for myosin than “normal” F-actin [11] and a lower affinity for cofilin [9]. This hypothesis can explain spectroscopic observations made from actin filaments under tension [43] and begins to address how allosteric relations in actin [14] mediate such conformational transitions. This hypothesis is testable, and we think that it will provide new understanding about why actin’s sequence has been anomalously conserved over one billion years of eukaryotic evolution.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478:260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Curr Biol. 2011;21:R560–R569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 5.Galkin VE, Orlova A, Schroder GF, Egelman EH. Structural polymorphism in F-actin. Nat Struct Mol Biol. 2010;17:1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egelman EH. Actin allostery again? Nat. Struct Biol. 2001;8:735–736. doi: 10.1038/nsb0901-735. [DOI] [PubMed] [Google Scholar]
- 7.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galkin V, Orlova A, Kudryashov D, Solodukhin A, Reisler E, Schroeder GN, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proc Nat Acad Sci, USA. 2011 doi: 10.1073/pnas.1110109108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195:721–727. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, Guerin C, Martiel JL, De La Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uyeda TQ, Iwadate Y, Umeki N, Nagasaki A, Yumura S. Stretching Actin Filaments within Cells Enhances their Affinity for the Myosin II Motor Domain. PLoS ONE. 2011;6:e26200. doi: 10.1371/journal.pone.0026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 13.Feng JJ, Marston S. Genotype-phenotype correlations in ACTA1 mutations that cause congenital myopathies. Neuromuscul Disord. 2009;19:6–16. doi: 10.1016/j.nmd.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 14.McKane M, Wen KK, Boldogh IR, Ramcharan S, Pon LA, Rubenstein PA. A mammalian actin substitution in yeast actin (H372R) causes a suppressible mitochondria/vacuole phenotype. J Biol Chem. 2005;280:36494–36501. doi: 10.1074/jbc.M506970200. [DOI] [PubMed] [Google Scholar]
- 15.Laing NG, Dye DE, Wallgren-Pettersson C, Richard G, Monnier N, Lillis S, Winder TL, Lochmuller H, Graziano C, Mitrani-Rosenbaum S, et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1) Hum Mutat. 2009;30:1267–1277. doi: 10.1002/humu.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galkin VE, Orlova A, Rivera C, Mullins RD, Egelman EH. Structural polymorphism of the ParM filament and dynamic instability. Structure. 2009;17:1253–1264. doi: 10.1016/j.str.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polka JK, Kollman JM, Agard DA, Mullins RD. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J Bacteriol. 2009;191:6219–6230. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popp D, Narita A, Maeda K, Fujisawa T, Ghoshdastider U, Iwasa M, Maeda Y, Robinson RC. Filament structure, organization, and dynamics in MreB sheets. J Biol Chem. 2010;285:15858–15865. doi: 10.1074/jbc.M109.095901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popp D, Narita A, Ghoshdastider U, Maeda K, Maeda Y, Oda T, Fujisawa T, Onishi H, Ito K, Robinson RC. Polymeric structures and dynamic properties of the bacterial actin AlfA. J Mol Biol. 2010;397:1031–1041. doi: 10.1016/j.jmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Popp D, Xu W, Narita A, Brzoska AJ, Skurray RA, Firth N, Ghoshdastider U, Maeda Y, Robinson RC, Schumacher MA. Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation. J Biol Chem. 2010;285:10130–10140. doi: 10.1074/jbc.M109.071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuno H, Higashida C, Yuan Y, Ishizaki T, Narumiya S, Watanabe N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 23.Papp G, Bugyi B, Ujfalusi Z, Barko S, Hild G, Somogyi B, Nyitrai M. Conformational changes in actin filaments induced by formin binding to the barbed end. Biophys J. 2006;91:2564–2572. doi: 10.1529/biophysj.106.087775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlova A, Prochniewicz E, Egelman EH. Structural dynamics of F-actin. II. Co-operativity in structural transitions. J Mol Biol. 1995;245:598–607. doi: 10.1006/jmbi.1994.0049. [DOI] [PubMed] [Google Scholar]
- 25.Prochniewicz E, Zhang Q, Janmey PA, Thomas DD. Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J Mol Biol. 1996;260:756–766. doi: 10.1006/jmbi.1996.0435. [DOI] [PubMed] [Google Scholar]
- 26.Schneider ME, Belyantseva IA, Azevedo RB, Kachar B. Rapid renewal of auditory hair bundles. Nature. 2002;418:837–838. doi: 10.1038/418837a. [DOI] [PubMed] [Google Scholar]
- 27.Prochniewicz E, Yanagida T. Inhibition of sliding movement of F-actin by crosslinking emphasizes the role of actin structure in the mechanism of motility. J Mol Biol. 1990;216:761–772. doi: 10.1016/0022-2836(90)90397-5. [DOI] [PubMed] [Google Scholar]
- 28.Prochniewicz E, Katayama E, Yanagida T, Thomas DD. Cooperativity in F-actin: chemical modifications of actin monomers affect the functional interactions of myosin with unmodified monomers in the same actin filament. Biophys J. 1993;65:113–123. doi: 10.1016/S0006-3495(93)81057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim E, Bobkova E, Hegyi G, Muhlrad A, Reisler E. Actin cross-linking and inhibition of the actomyosin motor. Biochemistry. 2002;41:86–93. doi: 10.1021/bi0113824. [DOI] [PubMed] [Google Scholar]
- 30.Kim E, Bobkova E, Miller CJ, Orlova A, Hegyi G, Egelman EH, Muhlrad A, Reisler E. Intrastrand cross-linked actin between Gln-41 and Cys-374. III. Inhibition of motion and force generation with myosin. Biochemistry. 1998;37:17801–17809. doi: 10.1021/bi981286b. [DOI] [PubMed] [Google Scholar]
- 31.Drummond DR, Peckham M, Sparrow JC, White DC. Alteration in crossbridge kinetics caused by mutations in actin. Nature. 1990;348:440–442. doi: 10.1038/348440a0. [DOI] [PubMed] [Google Scholar]
- 32.Schwyter DH, Kron SJ, Toyoshima YY, Spudich JA, Reisler E. Subtilisin cleavage of actin inhibits In vitro siding movement of actin filaments over myosin. J Cell Biol. 1990;111:465–470. doi: 10.1083/jcb.111.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson J. Axial period of actin filaments: electron microscope studies. Nature. 1967;213:353–356. [Google Scholar]
- 34.Egelman EH, DeRosier DJ. Image analysis shows that variations in actin crossover spacings are random, not compensatory. Biophys J. 1992;63:1299–1305. doi: 10.1016/S0006-3495(92)81716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431:104–107. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- 36.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egelman EH, Francis N, DeRosier DJ. Helical disorder and the filament structure of F-actin are elucidated by the angle-layered aggregate. J Mol Biol. 1983;166:605–629. doi: 10.1016/s0022-2836(83)80286-1. [DOI] [PubMed] [Google Scholar]
- 38.McEwen BF, Marko M, Hsieh CE, Mannella C. Use of frozen-hydrated axonemes to assess imaging parameters and resolution limits in cryoelectron tomography. J Struct Biol. 2002;138:47–57. doi: 10.1016/s1047-8477(02)00020-5. [DOI] [PubMed] [Google Scholar]
- 39.Greene GW, Anderson TH, Zeng H, Zappone B, Israelachvili JN. Force amplification response of actin filaments under confined compression. Proc Natl Acad Sci U S A. 2009;106:445–449. doi: 10.1073/pnas.0812064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orlova A, Egelman EH. A conformational change in the actin subunit can change the flexibility of the actin filament. J Mol Biol. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 41.McCullough BR, Blanchoin L, Martiel JL, De La Cruz EM. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J Mol Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozuka J, Yokota H, Arai Y, Ishii Y, Yanagida T. Dynamic polymorphism of single actin molecules in the actin filament. Nat Chem Biol. 2006;2:83–86. doi: 10.1038/nchembio763. [DOI] [PubMed] [Google Scholar]
- 43.Shimozawa T, Ishiwata S. Mechanical distortion of single actin filaments induced by external force: detection by fluorescence imaging. Biophys J. 2009;96:1036–1044. doi: 10.1016/j.bpj.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin Depolymerizing Factor Stabilizes an Existing State of F-Actin and Can Change the Tilt of F-Actin Subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. J Mol Biol. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 46.Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- 47.Huxley HE, Stewart A, Sosa H, Irving T. X-ray diffraction measurements of the extensibility of actin and myosin filaments in contracting muscle. Biophys J. 1994;67:2411–2421. doi: 10.1016/S0006-3495(94)80728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsaturyan AK, Koubassova N, Ferenczi MA, Narayanan T, Roessle M, Bershitsky SY. Strong binding of myosin heads stretches and twists the actin helix. Biophys J. 2005;88:1902–1910. doi: 10.1529/biophysj.104.050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pringle JW. The Croonian Lecture, 1977. Stretch activation of muscle: function and mechanism. Proc R Soc Lond B Biol Sci. 1978;201:107–130. doi: 10.1098/rspb.1978.0035. [DOI] [PubMed] [Google Scholar]
- 50.McKane M, Wen KK, Meyer A, Rubenstein PA. Effect of the substitution of muscle actin-specific subdomain 1 and 2 residues in yeast actin on actin function. J Biol Chem. 2006;281:29916–29928. doi: 10.1074/jbc.M602251200. [DOI] [PubMed] [Google Scholar]