Abstract
The emergence and spread of multidrug-resistant Plasmodium falciparum and recent detection of potential artemisinin-resistant strains in Southeast Asia highlight the importance of developing novel antimalarial therapies. Using a previously generated stable transgenic P. falciparum line with high-level firefly luciferase expression, we report the adaptation, miniaturization, optimization, and validation of a high-throughput screening assay in 384-well plates. Assay conditions, including the percentage of parasitemia and hematocrit, were optimized. Parameters of assay robustness, including Z′-value, coefficient variation (CV), and signal-to-background (S/B) ratio, were determined. The LOPAC1280 small-compound library was used to validate this assay. Our results demonstrated that this assay is robust and reliable, with an average Z′-value of >0.7 and CV of <10%. Moreover, this assay showed a very low background, with the S/B ratio up to 71. Further, identified hits were selected and confirmed using a SYBR Green I-based confirmatory assay. It is evident that this assay is suitable for large-scale screening of chemical libraries for antimalarial drug discovery.
Introduction
Malaria is a common and life-threatening disease in many tropical and subtropical areas. Globally, malaria affects about 5% of the world's population and brings a death toll of 0.5–2.5 million each year, despite the intensive international efforts fighting against malaria. Although the main impact of malaria is in sub-Saharan Africa, where at least 90% of deaths from malaria occur, it is also a major public health problem in Asia and South America.1–3 Malaria is caused by a blood parasite of the genus Plasmodium, which includes four species of Plasmodium that commonly infect humans: Plasmodium falciparum, Plasmodium vivax, Plasmodium ovale, and Plasmodium malariae.4 In the mid-50s, WHO launched a worldwide malaria eradication campaign using effective and inexpensive therapeutics and insecticides in designated malaria-infected areas. The program resulted in the elimination of endemic malaria in developed countries and a significant reduction of cases in developing parts of the world.3,5,6 The recent emergence and spread of P. falciparum parasite resistant to many antimalarial drugs, particularly chloroquine (CQ) and pyrimethamine (PY)-sulfadoxine, is largely responsible for the recent global resurgence of malaria, which severely hampers our capacity to roll back malaria.1,3,6 The prevalence of multidrug-resistant parasites has become a great challenge for malaria management in endemic regions. To avert this disaster, artemisinin and its derivatives, often in combination with more slowly eliminating antimalarial drugs, are being adopted in many countries for treating P. falciparum malaria.1,6,7 Artemisinin-based combination therapy has proven to be very effective in treating falciparum malaria without evident clinical resistance. However, the recent detection of artemisinin resistance at the Thai-Cambodian border, manifested by delayed parasite clearance, raised significant concerns.8,9 Therefore, there is a pressing need for discovering and developing novel chemotherapeutic agents against malaria parasites.1,6,10,11
Developing robust and reliable in vitro high-throughput screening (HTS) assays to evaluate the effect of a compound on the growth of malaria parasites is key to antimalarial drug discovery. Fluorescence-based assays such as SYBR Green I and DAPI are fast, relatively inexpensive, and as sensitive as standard radioactive assays and have been used in evaluating the antimalarial efficacy and drug screening.12–15 However, fluorescence-based assays often have high background readings.16,17 Advances in malaria parasite transfection technology have enabled the generation of transgenic parasite lines expressing different reporters such as green fluorescent protein (GFP), offering additional markers for HTS assays.18,19 Recently, we have designed a simple, robust, cell-based luminescent method for assaying antimalarial drugs based on a transgenic P. falciparum line that expresses high levels of firefly luciferase.20 Here, we report the further optimization, miniaturization, and validation of this assay for HTS purpose.
Materials and Methods
Parasites Culture and Synchronization
Both the wild-type (wt) and transgenic malaria parasite P. falciparum strain 3D7-Luciferase was maintained in complete medium (RPMI 1640 medium supplemented with 25 mM HEPES [pH 7.5], 25 mM sodium bicarbonate, 50 mg/L hypoxanthine, 0.5% Albumax II, and 40 μg/mL gentamicin sulfate) at 5% hematocrit in human O+ red blood cells (RBCs) and incubated in a gas mixture of 5% CO2, 3% O2, and 92% N2 at 37°C.21 Parasitemia was monitored daily using Giemsa staining of thin blood smears. For HTS, ring-stage parasites were synchronized twice by sorbitol treatment.
Reagents
To determine the level of luciferase expression in the transgenic P. falciparum line, parasites at different percentages of parasitemias and hematocrits were seeded in each well of a black 96-well plate with a total of 100 μL medium or in white 384-well plate with a total volume of 25 μL (Corning, Inc.) using the automated MicroFlo Select Dispenser (BioTek Instruments, Inc.). Luciferase assays were performed using either Steady-Glo® Luciferase Assay System or ONE-Glo™ Luciferase Assay System (Promega) following the manufacturer's instruction. An equal volume of the reconstituted reagent was added to the cultured parasites in each well. Relative luminescent unit (RLU) was measured using Synergy 2 Luminescence Microplate Reader (BioTek Instruments, Inc.).
Assay Optimization
Hematocrit and parasitemia were optimized in both 96- and 384-well plates using both luciferase assay reagents. Mixed stages of 3D7-luc transgenic parasites were robotically dispensed into culture plates in a combination of different hematocrit (0.5%–5%) and different parasitemia (0.5%–5%). For each combination of hematocrit, parasitemia, and incubation period, a total of eight replicates were included.
Assay Validation
The assay was validated in white 384-well plates (Corning) using the ONE-Glo luciferase assay system in a one-step “add-and-measure” procedure. The transgenic parasites 3D7-luc, synchronized at the ring stage, were seeded into culture plates at 15 μL culture/well. The Library of Pharmacologically Active Compounds (LOPAC1280; Sigma), a 1280-compound library for pilot testing, was used to validate this assay for HTS format. The compounds were dissolved in dimethyl sulfoxide (DMSO) at 2 mM and diluted in complete RPMI 1640 medium to a concentration of 25 μM. Ten microliters of each compound was loaded to each well and mixed with the 15 μL parasite culture to obtain a final concentration of 10 μM. The wt 3D7 parasites, 3% RBC media, and 2 μM CQ were also included as the negative signal controls and the transgenic 3D7-luc parasites without any compound treatment was included as the positive signal control. All procedures were operated under aseptic environment. Plates were incubated in a gas mixture of 5% CO2, 3% O2, and 92% N2 at 37°C for 72 h. Subsequently, 25 μL of ONE-Glo reagent was added to each well at room temperature following the manufacturer's instruction. After 5 min of incubation, RLU was determined. The assay validation experiment was repeated twice.
Secondary Assay
The SYBR Green I assay was used as a secondary assay to confirm the antimalarial efficacy of selected compounds from the validation assay. Briefly, 80 μL of wt 3D7 parasites at ring stage with 0.5% parasitemia and 1% hematocrit was seeded in a black 96-well plate. Selected compounds at fivefold serial dilutions in complete culture medium were added to each well to a total volume of 100 μL. The plates were then incubated for 72 h in a candle jar at 37°C. SYBR Green I assay was performed as previously described22 and fluorescence was quantified on a Tecan GENios microplate reader at an excitation wavelength of 485 nm and emission wavelength of 535 nm. The signal was expressed as relative fluorescent units (RFU). The inhibition of parasite growth was expressed as percentage of inhibition related to the RFU from the transgenic parasite control.
Data Analysis
During the validation process, wt 3D7 parasites, 3% RBC media, and 2 μM CQ were included as the assay's negative signal controls and the transgenic 3D7-luc parasites as the assay positive signal control in each assay plate. These controls were used to calculate Z′-value, signal-to-background (S/B) ratio, and coefficient variation (CV) for each plate and to normalize the data on a per-plate basis. All raw data were analyzed to determine the Z′-value, S/B, CV, and percentage of inhibition for assayed compounds. Results were expressed as percentage of inhibition of RLU at which 100% inhibition of RLU was equal to the mean of the media-only control. Compounds showing greater than average inhibition plus three times standard deviation of % inhibition of RLU of all compounds were considered “hits.” Statistical calculations of Z′-values were made as follows:
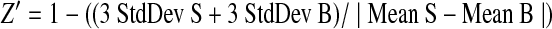 |
Here, Mean S is the mean RLU from the assay's negative controls, StdDev S is the standard deviation of the RLU from the assay's negative controls, Mean B is the mean RLU from the transgenic 3D7-luc parasites control, and StdDev B is the standard deviation of the RLU from the transgenic 3D7-luc parasites control. S/B is equal to “Mean S/Mean B.”23,24
Results
Assay Development and Adaptation in 96-Well Plates
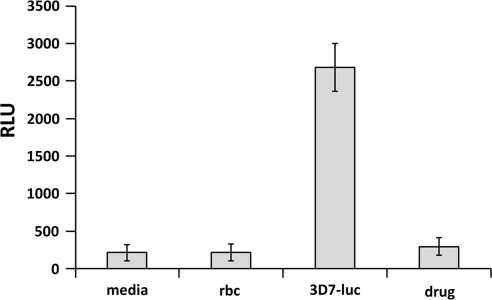
We have previously reported the generation of a stable transgenic 3D7 parasite expressing firefly luciferase under the P. falciparum heat shock protein 86 (hsp86) promoter to achieve high-level and constitutive expression of the luciferase.20 This transgenic P. falciparum line was used to develop a cell-based luminescent method for assaying antimalarial drugs, which generated similar sensitivity results for commonly used antimalarial drugs when compared with the standard [3H] hypoxanthine radioisotope method, suggesting a possibility for further miniaturization and automation of this assay. To develop this method into an automated HTS assay, we first automated and adapted this assay in the 96-well plate format. When medium containing 2.5% RBCs were infected with 2% parasitemia, the controls, including the media and RBC lysate, had 212 and 217 RLU, respectively, using the Steady-Glo Luciferase Assay System (Fig. 1). The level of luciferase in the 3D7-luc line was averaged at 2676 RLU, resulting in an S/B ratio above 12. The artemisinin (10 μM)-treated transgenic line showed a strong inhibition of parasite growth with luciferase expression at 294 RLU, with a 9.11 times reduction (Fig. 1).
Fig. 1.
Assay development and adaptation in 96-well plates. The transgenic Plasmodium falciparum line was used for assay adaptation in the 96-well plate and luciferase signals were determined using Steady-Glo Luciferase Assay System (Promega). Complete RPMI 1640 medium and RBCs were used to determine the background noise, and artemisinin at 10 μM was used as a positive drug control to inhibit the growth of the parasites. The experiment was repeated for three times and each data point represents the average value of eight replicates; error bar represents standard error of n=8 values. RBC, red blood cell.
Assay conditions, including DMSO tolerance, parasite developmental stages, parasitemia, hematocrit, and endpoint, were also optimized in 96-well plates in an automated setting. Similar to our previously reported results,20 the transgenic line could tolerate up to 3% of DMSO, and maximum expression of luciferase occurred at 2% parasitemia and 2% hematocrit at 72 h postseeding (results not shown).
Assay Miniaturization and Optimization in 384-Well Plates
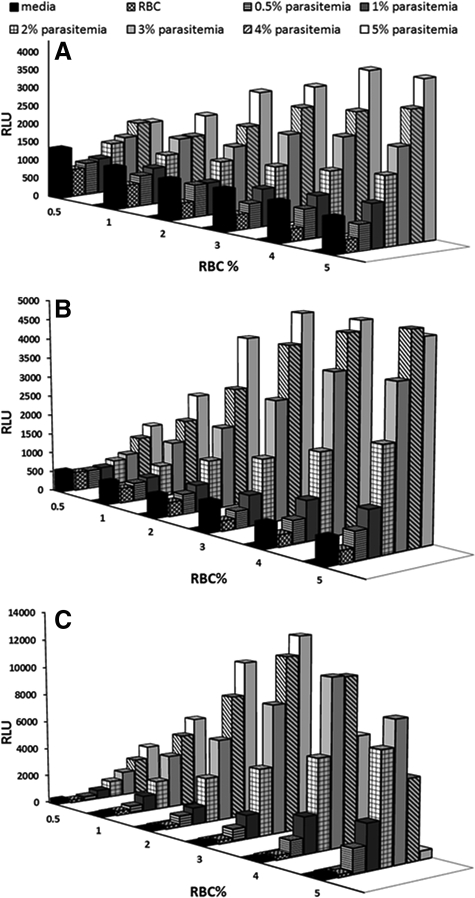
Configuring the assay in a 96-well plate format is a minimal requirement for the HTS of large-compound libraries, and a higher throughput format using 384-well or higher plate is highly desirable. Using the established optimum conditions in a 96-well plate as the starting point, the assay was miniaturized into 384-well plates. Because different plate formats hold different volumes of cell culture, the level of parasitemia will affect the amount of luciferase produced in each well and thus the robustness of the assay. To satisfy the requirement of a robust HTS assay, we first optimized hematocrit and parasitemia in 384-well plates and evaluated luciferase expression at different times after seeding the parasites. Using a combination of six fixed hematocrit and parasitemia concentrations, 0.5%, 1%, 2%, 3%, 4%, and 5%, we found that the luciferase signals were low at 24 h, with a maximum of 5,221 RLU being observed at 5% hematocrit and 5% parasitemia (Fig. 2A). At 48 h postseeding, the luciferase level increased and a maximum luciferase signal of 6,102 RLU was observed at 4% hematocrit and 5% parasitemia (Fig. 2B). Furthermore, at 72 h postseeding, the maximum luciferase signals of 13,269 RLU were observed at 3% hematocrit and 5% parasitemia (Fig. 2C), with CV at 9.1% and S/B ratio at 51. Considering the rigid requirements for an HTS assay for large-compound library screening, we opted to select the culture conditions of 3% hematocrit, 5% parasitemia, and 72 h incubation, which showed maximum expression of luciferase.
Fig. 2.
Assay miniaturization and optimization in 384-well plates. Different percentage of hematocrit (% RBC) and % parasitemia were seeded in 384-well plates and luminescent signals from the transgenic parasite line were determined using Steady-Glo Luciferase Assay System (Promega) at different hours postseeding, including 24 h (A), 48 h (B), and 72 h (C) postseeding. The experiment was repeated for three times and each data point represents the average value of eight replicates; error bar represents standard error of n=8 values.
Parasites Synchronization
We previously reported that the synchronized parasites at schizont stages express a higher level of luciferase when compared with the rings, early trophozoites, and unsynchronized parasites of mixed stages. To improve the robustness of the assay, we sought to determine the stage with the highest level of luciferase expression. Using 5% sorbitol treatment, ring-stage parasites were synchronized and the parasites were grown to different stages for the luciferase assay. Similar to our previous observation, the parasites at late trophozoite and the schizont stage produced comparably high levels of luciferase (result not shown). Thus, the late-stage trophozoite parasites were selected for future HTS assay validation. We also applied the ONE-Glo luciferase assay system, which provides a highly sensitive, robust, homogeneous assay for detection of firefly luciferase reporter gene expression and is ideally suited for high- and ultrahigh-throughput applications. The summary of the 384-well format luciferase-based assay protocol is listed in Table 1.
Table 1.
Summary of 384-Well Format Assay Protocols for Luciferase-Based Assay
| Step | Description | Volume or Time | Reagents and Conditions |
|---|---|---|---|
| 1 | Synchronizing Plasmodium falciparum | 5% sorbitol solution | |
| 2 | Plating P. falciparum-luc | 15 μL | 5% parasitemia, 3% RBC |
| 3 | Controls | 15 μL | Wild-type P. falciparum, media, 3% RBCs, P. falciparum-luc with CQ |
| 4 | Diluting Lopac1280 library compound | 80× dilution | |
| 5 | Loading Lopac1280 library compound | 10 μL | Final concentration at 10 μM |
| 6 | Incubation | 48 h | 37°C, 92% N2, 5% CO2, 3% O2 |
| 7 | Adding reporter reagent | 25 μL | OneGlo Promega |
| 8 | Incubation | 3 min | Room temperature |
| 9 | Assay readout | 0.5 s/well | BioTek, luminescent mode |
CQ, chloroquine; LOPAC, Library of Pharmacologically Active Compounds.
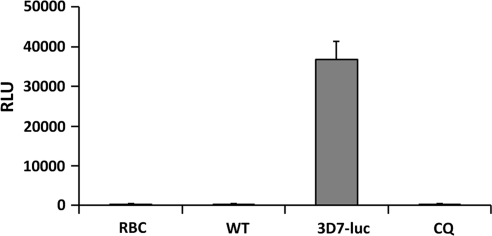
Using the synchronized parasites and the ONE-Glo luciferase assay system, we were able to achieve consistent and high levels of luciferase expression, with a maximum of 37,850 RLU, less deviation (CV of 7%), higher S/B ratio (139.81), and consistent Z′-values of 0.766 and 0.771, when using RBCs and wt-3D7 parasites as the negative signal controls (Table 2). When using CQ as the positive drug control at the optimal assay conditions, Z′-value, CV, and S/B ratio were determined to be 0.768±0.02, 9.5%±0.6%, and 70.9±11.4, respectively, indicating the robustness, reliability, and repeatability of the assay (Fig. 3).
Table 2.
Summary of the Assay Parameters During Assay Optimization and Validation
| Optimization in a 96-Well Plate | Optimization in a 384-Well Plate | First Pilot Screening | Second Pilot Screening | |
|---|---|---|---|---|
| Average Z'-value | 0.46±0.02 | 0.70±0.02 | 0.57±0.08 | 0.70±0.01 |
| Average S/B ratio | 9.1 | 50.9±11.4 | 60.6±12.80 | 70.9±19.6 |
| CV (RBCs only) | 12% | 9.1%±0.6% | 13.5%±3.0% | 9.5%±5.0% |
CV, coefficient variation; S/B, signal-to-background; RBC, red blood cell.
Fig. 3.
Parasites synchronization. Using the 5% sorbitol treatment, the ring-stage parasites were synchronized and grown to trophozoite stage, which was then seeded in the 384-well plate. At 72 h postseeding, luciferase signals were determined using the One-Glo Luciferase Assay System (Promega). CQ was used as the positive control drug to inhibit the growth of parasites, and RBCs and wild-type parasites were used as negative control for the background noises. The experiment was repeated for three times and each data point represents the average value of eight replicates; error bar represents standard error of n=8 values. CQ, chloroquine.
Assay Validation Using the LOPAC1280 Compound Library
The miniaturized assay was validated in a pilot screen using the LOPAC1280 compound library consisting of 1,280 structurally diverse small-molecule compounds. This is a collection of high-quality, innovative molecules that span a broad range of cell signaling and neuroscience areas, reflecting the most commonly screened targets in the drug discovery community. LOPAC1280 is designed for the validation of new drug discovery assays. In this pilot screening, all compounds were assayed at a 10 μM final concentration. RBCs, wt 3D7, and CQ were included in the assay for quality control. Based on the results from our duplicated screening, we obtained an average Z′-value of 0.57±0.08 for the first screening and 0.69±0.02 for the second screening, respectively (Table 2). The S/B ratio was high, at 60.6±12.8 for the first screening and 70.9±19.6 for the second screening, respectively (Table 2).
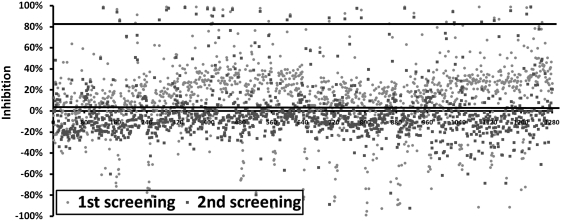
The percentages of inhibition of luminescent signals from each compound in the screening were plotted (Fig. 4). “Hits” are defined as compounds that display efficacy values greater than the mean of the negative control plus three times the standard deviation of the negative control mean, which was 90% in this duplicated screening. Of the 1,280 compounds screened, we identified 24 and 19 compounds with over 90% inhibition in first and second pilot screenings, respectively, including 18 compounds identified in both screenings. The results from this validation assay demonstrated the robustness and repeatability of the assay. It is noteworthy that we identified a number of positive hits in this LOPAC1280 compound library, including emetine dihydrochloride (EME), dequalinium dichloride, A-77636 hydrochloride, diphenyleneiodonium chloride, CGP-74514A hydrochloride, chloroquine diphosphate, ellipticine, mitoxantrone, brefeldin A, propylnorapomorphine hydrochloride, propafenone hydrochloride, and hexahydro-sila-difenidol hydrochloride.
Fig. 4.
The percentage of inhibition of parasite growth from each compound in the two pilot screenings of LOPAC1280. Each dot represents one compound from the library, and a total of two screenings were plotted. The higher line at 82.5% inhibition of parasite growth was calculated as the average inhibition (0.35%, the lower line) plus three times standard deviation of % inhibition of all compounds from the entire pilot screen. LOPAC, Library of Pharmacologically Active Compounds.
Confirmation of the Hits in the Secondary Assay
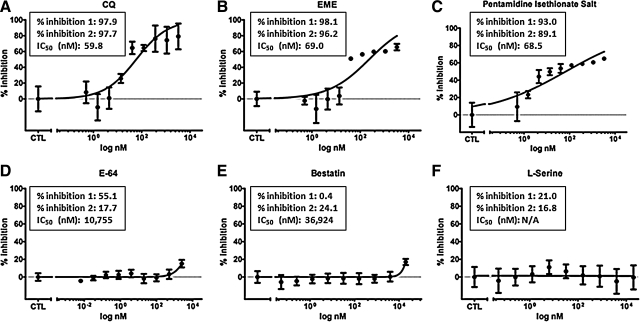
To further validate and confirm hits identified from the pilot screenings, we chose three positive hits, including EME, pentamidine isethionate salt, and CQ, as well as three negative hits, including E-64, destatin hydrochloride, and l-serine. In the repeated pilot screenings, EME, pentamidine isethionate salt, and CQ showed strong inhibition to the growth of transgenic 3D7 parasite indicated by the decrease of luminescent signals, including 98.1% and 96.2% inhibition by EME, 93.0% and 89.1% by pentamidine isethionate salt, and 97.9% and 97.7% by CQ, respectively. E-64 showed marginal effect with 55.1% and 17.7% inhibition rate at 10 μM in the first and second pilot screening, respectively. Both bestatin hydrochloride and l-serine showed <50% inhibitions (Fig. 5D–F). CQ has been known as an antimalarial drug for decades, whereas both pentamidine isethionate salt and EME are newly identified compounds. EME is the hydrochloride of an alkaloid found in the ipecac root and has been known to inhibit protein and nucleic acid syntheses.25–27 Using the SYBR Green-based assay as the secondary assay, we determined the 50% inhibitory concentration (IC50) of CQ, EME, pentamidine isethionate salt, E-64, and bestatin hydrochloride, which were 59.8, 69.0, 68.5, 10,755, and 36,924 nM (Fig. 5A–E). The IC50 of l-serine was undetermined because of its very weak inhibition. This result confirmed our hits and demonstrated that the HTS assay is reliable and ready to screen a large-compound library for the identification of novel antimalarial.
Fig. 5.
Hit validation in the secondary assay. The antimalarial efficacy of CQ (A), EME (B), pentamidine isethionate salt (C), E-64 (D), bestatin hydrochloride (E), and l-serine (F) were examined in the SYBR Green-based assay using the semilog dose–response assay, and 50% inhibitory concentrations were accordingly calculated. The % inhibition 1 and 2 represent the inhibition percentages obtained from the first and second repeated pilot screening. N/A, not available. The experiment was repeated for three times and each data point represents the average value of three replicates; error bar represents standard error of n=3 values. EME, emetine dihydrochloride.
Discussion
With the advancement in malaria parasite transgenics, we have developed a stable transgenic parasite line with high-level luciferase expression, which allows us to develop, optimize, and miniaturize a fully automated HTS-amenable, cell-based luminescent assay for antimalarial drug discovery. The assay was robust with Z′-value of 0.77±0.02, S/B ratio of 70.9±11.4, and CV of 9.5%±0.6%. This was further confirmed in the duplicated validation assay using the available LOPAC1280, with average Z′-values between 0.57±0.08 and 0.69±0.02, average CVs between 10.0%±0.6% and 13.0%±3.0%, and average S/B ratio of 60.6±12.8 and 70.9±19.6, respectively. All these results indicate that this cell-based luminescent assay against malaria is repeatable, reliable, and ready for the HTS to screen a large-compound library with assay robustness in line with HTS requirement. In addition, a secondary assay, the SYBR Green-based assay using a totally different detection method, is also available to confirm and prioritize hits and eliminate false-positives.
To develop an assay for a fully automated HTS environment, it is critical to develop the assay protocol with a single “mix-and-measure” step, using available reagents that can be easily read. There are several luciferase assay systems available, and we have chosen the ONE-Glo luciferase assay system, which provides a highly sensitive, robust, homogeneous assay for detection of firefly luciferase reporter gene expression in mammalian cells. Ideally suited for high- and ultrahigh-throughput applications, the ONE-Glo assay contains a new luciferase substrate, resulting in a reagent that is more stable and more tolerant to sample components when compared with standard luciferase assay reagents. The simple assay procedure (Table 1) involves a single step of adding the single ONE-Glo reagent directly to cells cultured in serum-supplemented medium. The simplicity, robustness, and reproducibility of the cell-based luminescent assay make it ideal for HTS. Comparing with other available HTS assays, including the SYBR Green-, the PicoGreen-, [3H]-hypoxanthine- and GFP-based assays,17,28–30 this assay showed much higher S/B ratio (>70) and lower CV (<10%), indicating the “signal strength” of the assay.
For decades, in vitro malaria drug assays have been performed by quantifying parasite uptake of radiolabeled substrates such as [H3] hypoxanthine and [H3] ethanolamine as a measure of growth and viability of the parasites.11,31 Apparently, these methods are not suitable for the HTS format. In recent years, the development of fluorescence-based antimalaria assay leads to the establishment of several high-throughput, nonradioactive assays, including the SYBR Green I22 and DAPI staining-based assays,29 as well as transgenic parasite-based assays using reporter genes such as luciferase20,32 and GFP.19 An antimalaria HTS assay has been developed and validated using transgenic 3D7 P. falciparum parasites that express firefly luciferase under the control of the histidine-rich protein II (HRPII) promoter.32 A pilot drug screen using this assay identified a total of 67 compounds that displayed over 85% inhibition of parasite growth in the primary HTS, and 19 of the hit compounds were confirmed as active using dose–response assays (two compounds were part of the NINDS library and the LOPAC1280 library).32 The transgenic parasites used in the HTS effort in this report utilized the hsp86 promoter controlling luciferase reporter gene expression.20 In our duplicated pilot drug screens of the LOPAC1280 library, we identified 18 compounds with over 90% inhibition in both screens. As the LOPAC1280 library has been also screened using the SYBR Green I assay,33 we compared hits identified from the library using these three assays. The SYBR Green I assay identified 155 compounds that inhibited growth in all seven parasite lines tested, including all hits identified from both HRPII- and hsp86-luciferase–based HTS assays. A total of 12 compounds that exhibited over 50% inhibition in our preliminary assay were also identified in the HRPII-luciferase and SYBR Green I assays,32,33 including quinacrine dihydrochloride, CQ diphosphate, EME hydrate, 3′,4′-dichlorobenzamil, R(−)-propylnorapomorphine hydrochloride, pentamidine isethionate, hexahydro-sila-difenidol hydrochloride, quinine sulfate, quinidine sulfate, benzamil hydrochloride, S(−)-UH-301 hydrochloride, and vincristine sulfate. Interestingly, 13 compounds with over 90% inhibition that were identified in our preliminary screen were also identified in the SYBR Green I assay with very reliable and consistent IC50s,33 indicating that this assay is very reliable. Interestingly, these hits were not identified in the HRPII-luciferase–based assay,32 including dequalinium dichloride, U-83836 dihydrochloride, A-77636 hydrochloride, naloxonazine dihydrochloride, dequalinium analog, mitoxantrone, CGP-74514A hydrochloride, SB 224289 hydrochloride, ellipticine, diphenyleneiodonium chloride, brefeldin A from Penicillium brefeldianum, U-74389G maleate, and (S)-(−)-propafenone hydrochloride. Also, seven hits identified in the HRPII-luciferase–based assay were not identified in our primary assay, including 5-(N-methyl-N-isobutyl) amiloride, vinblastine sulfate salt, SKF 95282 dimaleate, 5-(N,N-dimethyl)amiloride hydrochloride, 5-(N-ethyl-N-isopropyl)amiloride, amperozide hydrochloride, and BW 284c51. It is possible that these compounds could be PY analogs and/or might have similar mechanism of action like PY.
Although this assay showed many advantages when comparing with other available assays, it is notable that there are some limitations. As previously discussed, it requires a transgenic luciferase-expressing parasite line selected using PY. This may potentially cause cross-resistance to certain candidate drugs,20 as we did observe some PY analogs were not identified in the pilot screening. Although this problem could be easily resolved by developing another transgenic line using a different selection marker, it is possible that in an HTS screening, the PY analogs might not be identified when using this assay. This, in turn, might help one focus on “hits” with a novel antimalarial mechanism. Nevertheless, our study showed that a robust, fully automated HTS assay is ready for the screening of a large-compound library.
Abbreviations
- CQ
chloroquine
- CV
coefficient variation
- DMSO
dimethyl sulfoxide
- EME
emetine dihydrochloride
- GFP
green fluorescent protein
- HRPII
histidine-rich protein II
- hsp86
heat shock protein 86
- HTS
high-throughput screening
- IC50
50% inhibitory concentration
- LOPAC1280
Library of Pharmacologically Active Compounds
- PY
pyrimethamine
- RBC
red blood cell
- RLU
relative luminescent units
- S/B ratio
signal-to-background ratio
- wt
wild type
Acknowledgments
The authors thank Mr. Patrick W. Wright, who provided the technical support for drug diluting and loading using Precision Microplate Pipetting System. This research was supported by NIH grant 1R21NS061749-01 to L. Cui (PI) and Q. Li (co-PI) and by UAB IMPACT funds to Q. Li.
Disclosure Statement
No competing financial interests exist.
References
- 1.Cui L. Su XZ. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Rev Anti Infect Ther. 2009;7:999–1013. doi: 10.1586/eri.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenwood B. Intermittent preventive antimalarial treatment in infants. Clin Infect Dis. 2007;45:26–28. doi: 10.1086/518574. [DOI] [PubMed] [Google Scholar]
- 3.Snow RW. Guerra CA. Noor AM. Myint HY. Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White LJ. Maude RJ. Pongtavornpinyo W, et al. The role of simple mathematical models in malaria elimination strategy design. Malaria J. 2009;8:212. doi: 10.1186/1475-2875-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayton K. Su XZ. Genetic and biochemical aspects of drug resistance in malaria parasites. Curr Drug Targets Infect Disord. 2004;4:1–10. doi: 10.2174/1568005043480925. [DOI] [PubMed] [Google Scholar]
- 6.Travassos MA. Laufer MK. Resistance to antimalarial drugs: molecular, pharmacologic, and clinical considerations. Pediatr Res. 2009;65(5 Pt 2):64R–70R. doi: 10.1203/PDR.0b013e3181a0977e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley EA. White NJ. Artemisinin-based combinations. Curr Opin Infect Dis. 2005;18:531–536. doi: 10.1097/01.qco.0000186848.46417.6c. [DOI] [PubMed] [Google Scholar]
- 8.Dondorp AM. Nosten F. Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alker AP. Lim P. Sem R, et al. Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am J Trop Med Hyg. 2007;76:641–647. [PubMed] [Google Scholar]
- 10.Co EM. Dennull RA. Reinbold DD. Waters NC. Johnson JD. Assessment of malaria in vitro drug combination screening and mixed-strain infections using the malaria SYBR green I-based fluorescence assay. Antimicrob Agents Chemother. 2009;53:2557–2563. doi: 10.1128/AAC.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desjardins RE. Canfield CJ. Haynes JD. Chulay JD. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother. 1979;16:710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bacon DJ. Latour C. Lucas C. Colina O. Ringwald P. Picot S. Comparison of a SYBR green I-based assay with a histidine-rich protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob Agents Chemother. 2007;51:1172–1178. doi: 10.1128/AAC.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabre R. Berry A. Morassin B. Magnaval JF. Comparative assessment of conventional PCR with multiplex real-time PCR using SYBR Green I detection for the molecular diagnosis of imported malaria. Parasitology. 2004;128(Pt 1):15–21. doi: 10.1017/s0031182003004219. [DOI] [PubMed] [Google Scholar]
- 14.Izumiyama S. Omura M. Takasaki T. Ohmae H. Asahi H. Plasmodium falciparum: development and validation of a measure of intraerythrocytic growth using SYBR Green I in a flow cytometer. Exp Parasitol. 2009;121:144–150. doi: 10.1016/j.exppara.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Rason MA. Randriantsoa T. Andrianantenaina H. Ratsimbasoa A. Menard D. Performance and reliability of the SYBR Green I based assay for the routine monitoring of susceptibility of Plasmodium falciparum clinical isolates. Trans R Soc Trop Med Hyg. 2008;102:346–351. doi: 10.1016/j.trstmh.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JD. Dennull RA. Gerena L. Lopez-Sanchez M. Roncal NE. Waters NC. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob Agents Chemother. 2007;51:1926–1933. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vossen MG. Pferschy S. Chiba P. Noedl H. The SYBR Green I malaria drug sensitivity assay: performance in low parasitemia samples. Am J Trop Med Hyg. 2010;82:398–401. doi: 10.4269/ajtmh.2010.09-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadekoppala M. Kline K. Akompong T. Haldar K. Stable expression of a new chimeric fluorescent reporter in the human malaria parasite Plasmodium falciparum. Infect Immun. 2000;68:2328–2332. doi: 10.1128/iai.68.4.2328-2332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez BA. Varotti FP. Rodrigues FG. Carvalho LH. Validation of a Plasmodium falciparum parasite transformed with green fluorescent protein for antimalarial drug screening. J Microbiol Methods. 2007;69:518–522. doi: 10.1016/j.mimet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Cui L. Miao J. Wang J. Li Q. Plasmodium falciparum: development of a transgenic line for screening antimalarials using firefly luciferase as the reporter. Exp Parasitol. 2008;120:80–87. doi: 10.1016/j.exppara.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trager W. Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 22.Smilkstein M. Sriwilaijaroen N. Kelly JX. Wilairat P. Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh RN. DeBiasio R. Hudson CC. Ramer ER. Cowan CL. Oakley RH. Quantitative cell-based high-content screening for vasopressin receptor agonists using transfluor technology. J Biomol Screen. 2005;10:476–484. doi: 10.1177/1087057105274896. [DOI] [PubMed] [Google Scholar]
- 24.Zhang JH. Chung TD. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 25.Grollman AP. Inhibitors of protein biosynthesis. V. Effects of emetine on protein and nucleic acid biosynthesis in HeLa cells. J Biol Chem. 1968;243:4089–4094. [PubMed] [Google Scholar]
- 26.Moller M. Herzer K. Wenger T. Herr I. Wink M. The alkaloid emetine as a promising agent for the induction and enhancement of drug-induced apoptosis in leukemia cells. Oncol Rep. 2007;18:737–744. [PubMed] [Google Scholar]
- 27.Moller M. Wink M. Characteristics of apoptosis induction by the alkaloid emetine in human tumour cell lines. Planta Med. 2007;73:1389–1396. doi: 10.1055/s-2007-990229. [DOI] [PubMed] [Google Scholar]
- 28.Abiodun OO. Gbotosho GO. Ajaiyeoba EO, et al. Comparison of SYBR Green I-, PicoGreen-, and [3H]-hypoxanthine-based assays for in vitro antimalarial screening of plants from Nigerian ethnomedicine. Parasitol Res. 2010;106:933–939. doi: 10.1007/s00436-010-1743-z. [DOI] [PubMed] [Google Scholar]
- 29.Baniecki ML. Wirth DF. Clardy J. High-throughput Plasmodium falciparum growth assay for malaria drug discovery. Antimicrob Agents Chemother. 2007;51:716–723. doi: 10.1128/AAC.01144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cervantes S. Prudhomme J. Carter D, et al. High-content live cell imaging with RNA probes: advancements in high-throughput antimalarial drug discovery. BMC Cell Biol. 2009;10:45. doi: 10.1186/1471-2121-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elabbadi N. Ancelin ML. Vial HJ. Phospholipid metabolism of serine in Plasmodium-infected erythrocytes involves phosphatidylserine and direct serine decarboxylation. Biochem J. 1997;324(Pt 2):435–445. doi: 10.1042/bj3240435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucumi E. Darling C. Jo H, et al. Discovery of potent small-molecule inhibitors of multidrug-resistant Plasmodium falciparum using a novel miniaturized high-throughput luciferase-based assay. Antimicrob Agents Chemother. 2010;54:3597–3604. doi: 10.1128/AAC.00431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan J. Johnson RL. Huang R, et al. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum. Nat Chem Biol. 2009;5:765–771. doi: 10.1038/nchembio.215. [DOI] [PMC free article] [PubMed] [Google Scholar]