Abstract
Traditional drug discovery efforts have resulted in the approval of a handful of receptor tyrosine kinase (RTK) inhibitors; however, their discovery relied solely on screening recombinant kinases, often with poor cellular activity outcome. The ability to screen RTKs in their natural environment is sought as an alternative approach. We have adapted a novel strategy utilizing a green fluorescent protein–labeled SRC homology 2 domain–based biosensor as a surrogate reporter of endogenous epidermal growth factor receptor (EGFR) activity in A549 cells. Upon activation of the receptor, EGFR function in live cells is measured by the number of green granules that form. Here we describe assay miniaturization and demonstrate specificity for EGFR through its chemical inhibition and RNAi-dependent knockdown resulting in complete abrogation of granule formation. Gefitinib and PD 153035 were identified as hits in a pilot screen. This approach allows for the identification of novel EGFR modulators in high-throughput formats for screening chemical and RNAi libraries.
Introduction
Receptor tyrosine kinases (RTKs) constitute a subclass of signaling receptors anchored at the cell surface, with intrinsic tyrosine kinase activity triggering transduction signals in response to ligand binding. They regulate crucial cellular processes such as proliferation, differentiation, survival, migration, and metabolism, and for this reason RTK activity is tightly regulated in normal cells. In contrast, aberrant RTK activity resulting from receptor mutation is involved in oncogenesis and in the progression of many cancers.1 Therefore, RTKs constitute an important class of targets for cancer therapeutics and several small molecule RTK inhibitors (RTKis) have reached widespread use in the clinic. In particular, RTKis such as gefitinib targeting epidermal growth factor receptor (EGFR), which is commonly overexpressed and constitutively activated in human cancers, trigger a good initial response in patients for the treatment of non–small-cell lung cancer (NSCLC).2 However, resistance to RTKis has emerged as a major clinical problem, limiting the efficacy of currently available RTKis targeting kinase activity and their use in cancer patients.3
Novel inhibitors of RTKs are therefore needed because today's approaches to their identification rely on screening purified recombinant kinase domains in vitro against large kinase-focused or fragment-based chemical libraries. Though this traditional approach provided some success in identifying kinase inhibitors that later progressed to the clinic and were approved as drugs for the treatment of cancer, it suffers from several limitations, with the most detrimental one being the identification of potent hits in vitro with poor cellular activity. In addition, these in vitro assays allow for the identification of inhibitors of the recombinant kinase domain, while ignoring signaling events associated with RTK activation and internalization in cells. Therefore, it raises the question for finding alternative approaches to screen for novel potent and cell-permeable modulators.
Current approaches to quantify EGFR activation in cells mainly rely on immunoblots and microarrays. Detecting phosphorylated EGFR by immunoblotting is time consuming, labor intensive, and not easily amenable to large-scale studies.4 For this reason, micro-Western arrays have been developed, in theory allowing for much higher throughput;4 however, Western blotting inherently requires cell lysis and protein extraction, which prevents EGFR activity measurement directly in cells and precludes the method's use for live cell studies.
Another approach relies on microarray-based platforms, which aim at probing the abundance of phosphorylated EGFR on a solid substrate in a large number of samples in miniaturized formats.5,6 Microarray data, however, are prone to substantial noise, and side by side experiments with Western blots demonstrated poor correlation between lysate microarrays and immunoblots for the detection of phosphorylated EGFR.4,6 An alternative microarray method relying on fluorescence resonance energy transfer (FRET) measurement by fluorescence lifetime imaging microscopy was developed to detect phosphorylated tyrosines in a panel of proteins including EGFR.7 A limitation of this method lies in the requirement for fluorescent-tagged proteins for detection and therefore it is not amenable to measuring endogenous EGFR activity; in addition, it requires cell lysis similar to the antibody-based approaches already described and it is therefore not compatible with live-cell measurements.
Other reported cell-based assays in microtiter plate formats include immuno-histochemical methods such as the in-cell Western system,8 which requires a dedicated scanner employing two near-infrared lasers and detectors, or the use of automated fluorescence microscopy to measure EGFR phosphorylation.9 Such approaches rely on immunostaining and therefore do not allow measurements of EGFR activity in live cells. An alternative reported method provides an indirect measure of receptor activation by relying on measuring the translocation of a downstream effectors as surrogate assay receptors.10 To date, despite the large number of studies aiming at measuring EGFR activation in cells, there is no assay available that allows the identification of novel EGFR modulators in live cells in high-throughput format.
Ligand binding to RTKs triggers changes to the receptor conformation through homodimerization of the receptor, leading to the activation of intrinsic tyrosine kinase activity in cells. Subsequently, adaptor or signaling molecules with SRC homology 2 (SH2) bind to phosphotyrosines and recruit downstream signaling proteins. In order to develop an alternative screening method that would allow measurement of RTK activity in live cells, we devised a strategy taking advantage of the high affinity of SH2 domains to RTKs. It relies on the expression of an SH2 domain protein with high affinity for RTKs, fused to green fluorescent protein (GFP).11 We hypothesized that upon ligand binding, RTK activation and phosphorylation of tyrosine residues would lead to the recruitment of the fluorescent SH2 domain–based biosensor, followed by receptor endocytosis and recycling. Imaging of receptor clustering and endocytosis allowed by the recruitment of our fluorescent domain-based biosensor would enable us to visualize and quantify granule formation as a surrogate measure for endogenous RTK activity in live cells. If successful, it would offer significant advantages over existing assays in that it does not require EGFR modification but rather allows measurement of endogenous EGFR function in its natural environment, in real time, and in live cells.
To validate our strategy, we introduced the biosensor domain into the A549 cell line, resulting in an engineered and stable A549 EGFR biosensor cells (A549-EGFRB cells); we further demonstrated granule formation in live cells and their subsequent intracellular trafficking upon addition of epidermal growth factor (EGF) to resting cells. In this report, we describe assay miniaturization to 384-well format and its optimization for screening chemical libraries. We present the results of a study with a panel of 26 known effectors to validate our newly optimized assay and discuss the broad applications of the strategy for chemical and RNAi screening for modulators of endogenous RTK function in live cell models.
Materials and Methods
Reagents
A549-EGFRB cells11 derived from mutant KRAS and wild-type EGFR human lung adenocarcinoma cells, were obtained from Sigma-Aldrich and were cultured under a humidified atmosphere at 37°C/5% CO2–95% air in RPMI-1640 media (Sigma-Aldrich: R0883) containing 10% fetal bovine serum (PAA Laboratories GmbH: 100-125), 100 U/mL penicillin, 100 μg/mL streptomycin, and 1 μg/mL puromycin. The sources of RTK ligands and cytokines are described in Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/adt). Dharmafect-1 was purchased from Thermo Fisher Scientific. The anti-EGFR mouse monoclonal antibody (mAb) conjugated to AF647 was purchased from Santa Cruz Biotech (Santa Cruz, CA). EGFR siRNA (guide sequence: AUUUCUCAUGGGCAGCUCCTT; passenger sequence: GGAGCUGCCCAUGAGAAAUTT) was custom synthesized at the High-Throughput Screening (HTS) Core Facility, Memorial Sloan-Kettering Cancer Center (MSKCC). Silencer select negative control 112 referred to as “scrambled siRNA” in this article was purchased from Ambion. Paraformaldehyde was obtained as a 32% (v/v) aqueous solution (cat. no. 15714-S) from Electron Microscopy Sciences. Other reagents were obtained as previously described.13
EGFR Knockdown
A549-EGFRB cells were seeded either in 384-well microplates (#3985, Corning Life Sciences) at 2000 cells per well or in chambered coverglass (Labtek II chambered #1.5 german coverglass system, Nunc) at 40,000 cells per chamber in media without penicillin or streptomycin and incubated for 24 h at 37°C. Scrambled siRNA and EGFR siRNA complexes were preformed for 15 min at room temperature by mixing Dharmafect-1 transfection reagent and the siRNA in optiMEM prior to addition to cells at a final siRNA concentration of 100 nM and 0.1 μL Dharmafect-1 reagent per well or 0.5 μL per chamber. Four days post transfection, cells were fixed with 4% paraformaldehyde (v/v) in phosphate-buffered saline (PBS) for 20 min at room temperature and washed once with PBS, stained, and imaged 70 min following 500 nM EGF stimulation.
EGFR Immunostaining
A549-EGFRB cells were seeded either in 384-well microplates at 5000 cells per well or in chambered coverglass at 100,000 cells per chamber and incubated at 37°C. At 16 h post cell seeding, the media was aspirated, fresh media containing 500 nM EGF was added, and cells were incubated at room temperature for 70 min. Cells were then fixed with 4% paraformaldehyde (v/v) in PBS for 20 min at room temperature and washed once with PBS. Permeabilization and nuclei staining were then performed by incubating cells for 10 min at room temperature with 10 μM Hoechst in 0.05% (v/v) Triton X-100 in PBS. After two PBS washes, cells were blocked with 10% goat serum in PBS for 1.5 h at room temperature, followed by immunostaining of EGFR using mouse anti-EGFR conjugated to Alexa647 mAb diluted 1:20 in 1% goat serum (v/v) in PBS for 1 h. After two washes with PBS, cells were kept at 4°C in the dark before imaging.
Image Acquisition
Images of cells in 384-well microplates were acquired as previously described13 by using the following two IN Cell Analyzer platforms (GE Healthcare): the automated epifluorescence IN Cell Analyzer 2000 (INCA2000) and the line-scanning confocal automated microscope IN Cell Analyzer 3000 (INCA3000). Imaging on the INCA2000 was performed at 20×/0.45NA magnification. Images of nuclei stained with Hoechst in the blue channel were acquired using 350/50 nm excitation and 455/58 nm emission at an exposure time of 100 ms. Images of GFP in the green channel were acquired using 490/20 nm excitation and 525/36 nm emission, and GFP was imaged at an exposure time of 1.2 s. Imaging on the INCA3000 was performed at 40×/0.6NA objective magnification. Images of nuclei stained with Hoechst in the blue channel were acquired using 364-nm excitation and 450/65-nm emission in the blue channel at an exposure time of 1.5 ms. Images of GFP in the green channel were acquired using 488-nm excitation and 535/45-nm emission at an exposure time of 1.5 ms. Images of EGFR immunostaining in the red channel were acquired using by 647-nm excitation and 695/55-nm emission at an exposure time of 1.5 ms.
High resolution images of A549-EGFRB cells on chambered coverglass were acquired at MSKCC's Molecular Cytology Core Facility using a Leica TCS AOBS SP2 point-scanning confocal microscope (inverted stand) at 100× objective magnification using a Leica HCX PL APO oil immersion objective (1.4–0.7NA). Images of nuclei stained with Hoechst in the blue channel were acquired using 405-nm excitation and 410- to 475-nm emission. Images of GFP in the green channel were acquired using 488-nm excitation and 495- to 550-nm emission. Images of EGFR immunostaining in the red channel were acquired using 633-nm excitation and 640- to 750-nm emission.
Image Analysis
Images acquired by the INCA2000 were analyzed with the Developer Toolbox 1.7 software (GE Healthcare) by using a custom-developed image analysis protocol. GFP granules were identified using object-based segmentation on the green channel. Hoechst-stained nuclei were identified after post processing by using object-based segmentation on the blue channel. Automated image analysis using our custom-developed protocol allowed us to extract granule and nuclei counts, which were used for quantification of EGFR function and cytotoxicity, respectively. Granule count corresponds to the total granule count for imaged cells.
Assay Development and Validation
To optimize cell-seeding density, A549-EGFRB cells were seeded in 384-well microplates using an automated Multidrop 384 dispenser (Thermo Scientific) at a range of cell-seeding density covering 1000–10,000 cells per well. After 16 h, EGF was added at 100 nM final concentration in media, with media alone being added to control wells. After 70 min of incubation in a Cytomat automated temperature- and humidity-controlled incubator (Thermo Scientific) at 37°C and 5% CO2, GFP was imaged using the INCA2000, and granule count was quantified as previously described.
To optimize EGF concentration and induction time, A549-EGFRB cells were seeded in 45 μL of tissue culture media using Multidrop 384 in 384-well microplates at the optimal cell density of 5000 cells per well and incubated in Cytomat. After 16 h, EGF was added at 12 doubling dilutions from 1 μM final concentration as the upper limit. Plates were then incubated in Cytomat for 10, 30, or 70 min. At each time point, GFP was imaged using the INCA2000 and granule count was quantified as described above.
To assess the specificity of induction of GFP granule formation in our assay, A459-EGFRB cells were seeded at 2000 cells per well in 384-well microplates using Multidrop and transfected with scrambled or EGFR siRNA as already described. Four days post transfection, the media was aspirated using an automated plate washer ELx405 (Biotek Instruments) and replaced with media containing the tested ligands at three concentrations (Supplementary Table S1). After 70 min of incubation, cells were fixed with 4% paraformaldehyde (v/v) in PBS for 20 min at room temperature and washed once with PBS using the automated plate washer. GFP granules were imaged and quantified as previously described.
Assay Control Run
The optimized assay performance was assessed in a control run consisting of three 384-well microplates that contained 1% dimethylsulfoxide (DMSO) (v/v) for the high control and three 384-well microplates that contained 10 μM gefitinib in 1% DMSO (v/v) for the low control. Five microliters of 10% DMSO (v/v) were added to the high control plates and 5 μL of 100 μM gefitinib in 10% DMSO (v/v) were added to the low control plates with a custom-designed 384 head on a PP-384-M Personal Pipettor (Apricot Designs). A549-EGFRB cells were added to the plates in cell culture media using Multidrop 384 at the optimized density of 5000 cells per well and incubated in Cytomat for 16 h. The cell culture media was then aspirated using the automated plate washer and replaced with media containing 500 nM EGF. Plates were further incubated in Cytomat for 70 min as previously optimized, and cells were fixed and imaged as already described.
Pilot Screen
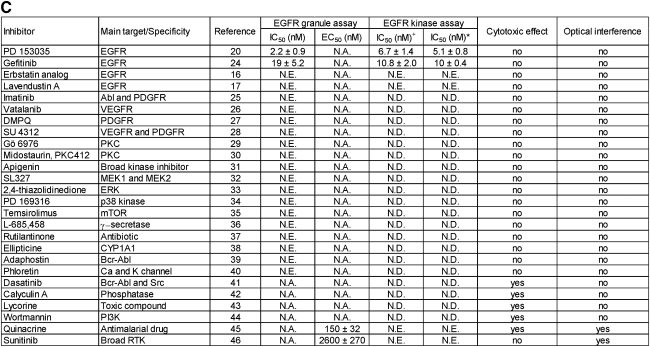
A pilot screen with the panel of 26 effectors was conducted by using 12-point doubling dilutions with 10 and 1 μM compound concentration in 1% DMSO (v/v) as the upper limit. The panel of effectors used in this study was assembled by purchasing 26 compounds with various known targets or specificity commercially available from various sources (Supplementary Table S2). Serial doubling dilutions of compounds were prepared as previously described.13 Controls consisted of 1% DMSO (v/v) (high control) and 10 μM gefitinib in 1% DMSO (v/v) (low control). The assay was performed according to our optimized workflow (Table 1). Dose–response curves were fitted using logistic four-parameter sigmoid regressions using SigmaPlot (Systat Software Inc.) to calculate half maximal inhibitory concentration (IC50) and half maximal effective concentration (EC50) values. Data from one representative experiment is presented for dose response in the EGFR granule assay, and each data point corresponds to the mean of two replicates for data from the EGFR kinase assay. The standard error corresponds to the standard error of the regression.
Table 1.
Workflow of the Optimized Epidermal Growth Factor Receptor Biosensor Assay
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Library compounds | 5 μL | 100 μM in 10% DMSO (v/v) |
| 2 | Low control | 5 μL | 100 μM gefitinib in 10% DMSO (v/v) |
| 3 | High control | 5 μL | 10% DMSO (v/v) |
| 4 | Cell plating | 45 μL | 5000 A549-EGFRB cells in cell culture RPMI 1640 media |
| 5 | Incubation time | 16 h | 37°C, 5% CO2 |
| 6 | Incubation time | 70 min | 500 nM EGF stimulation in cell culture RPMI 1640 media |
| 7 | Fix | 50 μL | 4% PFA for 20 min |
| 8 | Wash | 50 μL | Two times with 1× PBS |
| 9 | Nuclear staining | 50 μL | 10 μM Hoechst in 1× PBS for 10 min |
| 10 | Wash | 50 μL | Two times with 1× PBS |
| 11 | Assay readout | 350 nm/455 nm and 490 nm/525 nm (ex/em) | 20× objective imaging using IN Cell Analyzer 2000 microscope system |
| 12 | Image analysis | — | Multiparametric analysis using Developer Toolbox 1.7 software |
1–3. Dispensing with the PP-384-M Personal Pipettor using a custom 384 head.
4. Cells diluted in media and dispensed into 384-well assay plate with Multidrop 384.
5,6. 384-well assay plates stored in the Cytomat automated temperature-controlled incubator.
6–10. Aspirating with the ELx405 washer and dispensing with the Multidrop 384.
11. Readout performed on an automated platform; 19 s per well with total imaging time of 120 min per 384-well microtiter plate.
DMSO, dimethylsulfoxide; EGF, epidermal growth factor; PFA; PBS, phosphate-buffered saline; ex/em, excitation and emission.
ADP Glo EGFR Kinase Assay
Compounds displaying activity in the EGFR biosensor assay were confirmed by dose response against purified EGFR tyrosine kinase (Carna Biosciences) in the ADP Glo kinase assay (Promega Corporation). In vitro assays were carried out in 25 mM Hepes-NaOH, pH 7.5, containing 10 mM MgCl2, 2 mM tris(2-carboxyethyl)phosphine, 20 mM β-glycerol phosphate, and 100 mM NaVO4. EGFR kinase, ATP, and Poly(Glu,Tyr) substrate were present in all wells at 100 nM, 50 μM, and 50 μM, respectively. Compound dose response was assessed in duplicate by using 12-point doubling dilutions with 10 and 1 μM compound concentration in 1% DMSO (v/v) as the upper limit. Controls consisted of 1% DMSO (v/v) (fully uninhibited high control) and 10 μM gefitinib in 1% DMSO (v/v) (fully inhibited low control). Assays were carried out in white 384-well microtiter plates (Corning # 3570, Corning, NY), in which 1 μL of each compound dilution was initially transferred by using a PP-384-M Personal Pipettor. To this, 1 μl of a 1 μM solution of EGFR kinase was added and allowed to pre-incubate for either 10 or 60 min in the assay plate at 4°C prior to starting reactions via the addition of both substrates to a final assay volume of 10 μL per well. Reactions were incubated at 25°C for 30 min before stopping the assay through the addition of 10 μL of ADP Glo reagent from the ADP Glo Kinase Assay kit to each well. Assay plates were incubated at 25°C for an additional 40 min to fully deplete the residual ATP. Finally, 20 μL of kinase detection reagent was added to each well and the plates incubated at 25°C for a further 60 min. Luminescence signal was detected by using a LEADseeker™ Multimodality Imaging System (GE Healthcare) as previously described.14 IC50 values were calculated from a fit to a four-parameter logistic model for inhibition as already described. The mean data from duplicates are presented and the error bars correspond to the standard error of the regression for dose–response curves and to the standard error of duplicates for bar graphs.
Results
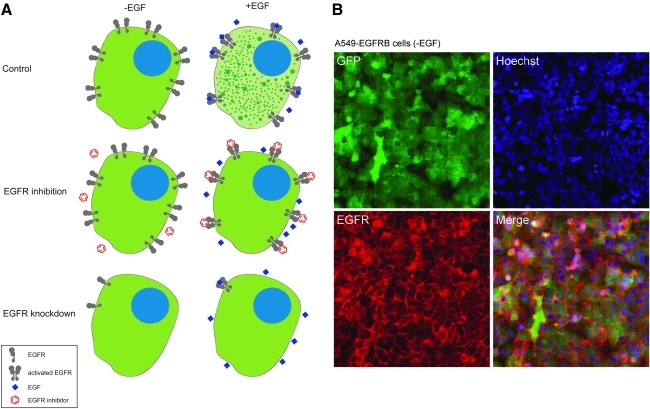
Our strategy for the development of a method enabling screening for RTK modulators in live cells relies on the expression of an SH2 domain protein with high affinity for a given RTK fused to a fluorescent protein. As a proof of concept, we developed an EGFR biosensor consisting of the TagGFP green fluorescent protein fused to two tandem SH2 domains from adapter protein Grb2 stably expressed in A549 human lung carcinoma cells. Upon ligand binding, EGFR homo- and heterodimerization induces EGFR activation and phosphorylation of tyrosine residues in the C-terminal cytoplasmic domain, which in turn leads to the recruitment of adaptor proteins such as Grb2 and phosphatidylinositol-3-kinase for signal transduction, followed by receptor clustering, endocytosis, recycling, and/or degradation. We hypothesized that in A549 cells stably expressing the SH2-TagGFP fusion protein (A549-EGFRB cells), addition of EGF would lead to the concomitant recruitment and clustering of the EGFR biosensor, enabling live imaging and quantification of granule formation as a surrogate for measuring endogenous EGFR activity. In contrast, in presence of an EGFR inhibitor or upon knockdown of EGFR expression, granule formation would be abrogated, allowing us to quantify the inhibition of EGFR function in live cells using automated image analysis (Fig. 1A).
Fig. 1.
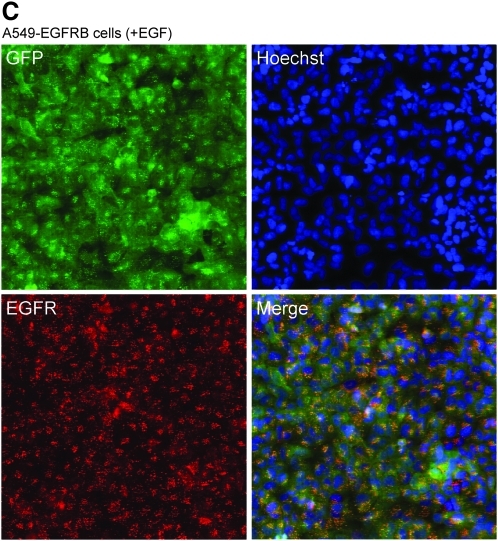
Epidermal growth factor receptor (EGFR) biosensor assay principle. (A) Schematics demonstrating the principle of the EGFR biosensor assay with A549 EGFR biosensor (A549-EGFRB) cells in the absence and presence of EGF stimulation. Green fluorescent protein (GFP) expression is diffuse in the cytoplasm in the absence of epidermal growth factor (EGF) stimulation, and EGF addition leads to the recruitment and clustering of the EGFR biosensor, enabling live imaging and quantification of granule formation as a surrogate for measuring endogenous EGFR activity. EGFR biosensor activation by EGF is prevented by small molecule EGFR inhibitor and by EGFR knockdown using RNAi. (B, C) EGFR biosensor activation is induced by EGF and reports EGFR activation. A549-EGFRB cells imaged with the confocal microscope INCA3000 at 40× objective magnification in the absence (B) or presence (C) of EGF stimulation. Green channel: EGF biosensor (GFP); blue channel: Hoechst staining of nuclei; red channel: immunostaining of EGFR and overlay of three channels.
To test our hypothesis, we imaged the A549-EGFRB cells stained for nucleus and EGFR using confocal microscopy (Figs. 1B, C). In the absence of EGF stimulation, the homogenous distribution of fluorescence in the green channel showed that the EGFR biosensor was evenly distributed throughout the cytoplasm, while EGFR was located at the cell surface as expected (Fig. 1B). In contrast, upon EGF addition, granule formation was observed in the GFP channel and co-localization of the GFP granules with EGFR indicated that the EGFR biosensor was recruited by EGFR upon receptor activation (Fig. 1C). This important result indicates that the EGFR biosensor accurately reports EGFR activation upon EGF addition.
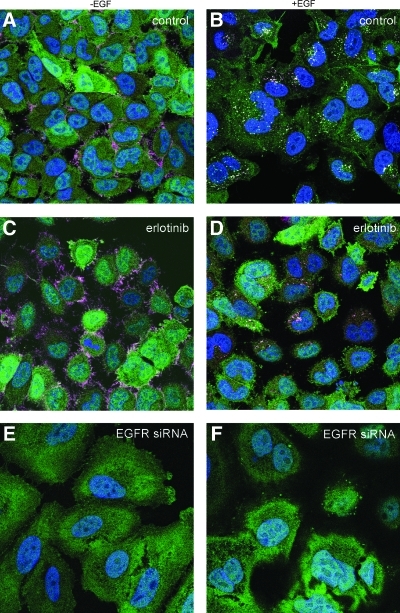
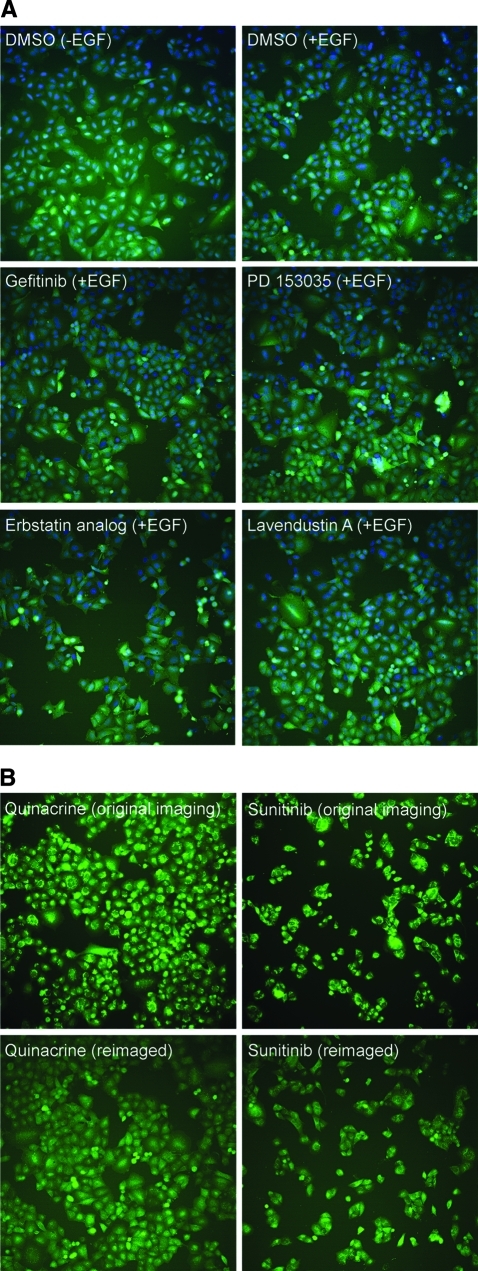
To further characterize our novel assay, we imaged A549-EGFRB cells stained for nucleus and EGFR at high resolution of 100× objective magnification (Fig. 2). As expected, cells treated with the DMSO control showed that the EGFR biosensor was evenly distributed in the cytoplasm in the absence of EGF stimulation (Fig. 2A). In contrast, upon EGF addition, granules in which GFP and EGFR co-localized indicated recruitment of the EGFR biosensor by EGFR upon receptor activation as previously observed (Fig. 2B). To assess the use of the EGFR biosensor cell line to identify modulators of EGFR function, we treated the A549-EGFRB cells with the EGFR kinase inhibitor erlotinib (Figs. 2C, D).15 As expected, and even in presence of EGF stimulation, the absence of receptor activation was observed by the absence of GFP granule formation and even distribution of the EGFR biosensor in the cytoplasm. In addition, in cells treated with DMSO control previously transfected with EGFR siRNA inducing EGFR knockdown as demonstrated by the absence of EGFR staining (Figs. 2E, F), no granule formation was observed in the GFP channel, either in the absence or presence of EGF. This result shows that the clustering of the EGFR biosensor in presence of EGF is dependent on EGFR expression and demonstrates that the EGFR biosensor specifically reports EGFR activation. Taken together, our findings show that the EGFR biosensor activation is abrogated both by inhibition with erlotinib and by RNAi-mediated EGFR knockdown, and strongly suggest that the observed granule formation is a true representation of EGFR activation and trafficking in live cells.
Fig. 2.
EGFR granule forming activity is inhibited by small molecules and RNAi knockdown. High resolution confocal imaging microscopy was performed at 100× objective magnification of A549-EGFRB cells in the absence and presence of EGF stimulation. Images of A549-EGFRB cells treated with 1% DMSO (v/v) control, 10 μM erlotinib in 1% DMSO (v/v), or 1% DMSO (v/v) control previously transfected with EGFR silencing siRNA. Each image is the overlay of the blue channel: nuclei; green channel: EGFR biosensor (GFP); and red channel: EGFR immunostaining.
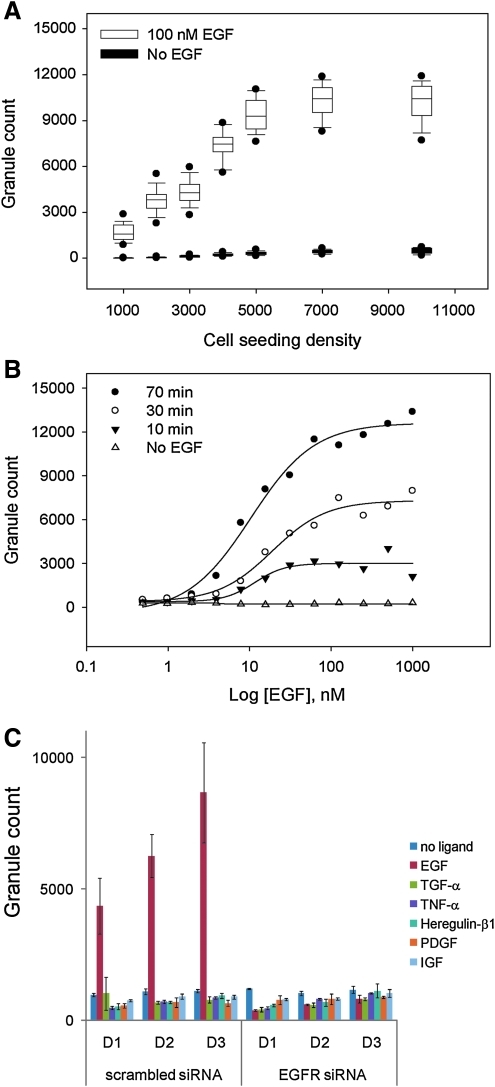
Having achieved a strong degree of validation as to the reporter activity and the EGFR status in cells, we miniaturized the assay into a 384-well format for high-content screening. In this format, microplates were fixed and imaged post activation with EGF using automated microscopy for the Hoechst and the GFP channels. Automated image analysis yielded nuclei count for the Hoechst channel and granule count readout for the GFP channel. Measuring granule count in the presence or absence of EGF stimulation for a cell density range of 1000 to 10,000 cells seeded per well led to the expected observation that granule count increased with the number of cells seeded until it reached maximum saturation at 7000 cells due most likely to cellular confluency at this density and beyond (Fig. 3A). Based on this result, we selected 5000 cells per well as the optimal cell seeding density. We assessed the effect of EGF concentration and duration of EGF stimulation on biosensor activation and we found that granule formation upon EGF addition is time and concentration dependent (Fig. 3B). This result further demonstrates the specificity of the EGFR biosensor for EGFR. We selected 500 nM EGF for an exposure time of 70 min as the optimal assay conditions, since this combination of EGF concentration and exposure time yielded the largest granule count for the assay. To test the specificity of induction of our EGFR biosensor, we treated A549-EGFRB cells with a panel of RTK ligands and cytokines (Supplementary Table S1). Significant granule formation was only induced by EGF and not by the ErbB3/4 ligand heregulin-β1 or by any of the tested RTK ligands and cytokines (Fig. 3C). In addition, granule formation induced by EGF was completely abrogated by RNAi-mediated EGFR knockdown, demonstrating that biosensor activation is highly specific to EGF. Altogether, our results demonstrate that the newly miniaturized and optimized assay specifically reports on the EGFR function and seems to be highly specific for EGF as the stimulant of activation.
Fig. 3.
EGFR biosensor characterization and assay miniaturization. (A) Optimization of A549-EGFRB cell seeding density leading to the selection of 5000 cells per well for the assay in 384-well format. In this box plot summarizing all replicate data (n=48), the bottom and top of the box represent the 25th and 75th percentiles, crossed by the median line. The end of whiskers below and above the box indicate the 10th and 90th percentiles. The outlying points show the 5th and 95th percentiles. White boxes correspond to 100 nM EGF stimulation and black boxes correspond to no EGF stimulation control. (B) Dose response of GFP granule count as a function of EGF concentration and duration of EGF stimulation in 384-well format, leading to the selection of 500 nM EGF stimulation for 70 min as the optimal conditions for the assay. The tested durations of EGF stimulation were 10 (▾), 30 (○), and 70 min (•), along with no EGF stimulation control (▴). The data presented correspond to one representative experiment. (C) Bar graph of granule count following treatment with RTK ligands and cytokines at three concentrations (D1, D2, D3), following transfection with scrambled or EGFR siRNA in a 384-well format. The data presented are means±standard error of replicate wells (n=8). TGF-α, transforming growth factor-α; TNF-α, tumor necrosis factor α; PDGF, platelet-derived growth factor; IGF, insulin-like growth factor.
To assess the robustness of our miniaturized assay, we performed a control run consisting of three 384-well microplates that contained 1% DMSO (v/v) measuring maximum granule formation as the high control of the assay and three 384-well microplates that contained 10 μM gefitinib in 1% DMSO (v/v) measuring complete inhibition of granule formation as the low control of the assay using our optimized assay workflow (Table 1). We observed an acceptable variability for both the high and low controls data points, with a coefficient of variation of 13% and 14% for the high and low controls, respectively. An average granule count of 10,700 for high controls compared to 500 for low controls for an average of 1200 imaged cells for both the high and low controls translated into a noise ratio of 21:1 and a calculated Z′ value of 0.56, indicative of good assay performance and robustness.
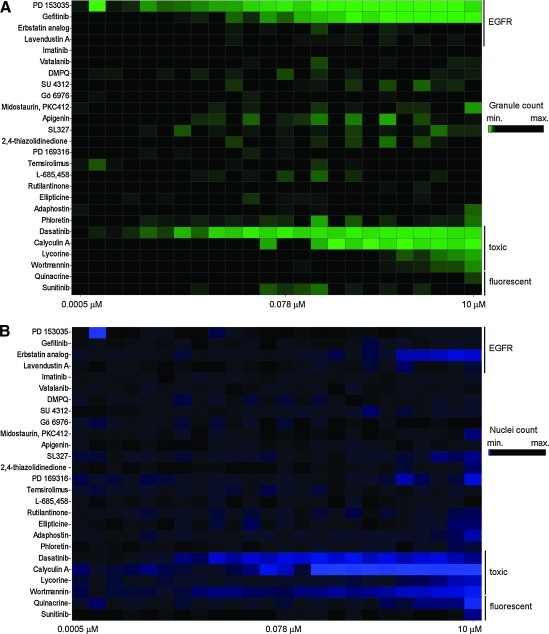
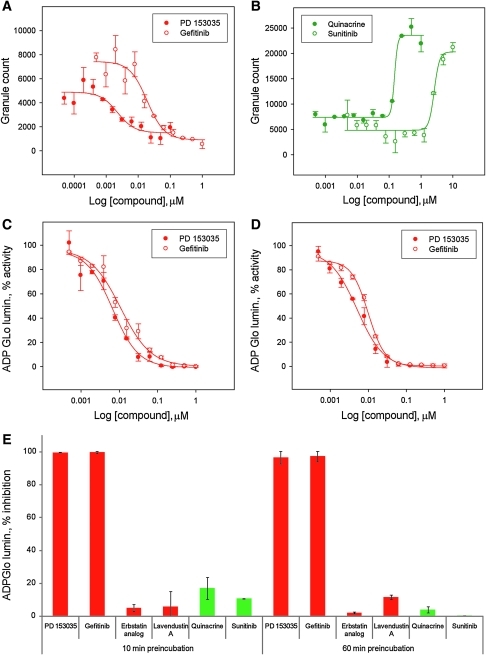
To further assess assay sensitivity in identifying modulators of EGFR function, we performed a pilot screen against a panel of 26 known effectors including EGFR inhibitors (Supplementary Table S2). Remarkably, out of the 26 compounds in the panel, only four compounds were found to significantly reduce granule formation; these were dasatinib, calyculin A, PD 153035, and gefitinib (Fig. 4A). Dasatinib and calyculin A were subsequently found to be very cytotoxic to the A549 cells as measured by the reduction in nuclei count (Fig. 4B), highlighting the advantages of this high-content assay and providing on-the-fly elimination of unwanted cytotoxic compounds. The EGFR inhibitors PD 153035 and gefitinib, on the other hand, inhibited granule formation with IC50 values of 2.2 and 19 nM, respectively, and without any observed cytotoxic effects on the A549 cells for the duration of the assay (Figs. 4C, 5A, 6A). Interestingly, two compounds were found to increase granule formation; namely, the antimalarial drug quinacrine and the pan-active RTKi sunitinib with EC50 values of 150 nM and 2.6 μM, respectively (Figs. 4C, 5B). Close inspection of their respective images revealed that their uptake and intracellular accumulation in the A549 cells appear to cause them to fluoresce in the GFP channel and display granule-like bodies in the cells, hence their high score in granule formation (Fig. 6B). Though not activators of EGFR and granule formation as such, they may potentially be used as probes to stain cells. Of note, upon re-imaging the plates after several weeks of storage at 4°C, fluorescence accumulation for both quinacrine and sunitinib was dimmer in intensity, most likely due to the leakage of those autofluorescent compounds outside the fixed cell and confirming that the earlier observed activation of granule formation was merely due to accumulation of fluorescence inside the A549 cells (Fig. 6B).
Fig. 4.
Pilot screen against a panel of 26 known effectors. (A) and (B) The results are presented as a heatmap to show the performance of the panel of 26 effectors that prevent granule formation (green) or are cytotoxic (blue) or inactive in the assay (black). Cytotoxic and fluorescent compounds are highlighted. (C) Summary table of the panel of 26 effectors tested in the EGFR biosensor assay summary of IC50 and EC50 data for dose–response curves fitted using logistic four parameter sigmoid regression. In the EGFR kinase assay, IC50 values were assessed following 10 min of pre-incubation of the compound with the enzyme (+) or 60 min (*). The standard error corresponds to the standard error of the regression. Cytotoxicity is assessed based on the nuclei count, and a compound was deemed cytotoxic if the imaged nuclei count was less than 50% of the DMSO control wells. 2,4-Thiazolidinedione is the abbreviated name for 3-(2-aminoethyl)-5-((4-ethoxyphenyl)methylene)-2,4-thiazolidinedione. NE, no effect; ND, not determined; NA, not applicable; PI3K, phosphatidylinositol-3-kinase; PDGFR, platelet-derived growth factor receptor; PKC, protein kinase C; VEGFR, vascular endothelial growth factor receptor.
Fig. 5.
Performance of selected effectors in the EGFR granule assay and EGFR kinase assay. Dose response curves in the EGFR granule assay of the EGFR inhibitors PD 153035 (•) and gefitinib (○) (calculated IC50 values: PD 153035 2.2±0.9 nM, gefitinib 19±5.2 nM) (A) and of the identified fluorescent compounds quinacrine (•) and sunitinib (○) (calculated apparent EC50 values: quinacrine 150±32 nM, sunitinib 2600±270 nM) (B). Dose–response curves in the EGFR kinase assay of the EGFR inhibitors PD 153035 (•) and gefitinib (○) following 10-min pre-incubation of the compound with the enzyme (calculated IC50 values: PD 153035 6.7±1.4 nM, gefitinib 10.8±2.0 nM) (C) or 60 min (calculated IC50 values: PD 153035 5.1±0.8 nM, gefitinib 10±0.4 nM) (D). Dose–response curves were fitted using logistic four parameter sigmoid regressions to calculate IC50 and EC50 values. Data from one representative experiment is presented for dose response in the EGFR granule assay and each data point corresponds to the mean of duplicates for data from the EGFR kinase assay (n=2). The standard error corresponds to the standard error of the regression. (E) Performance in the EGFR kinase assay of the reported EGFR inhibitors PD 153035, gefitinib, erbstatin analog, and lavendustin A as well as the fluorescent compounds quinacrine and sunitinib at 10 μM in 1% DMSO (v/v), following 10 or 60 min of pre-incubation with the enzyme. The data presented are means±standard deviation of duplicate wells (n=2).
Fig. 6.
Images of A549-EGFRB cells treated with selected compounds (A) INCA2000 imaging at 20× objective magnification of A549-EGFRB cells treated with 1% DMSO (v/v) control in absence or presence of 500 nM EGF stimulation for 70 min, and after EGF stimulation following pretreatment in 1% DMSO (v/v) with 1 μM gefitinib, 1 μM PD 153035, 10 μM erbstatin analog, or 10 μM lavendustin A. Overlay of blue channel: Hoechst staining of nuclei; green channel: EGFR biosensor (GFP). (B) INCA2000 imaging in the green channel: EGFR biosensor (GFP) at 20× objective magnification of A549-EGFRB cells treated with 1 μM quinacrine and 10 μM sunitinib in 1% DMSO (v/v) after 500 nM EGF stimulation for 70 min. Original images are compared to images acquired several weeks after the original imaging. Re-imaged fields of view may not be identical to those originally imaged but are representative of the whole well.
A somewhat surprising result from the pilot screen was the inability of the assay to identify erbstatin analog16 and lavendustin A,17 two reported EGFR inhibitors, as inhibitors of granule formation (Figs. 4C, 6A). Both compounds were originally included in the panel as EGFR inhibitors because of their chemical structure being completely different to known quinazoline-based scaffolds upon which gefitinib and PD 153035 were designed (Supplementary Table S2). For this purpose, we set up an in vitro EGFR kinase assay using ADP Glo as the readout to further interrogate whether erbstatin analog and lavendustin A do indeed inhibit the kinase function. No inhibitory activity was observed for either compound screened at concentrations up to 10 μM in 1% DMSO (v/v) with a 10-min pre-incubation with the kinase (Fig. 5E); moreover, a 1-h pre-incubation of both compounds yielded similar results (Fig. 5E). In contrast, assessment of PD 153035 and gefitinib in the same assay inhibited the EGFR activity with IC50 values of 5 nM and 10 nM, respectively (Figs. 4C, 5C, 5D), consistent with reported IC50 values of 1 and 33 nM, respectively.18,19 We further subjected quinacrine and sunitinib to the same in vitro EGFR assay, with no inhibitory activity observed at a concentration of up to 10 μM in 1% DMSO (v/v) with up to 1-h pre-incubation of the compounds with the kinase (Fig. 5E).
The remaining 20 compounds in the pilot screen were inactive at reducing granule formation and were found to induce less than 50% reduction in nuclei count at the screening concentration of 10 μM in 1% DMSO (v/v) and for the duration of the assay (Fig. 4C). Among them were inhibitors for vascular endothelial growth factor and platelet-derived growth factor receptor, which inherently had negative scores in the pilot study and provided the basis for referring to our optimized assay as selective, in that the assay has identified only those known and confirmed EGFR inhibitors.
Discussion
Despite the early success of RTKis in the clinic, the appearance of resistance to treatment significantly limits their efficacy, suggesting that RTKis with a novel mechanism of action are needed. Current RTKis in the clinic were identified in in vitro assays by targeting the kinase activity of RTKs. Important limitations to such an approach are (1) the high failure rate of such drug candidates due to lack of cell-based activity and (2) the restriction to finding inhibitors of kinase activity while inhibitors of receptor activation and trafficking in the cell are overlooked. To overcome these limitations, we sought to pursue a novel strategy for the development of cell-based assays for RTKs. Our strategy relies on a domain-based biosensor approach that would allow the identification of novel modulators of RTK function in live cells. As a proof of concept, we developed a novel assay for the identification of EGFR modulators. The characterization of A549-EGFRB cells stably expressing the EGFR biosensor using confocal imaging demonstrates that EGF addition leads to GFP granule formation, co-localizing with EGFR (Figs. 1, 2). Furthermore, RNAi-mediated EGFR knockdown abrogated granule formation and demonstrated that granule formation is dependent on EGFR expression. In addition, granule formation was specifically observed upon EGF addition and was not induced by any other tested RTK ligands (Fig. 3C). Altogether, our results demonstrate the specificity of our novel assay for measuring EGFR function in live cells.
Both gefitinib, an EGFR inhibitor approved by the U.S. Food and Drug Administration for the treatment of various cancers,2 and PD 153035, a well-characterized EGFR inhibitor,20 scored as hits in our pilot study against 26 known effectors, demonstrating that our assay can easily identify potent cell-permeable EGFR inhibitors (Figs. 4, 5). The two other reported EGFR inhibitors present in our panel of effectors, namely the erbstatin analog16 and lavendustin A,17 did not score in our assay and failed to inhibit granule formation, suggesting that they may fall into the class of kinase inhibitors with poor cellular activity (Fig. 4). Follow-up studies, however, using an in vitro ADP Glo kinase assay found both compounds inactive against the target even with prolonged pre-incubation time of up to one hour typically used to investigate slow binding characteristics of inhibitors. Hsu and co-workers21,22 were the first group to report on the inhibitory activities of lavendustin A and the erbstatin analog back in the 1990s, but no follow-up studies have been published, making it difficult to ascertain whether their observations are indeed reproducible. Though sunitinib has been reported by many groups as a broad RTK inhibitor,23 it did not score in our assay and was subsequently found to be inactive against the EGFR kinase activity in vitro (Fig. 5). We could not find any published reference demonstrating its inhibitory activity against EGFR. Thus, it is likely that sunitinib does not significantly inhibit EGFR activity. Sunitinib, on the other hand, did initially score as a potential activator of granule formation together with quinacrine but further inspection of both their images revealed that the observed effects were merely due to accumulation of fluorescent compounds in the A549 cells mimicking granule-like structures, hence the increase in granule formation count. Quinacrine, not surprisingly, has previously been reported as an autofluorescent compound in high-content screening,* whereas it is the first observation for sunitinib being a fluorescent compound in cells. Interestingly, the observed enhancement in granule count faded away several weeks after the initial imaging was performed. Despite the intended enrichment in the panel of other well-known kinase inhibitors, an important observation from our pilot screen was the fact that only gefitinib and PD 153035 scored as inhibitors of granule formation, suggesting that the selective nature of our optimized assay makes it ideal for screening large chemical libraries.
Taken together, we report on the first biosensor domain-based high-content assay to screen for modulators of RTK function in live cells amenable to both chemical and RNAi screens. We provide a concise proof of concept study using the A549 cells harboring the biosensor and monitoring EGFR function, with a high-content assay miniaturized to a 384-well format, validated by using known EGFR inhibitors and RNAi-mediated EGFR knockdown. We further report on the assay performance in screening against a panel of 26 known effectors resulting in the identification of only EGFR inhibitors as hits. The ability to perform such screens has tremendous implications for the identification of novel agents to study the regulation of RTK function as well as for the identification of cancer therapeutics overcoming resistance to current treatments.
Supplementary Material
Glossary
Abbreviations
- A549-EGFRB cells
A549 EGFR biosensor cells
- DMSO
dimethylsulfoxide
- EC50
half maximal effective concentration
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- HTS
high-throughput screening
- IC50
half maximal inhibitory concentration
- INCA
IN Cell Analyzer
- KRAS
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
- mAb
monoclonal antibody
- MSKCC
Memorial Sloan-Kettering Cancer Center
- NSCLC
non–small-cell lung cancer
- PBS
phosphate-buffered saline
- RNAi
RNA interference
- RTK
receptor tyrosine kinase
- RTKi
receptor tyrosine kinase inhibitor
- SH2
SRC homology 2 domain.
Footnotes
Shum D, Bhinder B, Bermingham A, Calder P, Radu C, Djaballah H: A high-content cell-based assay to identify modulators of miRNA biogenesis in the screening of chemical libraries. Memorial sloan-kettering Cancer Center, New York, 2011; manuscript in preparation.
Acknowledgments
The authors wish to thank members of the HTS Core Facility for their help during the course of this study, especially Irena Ivnitski-Steele and members of the Molecular Cytology Core Facility for their help with high resolution confocal imaging. The HTS Core Facility is partially supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of MSKCC, the William Randolph Hearst Fund in Experimental Therapeutics, the Lillian S. Wells Foundation, and by a NIH/NCI Cancer Center Support Grant 5 P30 CA008748-44.
Disclosure Statement
Dmitry Malkov, Keming Song, and John Fetter are employees of Sigma-Aldrich, the company selling the A549 EGFR biosensor cells.
References
- 1.Gschwind A. Fischer OM. Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 2.Pao W. Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hynes NE. Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 4.Ciaccio MF. Wagner JP. Chuu CP. Lauffenburger DA. Jones RB. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat Methods. 2010;7:148–155. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botvinnik A. Wichert SP. Fischer TM. Rossner MJ. Integrated analysis of receptor activation and downstream signaling with EXTassays. Nat Methods. 2010;7:74–80. doi: 10.1038/nmeth.1407. [DOI] [PubMed] [Google Scholar]
- 6.Sevecka M. MacBeath G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat Methods. 2006;3:825–831. doi: 10.1038/NMETH931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grecco HE. Roda-Navarro P. Girod A, et al. In situ analysis of tyrosine phosphorylation networks by FLIM on cell arrays. Nat Methods. 2010;7:467–472. doi: 10.1038/nmeth.1458. [DOI] [PubMed] [Google Scholar]
- 8.Olive DM. Quantitative methods for the analysis of protein phosphorylation in drug development. Expert Rev Proteomics. 2004;1:327–341. doi: 10.1586/14789450.1.3.327. [DOI] [PubMed] [Google Scholar]
- 9.Pennucci J. Swanson S. Kaliyaperumal A. Gupta S. Multiplexed evaluation of a cell-based assay for the detection of antidrug neutralizing antibodies to panitumumab in human serum using automated fluorescent microscopy. J Biomol Screen. 2010;15:644–652. doi: 10.1177/1087057110370893. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh RN. Grove L. Lapets O. A quantitative cell-based high-content screening assay for the epidermal growth factor receptor-specific activation of mitogen-activated protein kinase. Assay Drug Dev Technol. 2004;2:473–481. doi: 10.1089/adt.2004.2.473. [DOI] [PubMed] [Google Scholar]
- 11.Sigma-Aldrich: EGFR biosensor cell line. www.sigmaaldrich.com/life-science/cells-and-cell-based-assays/egfr-biosensor-cell-line.html. [Oct 25;2011 ]. www.sigmaaldrich.com/life-science/cells-and-cell-based-assays/egfr-biosensor-cell-line.html
- 12.Ambion: Silencer select negative control #1. www.ambion.com/catalog/CatNum.php?4390843. [Oct 26;2011 ]. www.ambion.com/catalog/CatNum.php?4390843
- 13.Ramirez CN. Ozawa T. Takagi T, et al. Validation of a high-content screening assay using whole-well imaging of transformed phenotypes. Assay Drug Dev Technol. 2011;9:247–261. doi: 10.1089/adt.2010.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagi T. Shum D. Parisi M, et al. Comparison of luminescence ADP production assay and radiometric scintillation proximity assay for Cdc7 kinase. Comb Chem High Throughput Screen. 2011;14:669–687. doi: 10.2174/138620711796504442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moyer JD. Barbacci EG. Iwata KK, et al. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res. 1997;57:4838–4848. [PubMed] [Google Scholar]
- 16.Umezawa K. Hori T. Tajima H, et al. Inhibition of epidermal growth factor-induced DNA synthesis by tyrosine kinase inhibitors. FEBS Lett. 1990;260:198–200. doi: 10.1016/0014-5793(90)80102-o. [DOI] [PubMed] [Google Scholar]
- 17.Onoda T. Iinuma H. Sasaki Y, et al. Isolation of a novel tyrosine kinase inhibitor, lavendustin A, from Streptomyces griseolavendus. J Nat Prod. 1989;52:1252–1257. doi: 10.1021/np50066a009. [DOI] [PubMed] [Google Scholar]
- 18.Blencke S. Ullrich A. Daub H. Mutation of threonine 766 in the epidermal growth factor receptor reveals a hotspot for resistance formation against selective tyrosine kinase inhibitors. J Biol Chem. 2003;278:15435–15440. doi: 10.1074/jbc.M211158200. [DOI] [PubMed] [Google Scholar]
- 19.Wakeling AE. Guy SP. Woodburn JR, et al. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
- 20.Fry DW. Kraker AJ. McMichael A, et al. A specific inhibitor of the epidermal growth factor receptor tyrosine kinase. Science. 1994;265:1093–1095. doi: 10.1126/science.8066447. [DOI] [PubMed] [Google Scholar]
- 21.Hsu CY. Jacoski MV. Maguire MP. Spada AP. Zilberstein A. Inhibition kinetics and selectivity of the tyrosine kinase inhibitor erbstatin and a pyridone-based analogue. Biochem Pharmacol. 1992;43:2471–2477. doi: 10.1016/0006-2952(92)90327-f. [DOI] [PubMed] [Google Scholar]
- 22.Hsu CY. Persons PE. Spada AP, et al. Kinetic analysis of the inhibition of the epidermal growth factor receptor tyrosine kinase by Lavendustin-A and its analogue. J Biol Chem. 1991;266:21105–21112. [PubMed] [Google Scholar]
- 23.Patyna S. Laird AD. Mendel DB, et al. SU14813: a novel multiple receptor tyrosine kinase inhibitor with potent antiangiogenic and antitumor activity. Mol Cancer Ther. 2006;5:1774–1782. doi: 10.1158/1535-7163.MCT-05-0333. [DOI] [PubMed] [Google Scholar]
- 24.Ciardiello F. Caputo R. Bianco R, et al. Antitumor effect and potentiation of cytotoxic drugs activity in human cancer cells by ZD-1839 (Iressa), an epidermal growth factor receptor-selective tyrosine kinase inhibitor. Clin Cancer Res. 2000;6:2053–2063. [PubMed] [Google Scholar]
- 25.Druker BJ. Tamura S. Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 26.Wood JM. Bold G. Buchdunger E, et al. PTK787/ZK 222584, a novel and potent inhibitor of vascular endothelial growth factor receptor tyrosine kinases, impairs vascular endothelial growth factor-induced responses and tumor growth after oral administration. Cancer Res. 2000;60:2178–2189. [PubMed] [Google Scholar]
- 27.Dolle RE. Dunn JA. Bobko M, et al. 5,7-Dimethoxy-3-(4-pyridinyl)quinoline is a potent and selective inhibitor of human vascular beta-type platelet-derived growth factor receptor tyrosine kinase. J Med Chem. 1994;37:2627–2629. doi: 10.1021/jm00043a002. [DOI] [PubMed] [Google Scholar]
- 28.Zaman GJ. Vink PM. van den Doelen AA. Veeneman GH. Theunissen HJ. Tyrosine kinase activity of purified recombinant cytoplasmic domain of platelet-derived growth factor beta-receptor (beta-PDGFR) and discovery of a novel inhibitor of receptor tyrosine kinases. Biochem Pharmacol. 1999;57:57–64. doi: 10.1016/s0006-2952(98)00271-8. [DOI] [PubMed] [Google Scholar]
- 29.Martiny-Baron G. Kazanietz MG. Mischak H, et al. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J Biol Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 30.Fabbro D. Ruetz S. Bodis S, et al. PKC412—a protein kinase inhibitor with a broad therapeutic potential. Anticancer Drug Des. 2000;15:17–28. [PubMed] [Google Scholar]
- 31.Sato F. Matsukawa Y. Matsumoto K. Nishino H. Sakai T. Apigenin induces morphological differentiation and G2-M arrest in rat neuronal cells. Biochem Biophys Res Commun. 1994;204:578–584. doi: 10.1006/bbrc.1994.2498. [DOI] [PubMed] [Google Scholar]
- 32.Atkins CM. Selcher JC. Petraitis JJ. Trzaskos JM. Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- 33.Hancock CN. Macias A. Lee EK, et al. Identification of novel extracellular signal-regulated kinase docking domain inhibitors. J Med Chem. 2005;48:4586–4595. doi: 10.1021/jm0501174. [DOI] [PubMed] [Google Scholar]
- 34.Gallagher TF. Seibel GL. Kassis S, et al. Regulation of stress-induced cytokine production by pyridinylimidazoles; inhibition of CSBP kinase. Bioorg Med Chem. 1997;5:49–64. doi: 10.1016/s0968-0896(96)00212-x. [DOI] [PubMed] [Google Scholar]
- 35.Wu C. Wangpaichitr M. Feun L, et al. Overcoming cisplatin resistance by mTOR inhibitor in lung cancer. Mol Cancer. 2005;4:25. doi: 10.1186/1476-4598-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shearman MS. Beher D. Clarke EE, et al. L-685,458, an aspartyl protease transition state mimic, is a potent inhibitor of amyloid beta-protein precursor gamma-secretase activity. Biochemistry. 2000;39:8698–8704. doi: 10.1021/bi0005456. [DOI] [PubMed] [Google Scholar]
- 37.Yoshimoto A. Matsuzawa Y. Oki T, et al. Microbial conversion of epsilon-pyrromycinone and epsilon-isorhodomycinone to 1-hydroxy-13-dihydrodaunomcyin and N-formyl-1-hydroxy-13-dihydrodaunomycin and their bioactivities. J Antibiot (Tokyo) 1980;33:1150–1157. doi: 10.7164/antibiotics.33.1150. [DOI] [PubMed] [Google Scholar]
- 38.Tassaneeyakul W. Birkett DJ. Veronese ME, et al. Specificity of substrate and inhibitor probes for human cytochromes P450 1A1 and 1A2. J Pharmacol Exp Ther. 1993;265:401–407. [PubMed] [Google Scholar]
- 39.Svingen PA. Tefferi A. Kottke TJ, et al. Effects of the bcr/abl kinase inhibitors AG957 and NSC 680410 on chronic myelogenous leukemia cells in vitro. Clin Cancer Res. 2000;6:237–249. [PubMed] [Google Scholar]
- 40.Fan HT. Morishima S. Kida H. Okada Y. Phloretin differentially inhibits volume-sensitive and cyclic AMP-activated, but not Ca-activated, Cl(-) channels. Br J Pharmacol. 2001;133:1096–1106. doi: 10.1038/sj.bjp.0704159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombardo LJ. Lee FY. Chen P, et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 42.Ishihara H. Martin BL. Brautigan DL, et al. Calyculin A and okadaic acid: inhibitors of protein phosphatase activity. Biochem Biophys Res Commun. 1989;159:871–877. doi: 10.1016/0006-291x(89)92189-x. [DOI] [PubMed] [Google Scholar]
- 43.Kukhanova M. Victorova L. Krayevsky A. Peptidyltransferase center of ribosomes. On the mechanism of action of alkaloid lycorine. FEBS Lett. 1983;160:129–133. doi: 10.1016/0014-5793(83)80951-x. [DOI] [PubMed] [Google Scholar]
- 44.Arcaro A. Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J. 1993;296(Pt 2):297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duncan GG. Quinacrine hydrochloride (atabrine) as a malaria suppressive agent. Air Surg Bull. 1945;2:413. [PubMed] [Google Scholar]
- 46.O'Farrell AM. Abrams TJ. Yuen HA, et al. SU11248 is a novel FLT3 tyrosine kinase inhibitor with potent activity in vitro and in vivo. Blood. 2003;101:3597–3605. doi: 10.1182/blood-2002-07-2307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.