Abstract
Affinity purification (AP) of protein complexes combined with LC-MS/MS analysis is the current method of choice for identification of protein-protein interactions. Their interpretation with respect to significance, specificity, and selectivity requires quantification methods coping with enrichment factors of more than 1000-fold, variable amounts of total protein, and low abundant, unlabeled samples. We used standardized samples (0.1–1000 fmol) measured on high resolution hybrid linear ion trap instruments (LTQ-FT/Orbitrap) to characterize and improve linearity and dynamic range of label-free approaches. Quantification based on spectral counts was limited by saturation and ion suppression effects with samples exceeding 100 ng of protein, depending on the instrument setup. In contrast, signal intensities of peptides (peak volumes) selected by a novel correlation-based method (TopCorr-PV) were linear over at least 4 orders of magnitude and allowed for accurate relative quantification of standard proteins spiked into a complex protein background. Application of this procedure to APs of the voltage-gated potassium channel Kv1.1 as a model membrane protein complex unambiguously identified the whole set of known interaction partners together with novel candidates. In addition to discriminating these proteins from background, we could determine efficiency, cross-reactivities, and selection biases of the used purification antibodies. The enhanced dynamic range of the developed quantification procedure appears well suited for sensitive identification of specific protein-protein interactions, detection of antibody-related artifacts, and optimization of AP conditions.
Antibody-based affinity purification (AP)1 of protein assemblies from biological samples followed by mass spectrometric analysis represents an increasingly popular approach for identification of protein-protein interactions (AP-MS) (1–3). Despite the exquisitely high and specific enrichment theoretically obtainable with antibodies (Abs), this approach faces a number of technical and intrinsic challenges in practice. Target protein complexes typically suffer from poor solubility, instability, and low abundance, particularly when associated with lipid membranes. Moreover, various antibody-related properties such as target selectivity, cross-reactivity, and interference with protein-protein interactions may lead to false-positive and false-negative results (4). Finally, biological protein-protein interactions may have a more dynamic character, may depend on regulated modifications, or may involve rare protein partners. Together, these effects lead to a significant reduction of AP signal to noise, i.e. low co-enrichment efficiency of interaction partners and significant overlap with background or nonspecific proteins.
Classically, AP specificity has been addressed by visualization of purified proteins on one- or two-dimensional gels and comparison of band patterns or spots with those obtained in controls (5, 6). However, nano-flow liquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS) has eliminated the need for protein separation and opened new possibilities for protein quantification (1, 3). A number of protein quantification techniques have been established and successfully applied, most of them based on chemical or metabolic labeling of proteins or peptides (as reviewed by (7, 8)). Notwithstanding, in functional proteomic studies, label-free quantification methods are becoming increasingly popular because the use of native source material often precludes metabolic isotope labeling, and chemical derivatization tends to introduce biases and to reduce sensitivity (7). In addition, label-free approaches do not suffer from multiplexing restrictions or dynamic range limitations caused by limited isotopic purity of labels (9).
Label-free LC-MS/MS quantification can be based on two different types of data: MS/MS (peptide fragment) spectra generally acquired in data-dependent mode have been used to calculate rough quantitative parameters, like the exponentially modified protein abundance index score (exponentially modified protein abundance index (10)), the rPQ (relative peptide query count (11)), or the relative protein sequence coverage. Preferably, LC-MS data can be used to extract peak volumes (PVs) as the integrated m/z intensities (extracted ion currents (XICs)) over elution time for all peptide ions (12). Because of the high complexity of peptide samples and resulting m/z signals, the applicability of PV-based quantification critically depends on the performance of the LC-MS instrument setup and requires rather sophisticated software tools (13). High m/z resolution and mass accuracy in the low ppm range as recently achieved on an LTQ-Orbitrap with the newly developed MaxQuant software (14, 15) provides an excellent basis for reliable large scale quantification of proteins. In fact, such high resolution LC-MS PV-based methods have recently been used for identification of novel membrane protein interaction partners (16–18) and associated protein networks (19).
An important parameter for quantitative evaluation of native source AP samples is the dynamic range. Antibodies are known to enrich their target proteins by more than 10000–100000-fold (20, 21), suggesting that the differences in protein abundance between AP samples and controls exceed by far the range of protein changes observed in typical proteomic studies. In fact, established label-free (PV-based) quantification methods have a reported dynamic range of just 2–3 orders of magnitude, slightly higher than that achieved with isotopic labeling techniques (7). Moreover, a broad standardized study conducted by the Association of Biomolecular Resource Facilities Proteomics Research Group with 52 participating laboratories revealed rather large errors in the relative quantification of proteins differing by more than 1 order of magnitude (22). The factors contributing to quantification errors and dynamic range limitations have so far been barely studied. Liu et al. (23) recently took a first step to explore the accuracy and linearity of peptide identification and PVs over a broader abundance range. They observed strong saturation of MS/MS identification and nonlinear behavior of distinct groups of peptide PVs. However, it remained open how their suggested method for selection of suitable peptide PVs might translate into improved quantification of proteins in real samples.
We therefore conducted a systematic study over an extended dynamic range on three different MS instrument setups starting with standardized protein samples designed to reflect the characteristics of typical AP probes. A derived label-free quantification method with improved linearity was then validated and exemplarily applied to evaluation of APs and controls of a model membrane protein complex.
EXPERIMENTAL PROCEDURES
AP of Kv1.1 and Associated Proteins
Plasma membrane enriched fractions were prepared from pools of freshly isolated rat and mouse brains as described (31). Control brains from mice carrying homozygous deletions of the Kv1.1 gene (Kv1.1−/−) were kindly provided by J. S. Trimmer (University of California, Davis) and processed accordingly. The membranes were solubilized for 30 min on ice by mixing with ComplexioLyte 80 (neutral detergent buffer, Logopharm GmbH), supplemented with protease inhibitors (1 mM PMSF (Roth), 1 μg/ml aprotinin (Roth), 1 μg/ml pepstatin A (Sigma), and 1 μg/ml leupeptin (Sigma)), at a protein/buffer ratio of 1.25 mg/ml. Solubilization buffers with different ionic strengths (experiments in Fig. 5) were generated by adjusting the salt concentration of ComplexioLyte 80. After removal of nonsolubilized material by ultracentrifugation at 100,000 × g for 10 min, solubilisates were incubated with immobilized antibodies (1.0 ml/15 μg of antibody) for 3 h on ice and washed two times with solubilization buffer. Bound proteins were eluted with nonreducing Laemmli buffer (10 mM DTT added after elution). The following antibodies were used for immunoaffinity purification: anti-Kv1.1A (rabbit polyclonal, directed against amino acids 458–475 of rat Kv1.1; gift of H.G. Knaus, Medical University of Innsbruck, Austria (26)), anti-Kv1.1B (rabbit polyclonal; Alomone Labs APC-009), anti-Kv1.1C (mouse monoclonal K36/15; Neuromab 73–105), and anti-Kv1.1D (mouse monoclonal K20/78; Neuromab 73–007); preimmune control IgG was from rabbit (Millipore/Upstate, catalog number 12-370). All of the affinity purifications were prepared and analyzed at least two times. Aliquots of solubilisates, unbound material, and antibody eluates were resolved by SDS-PAGE (7.5% (w/v) acrylamide) and blotted onto PVDF membrane. Kv1.1 and LGI1 proteins were detected by successive incubation with anti-Kv1.1C and anti-LGI1 antibodies (gift of H.G. Knaus, Medical University of Innsbruck, Austria (26); diluted 1:10000 and 1:500, respectively, in 2% skim milk powder in phosphate-buffered saline + 0.05% Tween), HRP-coupled anti-mouse or rabbit IgG (Santa Cruz Biotechnology) and ECL Plus (GE Healthcare).
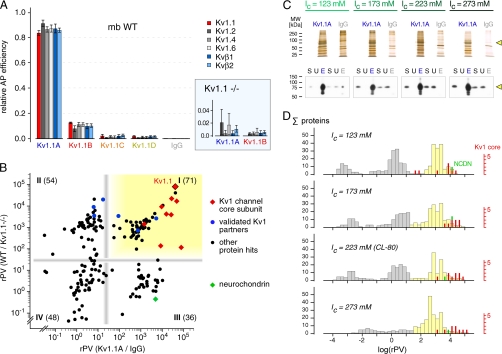
Fig. 5.
Quantitative evaluation of Kv1.1 APs by the TopCorr-PV method. A, relative amounts of the core subunits of Kv1 channels as obtained in APs with the indicated anti-Kv1.1 Abs from CL-80 solubilized mouse brain membranes. The bars are the means ± S.D. of four to six specific peptide PVs normalized to their sum over all four APs; order of AP efficiency: anti-Kv1.1A (86.7%), anti-Kv1.1B (10.9%), and anti-Kv1.1C and anti-Kv1.1D (< 3%). Inset, relative quantification of the indicated proteins in APs from mouse brain membranes lacking Kv1.1α at 5-fold enlarged scale. Note subtype specificity of both anti-Kv1.1A and anti-Kv1.1B (cross-reactivity with other Kv1α subtypes ≤2%). B, rPVs obtained in APs from mouse brain membrane with anti-Kv1.1A versus IgG (x axis) and in anti-Kv1.1A APs from membranes of wild type versus Kv1.1α knockout mice (y axis). The red diamonds represent Kv1 α and β subunits (Kv1.1–6, Kvβ 1–3), blue dots highlight known interaction partners of Kv1 channels, and black dots symbolize all other proteins. Gray bars (rPV of 25) are thresholds suggested by the clustering of the proteins; clusters (quadrants) are numbered I–IV and contain the indicated number of proteins (supplemental Fig. S3A; details given in the text). C, series of anti-Kv1.1A APs from mouse brain membranes solubilized with CL-80 buffer adjusted to the indicated ionic strengths (Ic). Upper panel, silver staining of the APs resolved by SDS-PAGE; loaded amounts correspond to 15 μg of purification Ab incubated with 1 mg of solubilized rat brain membrane. Lower panel, Western blot (anti-Kv1.1C) probing gel separated inputs, breakthroughs, and eluates of the AP series; equivalent amounts of input (S) and breakthrough (U) were loaded as well as 10% of each AP eluate (E). D, distribution histograms of rPVs obtained in the AP series in C. The filled bars represent the number of proteins (calculated as in B) per logarithmically binned rPV interval. Gray bars, background proteins; yellow bars, proteins significantly enriched by anti-Kv1.1A versus IgG. The positions of the nine Kv1 core subunits and of the cross-reactive neurochondrin (NCDN) are marked by red and green bars, respectively (scale on the right). Note the changes in number and distribution of proteins with increasing Ic (details given in the text).
Preparation of Samples for MS Analysis
Protein samples were shortly run on SDS-PAGE gels and silver-stained (without enhancement by cross-linkers; supplemental Fig. S2). Gel lanes were excised and subjected to tryptic digestion with sequencing grade modified trypsin (Promega) as described in Ref. 32.
Standard proteins were formulated as a stock mix of 10 different commercially available protein preparations (each adjusted to 1 μm) covering a broad molecular mass range: laminin I (α1/β1/γ1; murine EHS sarcoma; Trevigen 3400-010-01; UniProtKB accessions P19137/P02469/P02468; 338 kDa/197 kDa/177 kDa); myosin heavy chain (rabbit muscle; Sigma M7659; Q28641; 223 kDa); β-galactosidase (Escherichia coli, Merck/Calbiochem 345788; P00722; 116 kDa); transferrin (human plasma; Merck/Calbiochem 616419; P02787; 77 kDa); albumin (bovine serum; Sigma A7517; P02769; 69 kDa); glutamate dehydrogenase (bovine liver; Sigma G2626; P00366; 62 kDa); aldolase (rabbit muscle; Roche 102644; P00883; 39 kDa); glyceraldehyde-3-phosphate dehydrogenase (rabbit muscle; Sigma G5262; P46406; 36 kDa); carbonic anhydrase (bovine erythrocytes; Sigma C2273; P00921; 29 kDa); and lysozyme (chicken egg white; Sigma L4631; P00698; 14 kDa).
A mixture containing 3 pmol of each protein was in-gel digested, and the obtained peptide extracts were used to prepare dilution series 1 (protein abundances ranging from 0.1 to 1000 fmol) for studying linearity and sensitivity of different mass spectrometric setups. A second set of samples (series 2) simulating the protein composition of typical APs was generated by spiking defined amounts of standard proteins (1, 10, 100, 1000, and 10000 fmol) into a complex mixture of different rat brain AP eluates. A series of more complex samples (series 3) was generated by mixing 1 μg of E. coli membrane with 0.1, 1, 10, or 100 μg of rat brain membrane (IF), both solubilized with ComplexioLyte 80 (supplemental Fig. S2). All of the series were prepared as triplicate samples and analyzed on the LTQ-FT or the LTQ-FT Ultra and the LTQ-Orbitrap XL instrument, unless indicated otherwise.
Mass Spectrometric Analysis
Nano-LC-MS/MS analysis of in-gel digested samples was performed on three different high resolution hybrid ion trap instruments: the LTQ-FT, the upgraded LTQ-FT Ultra, and the LTQ-Orbitrap XL (all Thermo Scientific with Proxeon nano-electrospray ion source). Extracted peptides were dissolved in 0.5% (v/v) trifluoroacetic acid, trapped on a C18 PepMap100 precolumn (5 μm; Dionex) of an UltiMate 3000 HPLC system (Dionex), and separated with an aqueous-organic gradient (solution A: 0.5% (v/v) acetic acid; solution B: 0.5% (v/v) acetic acid in 80% (v/v) acetonitrile; flow 300 nl/min) on a 10 cm C18 column (PicoTipTM Emitter, 75 μm, tip: 8 ± 1 μm, New Objective; self-packed with ReproSil-Pur 120 ODS-3, 3 μm, Dr. Maisch). Calculated amounts and 38% of the peptide extracts from the standardized samples and the AP eluates, respectively, were used for each nano-LC-MS/MS analysis. Elution gradient settings were (i) for standard protein samples: 5 min 3% B, 30 min from 3% B to 30% B, 10 min from 30% B to 100% B, 5 min 100% B, 5 min from 100% B to 3% B, 15 min 3% B and (ii) for AP eluate samples: 5 min 3% B, 60 min from 3% B to 30% B, 15 min from 30% B to 100% B, 5 min 100% B, 5 min from 100% B to 3% B, 15 min 3% B.
MS instrument settings were as follows: spray voltage, 2.3 kV; capillary temperature, 125 °C; FT full MS target, 500000 (maximum injection time, 1000 ms; full scan injection waveforms enabled); IT MSn target, 10000 (maximum injection time, 400 ms; injection waveforms enabled). Each scan cycle consisted of one FTMS full scan (resolution, 100000 for LTQ-FT (Ultra) and 60000 for LTQ-Orbitrap XL; positive polarity; profile data; scan range, 370–1700 m/z) and up to five ion trap data-dependent scans (centroid data; five most intense ions from scan event 1) with dynamic exclusion (repeat count, 1; exclusion list size, 500; repeat/exclusion duration, 30 s; exclusion mass width, ±20 ppm), preview mode for FTMS master scans, charge state screening, monoisotopic precursor selection, and charge state rejection (charge state 1) enabled. Activation type was CID with default settings.
Database Search and Quantification Procedures
LC-MS/MS data was extracted using the extract_msn utility (grouping tolerance, 0.05) and searched against the UniProtKB/Swiss-Prot database (release 2010_08) using the Mascot search engine (version 2.3.01; Matrix Science). The search space was restricted to all mouse, rat, and human entries including P00761, P00766, and P02769, for standardized samples supplemented with the standard proteins from other species and their major contaminants (extracted from a search against all entries). Common variable modifications (carbamidomethyl (C), deamidated (NQ), Gln → pyro-Glu (N-term Q), Glu → pyro-Glu (N-term E), oxidation (M), and propionamide (C)) were allowed as well as one missed cleavage; monoisotopic peptide mass tolerance was ±5 ppm (after linear recalibration), and fragment mass tolerance was ±0.8 Da. Proteins identified by only one specific MS/MS spectrum or representing exogenous contaminations such as keratins or immunoglobulins were eliminated.
Relative amino acid sequence coverage of proteins (Fig. 1C and supplemental Fig. S1) was calculated as SC = Ni/(Ni + Nan), where Ni is the number of amino acid residues covered by identified peptides (with Mascot Score ≥ 20), and Nan is the number of MS-accessible (peptides within 740 < molecular mass < 3000 with trypsin cleavage C-terminal to the basic amino acids, but not N-terminal to proline; missed cleavages were not considered) but not identified amino acids in the respective SwissProt sequence.
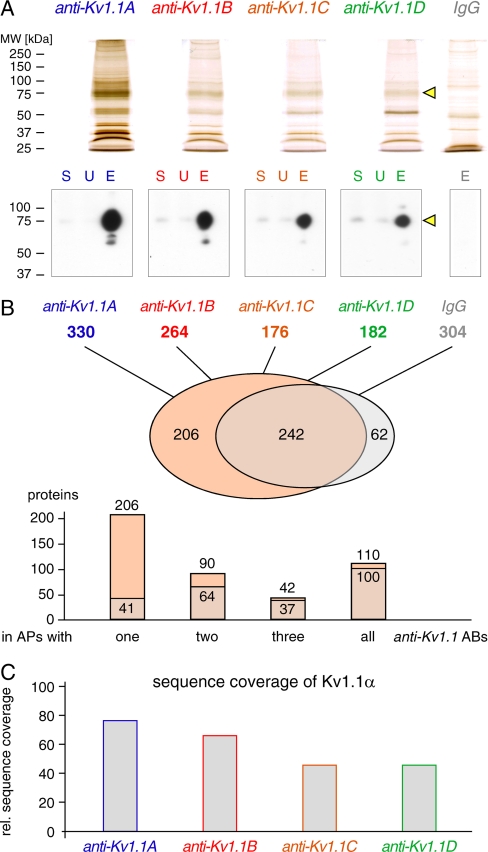
Fig. 1.
APs of Kv1 channel complexes with distinct anti-Kv1.1 Abs. A, upper panel, silver-stained SDS-PAGE gel separations of APs obtained with the indicated anti-Kv1.1 Abs or control IgGs. The loads correspond to 15 μg of Ab incubated with 1 mg of solubilized rat brain membrane. Lower panel, Western blot of the indicated APs using anti-Kv1.1C; equivalent amounts of solubilisate (S) and unbound (U) material were loaded, as well as 10% of each AP (E). Note the different purification efficiencies of the anti-Kv1.1 Abs (yellow arrowheads refer to Kv1.1α subunit). B, MS identification of Kv1.1 and co-purified proteins (SwissProt database; mouse matches); from top to bottom, the total number of significant protein hits in each purification data set, overlap of proteins identified in Kv1.1 APs (red) and IgG control (gray) and consistency of protein hits among the four Kv1.1 APs and IgG control. C, relative amino acid sequence coverage of Kv1.1α deduced from peptides retrieved by MS analysis of APs with the indicated anti-Kv1.1 Abs (see “Experimental Procedures”). MW, molecular mass.
Total MS/MS count information and total ion currents (TICs) during the gradient elution phase (Fig. 2) were extracted using Xcalibur and an in-house script. For evaluation of protein spectral counts, the number of total MS/MS matches (score ≥ 20; without miscleaved peptides) assigned to each protein by Mascot was determined by an in-house written software tool; exponentially modified protein abundance index was calculated as described (10).
Fig. 2.
Properties of LC-MS and MS/MS data recorded from the reference sample. Dilution series of the in-gel digested reference sample analyzed on three hybrid MS instruments (LTQ-FT, LTQ-FT Ultra, and LTQ-Orbitrap XL); data points in A–C are either the means ± S.D. of three (0.1–100 fmol) or two (300 fmol) experiments or represent a single measurement (1000 fmol), respectively. A, number of fragment ion spectra recorded along a 30-min aqueous-organic nano-HPLC gradient as a function of loaded reference protein. Note the pronounced saturation of MS/MS spectra recording with both LTQ-FT configurations starting at 100 fmol. B, number of MS/MS spectra (recorded on LTQ-Orbitrap XL) assigned to indicated protein components of the reference sample. Note the marked variation among individual proteins, as well as pronounced discontinuity and saturation. C, TICs recorded in the experiments in A. The dashed line represents result of a linear regression (m = 1.25) fitted to the slope of the TICs at loads ≥10 fmol; asymptotes of TICs at loads below ∼3 fmol reflect nonpeptide background signals. D, XICs determined for individual laminin A precursor ions on the LTQ-Orbitrap XL plotted over the load (S.D. omitted for clarity). Signal intensities span more than 6 orders of magnitude with an apparent detection threshold of ∼1000 intensity units (dashed green line). The top 20% of precursor ions selected by linear correlation ranking of their XICs (see text and “Experimental Procedures”) are colored in pink. Of the remaining precursor ion profiles, the 10 most intense ones are depicted in black.
For peak volume-based quantification, m/z features along LC-MS scans were detected and quantified (as intensity × retention time × m/z width) using msInspect (Computational Proteomics Laboratory, Fred Hutchinson Cancer Research Center, Seattle, WA; also tested were OpenMS, MZMine 1, XCMS, and IDEAL-Q). After correction of m/z shifts (based on MS-sequenced peptides using an in-house written script), features were aligned between different LC-MS/MS runs and assigned to the peptides identified by Mascot using an in-house written software tool (retention time tolerance, 3% or 1 min; m/z difference threshold, ± 5 ppm). The resulting peptide PVs were used for two different quantification procedures (carried out by an in-house-written software tool).
The first procedure quantified the relative abundance of a protein in different samples. Protein (isoform)-specific peptide peak volumes across the evaluated data set were ranked by their consistency determined by pair-wise linear correlation analysis (Pearson correlation); this reduced the influence of run-to-run variations in ionization, chemical modification, and peak detection. Starting with the top 20% of these peptides, a maximum of six to a minimum of two peptide PVs were selected for relative quantification from the best correlating PVs (sequenced peptides without an assigned PV were omitted). In standard protein dilution measurements (Figs. 3B and 4A), relative amounts of the indicated proteins were determined as medians of selected PVs, each normalized to the linear regression fit to its slope (i.e. fitted to PV versus amount of loaded protein (0.1–1000 fmol in Fig. 3B and 0.1–100 fmol in Fig. 4A)), reflecting the specific MS signal intensity of each peptide. Relative amounts of a protein in a sample versus control (Figs. 4C, and 5 and supplemental Figs. S3 and S4) were determined as medians of the respective PV ratios (rPV score) or means of all available PV ratios (supplemental Fig. S3B); to ensure validity, at least two peptide ratios with assigned PVs of 100000 volume units were required; if no PV could be assigned to a peptide in the AP controls, the detection limit of the spectrometer (3000 volume units with the settings used here) was inserted as a minimum estimate; the number of insertion-based ratios used for TopCorr-PV protein quantification was limited to three. For relative quantification of protein subtypes (Fig. 5A), subtype-specific peptides were manually inspected and means ± S.D. of selected PVs (4–6) normalized to their total across the four AP data sets were determined. For the second procedure, the relative molar amounts of different proteins (Fig. 4B) were estimated based on abundancenorm values (calculated as the sum of PVs divided by the number of MS-accessible amino acids (33)). Diagrams, calculation of means, medians, and standard deviations as well as linear or Gaussian fits to data were made in Excel (Microsoft) and IGOR Pro (Wavemetrics), respectively.
Fig. 3.
Quantification of the reference sample by the TopCorr-PV method. Quantification of proteins in the post-digest dilution series of the reference sample (Fig. 2) analyzed on the LTQ-Orbitrap XL. A, the top three to six peptide PVs (means ± S.D. as in Fig. 2) selected by linear correlation ranking for the indicated proteins; the dashed lines indicate the theoretically expected slope of 1. Beta-gal., β-galactosidase. B, relative PVs (medians of the three to six top-ranked peptide PVs individually normalized to their ionization efficiency as determined by linear regression) of the indicated proteins. Note the linear increase of the medians (slope factors ≈ 1.2) over the entire abundance range.
Fig. 4.
Validation of the TopCorr-PV method. Dilutions of the reference sample (amounts of 0.1, 1, 10, 100, and 1000 fmol for each protein) were spiked into a constant protein background and analyzed on an LTQ-Orbitrap XL (1/10 of each sample, three independent sample series). A, relative amounts of the indicated reference proteins determined with the TopCorr-PV method (medians of one to six top-ranked peptide PVs individually normalized to their total sum; means ± SDs of three (six for 100 fmol) replicate samples). The dashed lines indicate expected protein amounts, and the asterisks refer to values based on just one experiment or PV value. B, abundancenorm values (33) of the reference proteins shown in A (means ± S.D. as in A); respective values of consistently identified and quantified proteins of the background are shown as gray lines (protein profiles, S.D. values omitted for clarity; profile for trypsin is shown in black). Background proteins cover an abundance range of ∼104 corresponding to ∼0.1–100 fmol of the reference proteins. The high values obtained for albumin at 0.1 fmol result from background contamination in the samples. C, histogram plots of rPVs as obtained from the M1–M4 mixtures of solubilized E. coli and rat brain membranes (supplemental Fig. S2). The histogram bars reflect rPVs obtained from the medians of the three to six top-ranked peptide PVs averaged from three replicate samples. Left panel, summary of all E. coli protein rPVs (total of 311 values) from M1:M2, M1:M3, and M1:M4; three right panels, rat, mouse, or human protein rPVs from the indicated mixture ratios. The lines are the results of a Gaussian function fitted to the data; fit parameters peak and half-width (nonlogarithmic values) were 1.02 and 1.29 (E. coli 1:1; left panel), 0.083 and 1.53, 0.0056 and 2.00, and 0.0012 and 3.25 for rat brain 1:10, 1:100, and 1:1000, respectively.
RESULTS
Properties of Samples Obtained from Native Source APs
Kv1 voltage-gated potassium channels are multi-component membrane protein complexes that are predominantly found in axons and presynaptic boutons of central nervous system neurons (reviewed in Ref. 24) and that are readily accessible to Ab-based APs (25–28). Fig. 1A illustrates results from APs from solubilized mouse brain membranes using four different antibodies targeting Kv1.1 (anti-Kv1.1A–D), the most abundant pore-forming subunit of neuronal Kv1 channels (Kv1.1α), and a pool of preimmunization IgGs as a control. Isolated proteins resolved by silver-stained SDS-PAGE (Fig. 1A, upper panel) displayed complex band patterns dominated by a few abundant protein species and variable amounts of total protein estimated to range from 0.5 to 10 μg. Western blot analysis of AP aliquots confirmed high enrichment of Kv1.1α with all four Abs, albeit with different efficiency (Fig. 1A, lower panel). Although anti-Kv1.1A largely depleted the source material of its target, the other three Abs only purified fractions of the solubilized Kv1.1α. These differences between individual anti-Kv1.1 APs were recapitulated in subsequent LC-MS/MS analyses (Fig. 1B). Thus, all APs with the anti-Kv1.1 Abs displayed considerable complexity of between 176 and 330 proteins unambiguously identified (Fig. 1B, top panel) and offered a distinct coverage of the Kv1.1α primary sequence ranging from 46 to 76% (Fig. 1C and supplemental Fig. S1). Together, all anti-Kv1.1 APs co-purified 448 different proteins; 242 of those, however, were also retrieved by the control IgGs (Fig. 1B, bottom panel). Of the remaining 206 proteins, the majority was identified with only one of the anti-Kv1.1 Abs; only 10 proteins were retrieved by all four Abs (Fig. 1B, middle panel).
These results demonstrate that individual APs may exhibit complex and largely divergent protein patterns typically resulting from differences in both efficiency of target purification and background of Abs. As a consequence, any identification of proteins that are specifically co-purified with the target, such as Kv1.1 channels, requires an evaluation procedure providing reliable quantification of protein amounts from MS data on a broad abundance range and independent of composition and the total amount of protein in the AP sample.
LC-MS/MS Data Acquired from Hybrid Linear Ion Trap Instruments
For setting up such a quantification procedure, we used a well defined reference sample composed of equimolar amounts of 12 proteins with molecular masses ranging from 14 to 340 kDa (see “Experimental Procedures”) measured with three high resolution ion trap MS instruments, LTQ-FT, LTQ-FT Ultra, and LTQ-Orbitrap XL (each coupled to an UltiMate 3000 nano-HPLC system). In a first set of experiments, sensitivity and dynamic range of MS raw data (LC-MS and MS/MS data sets) were explored by measuring a triplicate dilution series of the in-gel digested reference sample (see series 1 in supplemental Fig. S2). Fig. 2A shows the total number of MS/MS spectra recorded with either instrument and plotted against the amount of each protein in the sample load (0.1–1000 fmol). Up to 100 fmol, spectral counts increased steadily with the load, whereas above 100 fmol further increase was only observed with the LTQ-Orbitrap XL, although there was some variation between the individual sample components (Fig. 2B). In contrast to MS/MS fragmentation spectra, TIC (sum of all XICs) displayed linear increase with the sample load on all three instruments (slope of 1.25); significant saturation was not observed even at higher sample loads (Fig. 2C). For individual peptides, however, abundance-intensity relations were surprisingly divergent, as exemplified for laminin A (Fig. 2D). Thus, the XIC values of the 277 m/z species identified with laminin A ranged from 103 to 108 intensity units (line profiles in Fig. 2D), but only a small fraction (highlighted in pink) showed the expected linear increase over the entire abundance range. The majority of peptide profiles exhibited more irregular shapes with a strong tendency of signal loss at lower sample loads (Fig. 2D).
This nonlinear behavior was neither a result of poor LC-MS data processing (LC-MS spectra of more than 100 standard protein peptides were manually verified in Xcalibur) nor a consequence of absolute signal intensity. Some of the most intense peptides (at 1 pmol, black lines) declined in a strongly nonlinear fashion or became undetectable below 33 fmol. Obviously, this heterogeneity might lead to significant errors and biases in relative quantification (based on peptide intensity ratios), especially if protein abundances are largely different.
Establishment of an Improved PV-based Quantification Method
For eliminating the influence of nonlinear peptide signals, we set forth a quantification method that uses the mean (or median) of the PVs of a subset of two to six protein-specific peptides selected among all peptides of an individual protein according to their consistency across the entire set of samples (TopCorr-PV). The consistency of a particular peptide was determined by pair-wise correlation of its PV with the PVs of all other peptides of a given protein across the entire set of samples (see “Experimental Procedures”).
When applied to the dilution series of the aforementioned reference sample, the TopCorr-PV method provided linear concentration-intensity relations for all protein components over the entire range of sample loads (Fig. 3). More specifically, the vast majority of PVs of the individual peptides selected by the correlation analyses increased linearly with the load displaying slopes of between 1.0 and 1.3 (slightly more than the expected slope of 1; Fig. 3A). This increase was essentially independent of the type and molecular mass of the source protein in the reference sample, including the 14-kDa lysozyme that offered only eight peptides for MS analysis.
The consistent peptide PVs were used for determining protein amounts by calculating the medians of the PVs after normalization to the slopes of their corresponding peptide profiles (see also “Experimental Procedures”). As shown in Fig. 3B, the resulting normalized PVs showed a linear increase with sample load across the tested range of 4 orders of magnitude, albeit with slight variations at the lowest sample amount of 0.1 fmol. Overall, the relative protein amounts determined from the normalized PV values closely matched the quantities predicted from the dilution steps of the reference sample (Fig. 3B, dashed lines). Thus, the newly established peak volume-based quantification procedure is able to provide reliable results for protein amounts over at least 3 orders of magnitude (1–1000 fmol).
Validation of the TopCorr-PV Method
The validity of the TopCorr-PV method was further probed with a series of samples where the reference stock and dilutions thereof were added to a constant protein background to mimic the conditions typically faced in APs (series 2 in supplemental Fig. S2 and “Experimental Procedures”). In contrast to the experiments above, the three replicate MS samples were directly obtained from in-gel digested protein mixtures rather than from post-digest dilutions. Fig. 4A summarizes the medians of normalized PVs as determined for samples containing 0.1, 1, 10, 100, or 1000 fmol of the reference proteins (Figs. 2 and 3). At amounts of 1, 10, 100, and 1000 fmol, all protein components of the reference were reliably identified and quantified (Fig. 4A), despite the complex background that contained a large number of distinct proteins covering an abundance range very similar to that of the added reference proteins (Fig. 4B, proteins quantified with the abundancenorm score (33) and “Experimental Procedures”). In the 0.1-fmol sample, laminin A was successfully evaluated, whereas other components such as lysozyme and transferrin failed to yield sufficient MS data for unambiguous identification and quantification (i.e. less than two specific peptides or PVs were obtained; Fig. 4A).
Next we determined the accuracy of the TopCorr-PV method by using it on distinct mixtures of E. coli membrane lysates (1 μg) and solubilized membrane fractions from rat brain (0.1, 1, 10, and 100 μg) (series 3 in supplemental Fig. S2). Triplicate mixtures M1 (1 μg of E. coli + 0.1 μg of rat brain), M2 (1 + 1), M3 (1 + 10), and M4 (1 + 100) were MS-analyzed, quantified with the TopCorr-PV method, and evaluated by calculating the PV ratios (rPVs) of individual E. coli proteins between mixtures M1:M2 + M1:M3 + M1:M4 (311 protein ratios) and individual rat proteins M1:M2 (72 proteins), M1:M3 (71 proteins), and M1:M4 (71 proteins). Fig. 4C illustrates the respective histogram plots with rPV values binned in logarithmic intervals and the distributions fitted with Gaussian functions. Bacterial proteins (Fig. 4C, black bars, left panel) are clustered at rPVs of ∼1 (fit value of 1.02), whereas rat proteins (Fig. 4C, three right panels) were distributed at rPVs close to the expected ratios of 1:10, 1:100, and 1:1000 (fit values of 0.083, 0.0056, and 0.0012, respectively). The half-widths of the Gaussian functions corresponded to factors of 1.29 (1:1), 1.53 (1:10), 2.00 (1:100), and 3.25 (1:1000). Together, these results indicated that the TopCorr-PV method allows for reliable and reasonably accurate quantification of protein amounts in complex samples with a sensitivity threshold of ∼0.1 fmol and over at least 3 orders of magnitude.
Application of the TopCorr-PV Method to Complex APs of Kv1.1
Finally, the newly developed TopCorr-PV method was applied to the APs of Kv1.1α (Fig. 1) to address three fundamental issues of AP-based functional proteomics: efficiency and selectivity of the Abs, specificity of co-purified proteins, and stability of protein-protein interactions under various experimental conditions. Purification efficiency and subtype selectivity of the anti-Kv1.1 Abs (Fig. 1A) was explored by relative quantification of the well known core subunits (Kv1.1, 1.2, 1.4, 1.6, Kvβ1, and β2) in the four anti-Kv1.1 APs (see also “Experimental Procedures”). Fig. 5A compares the abundance of these subunits obtained by each Ab relative to their sum in all four APs, with anti-Kv1.1A being the most efficient Ab, followed by anti-Kv1.1B and anti-Kv1.1C/D (Fig. 1A). Notably, all the aforementioned Kv1 α and β subunits were retrieved in similar amounts in each AP, indicating that they were co-purified through Kv1.1α rather than being targeted directly by the Abs. Accordingly, control APs performed with mouse brain membranes lacking Kv1.1α (Kv1.1−/− mice) failed to purify significant amounts of these core subunits (Fig. 5A, inset, at 5-fold enlarged scale). Based on these results, anti-Kv1.1A was selected for further purification experiments and evaluations.
The large number of 330 proteins identified with anti-Kv1.1A and their overlap with proteins identified in IgG controls (Fig. 1B) suggested that a significant portion may actually represent nonspecific or Ab-related background. To carve out those proteins specifically co-purified with Kv1.1, we combined relative quantification of target APs versus two different controls, APs with preimmunization IgGs and APs from target knockout source. Accordingly, the results may be displayed in a two-dimensional plot (Fig. 5B) distributing proteins by their rPV values as determined in APs from wild type versus Kv1.1 knockout (y axis in Fig. 5B) and in APs with anti-Kv1.1A versus control IgGs (x axis in Fig. 5B). The total of 209 proteins successfully quantified in these anti-Kv1.1A APs distributed over a wide range of relative abundances but sorted into four apparent clusters (Fig. 5B, panels I–IV). Proteins in the upper right field (Fig. 5B, panel I, shaded yellow) displayed strong enrichment with respect to both controls and included Kv1.1α, as well as the aforementioned core subunits and some proteins known to tightly interact with Kv1 channels (Fig. 5B and supplemental Fig. S3A). The 54 proteins found in the upper left quarter are robustly enriched relative to Kv1.1 knockout membranes but are also retrieved with control IgGs. Accordingly, these proteins may either represent Kv1.1-associated partners with promiscuous binding to IgG as suggested by the three well known Kv1.1 protein partners (Fig. 5B, blue dots), or, alternatively, they could form part of the background proteins whose expression was reduced in the knockout membranes. The lower right cluster is formed by proteins that are strongly enriched versus IgG, although in a target-independent manner (such as neurochondrin; Fig. 5B, green diamond) as expected for direct or indirect anti-Kv1.1A cross-reactivities. Proteins located in the lower left quarter lack any significant enrichment and therefore are considered nonspecific background of the Kv1.1 AP. In line with these interpretations, the gray bars separating the four clusters may define threshold rPVs that discriminate target candidate interaction partners. Accordingly, 71 of the 209 proteins (panel I, 34.0%) were specifically co-purified with Kv1.1, whereas 84 (panels III and IV, 40.2%) lacked any specific association with Kv1.1; the 54 proteins of panel II (25.8%) represent potential interactors or knockout related background (supplemental Fig. S3A). Kv1.1 AP-MS analysis and TopCorr-PV-based quantification appear to be robust and well reproducible. Thus, repeated MS analysis of the samples in Fig. 5B (supplemental Fig. S4A) and analysis of an independent set of Kv1.1 APs (supplemental Fig. S4B) both show similar distributions of identified proteins, particularly of the known Kv1 interaction partners. The significance of correlation-based peptide selection for protein quantification was verified by re-evaluation of the data in Fig. 5B using means of all peptide PV ratios (supplemental Fig. S3B). The resulting two-dimensional log/log plot shows a significant accumulation of proteins in the upper right field (144 of 235, 61.3%) with strong shifts of individual proteins to higher ratios (three examples marked by arrows), as actually expected from the nonlinear bias of peptide PVs (Fig. 2D). Two lines of experimental evidence suggest that these increases in rPV may be largely artificial. First, assessment of the relative abundance of the Kv1.1 interactor LGI1 in Kv1.1 AP versus IgG control by Western blot analysis was much more consistent with the TopCorr-PV Method in Fig. 5B (rPV = 17.1 compared with rPVmeanall = 937.9). Second, the ratio shifts observed for Kv1 core subunits are inconsistent with the (low) anti-Kv1.1A cross-reactivity for Kv1.2 (Fig. 5A, inset). Together, these results indicate that both types of control, nonspecific IgG and target knockout tissue, may be equally important and, when combined with PV-based quantification using correlation-based peptide filtering, can effectively eliminate false-positives from AP-based protein-protein interaction studies.
Finally, rPV-based quantification was used to investigate the influence of solubilization conditions on the stability of protein interactions and on background. For this purpose, anti-Kv1.1A APs (together with IgG controls) were performed with membrane fractions solubilized at different ionic strength (Ic). Silver staining of the SDS-PAGE separated AP eluates (Fig. 5C, upper panel) suggested a marked decrease of proteins in the IgG controls with increasing ionic strength and largely unaffected signal intensities for the dominant bands in the anti-Kv1.1A APs (Kv1.1 α-subunit band marked by an arrowhead). In line with this observation, Western blot analysis (Fig. 5C, lower panel) showed that the Kv1.1 signals observed in IgG controls at low ionic strength disappeared when Ic was increased to 173 mm; at the same time, anti-Kv1.1A retained its purification efficiency and fully depleted the source material of Kv1.1 over the entire Ic range. For a more comprehensive and unbiased view of the Ic effects, rPV values (relative to IgG controls) were determined for all MS-identified proteins in anti-Kv1.1A APs and plotted as a histogram (Fig. 5D). These plots revealed three distinct groups of proteins at any of the Ic conditions: proteins showing strong enrichment versus IgG (log(rPV) > 1.5; Fig. 5D, yellow bars), including all known Kv1 channel core subunits (red bars), as well as proteins with target-independent binding to anti-Kv1.1A such as neurochondrin, proteins representing nonspecific background (at −1.5 < log(rPV) < 1.5), and proteins that display stronger binding to IgG than to anti-Kv1.1A (log(rPV) < −1.5). Comparison of the histogram plots also demonstrated that increasing ionic strength from 123 to 273 mM resulted in a marked enhancement of the AP signal-to-noise ratio: rPVs of Kv1 channel core subunits and strongly enriched proteins either shifted to higher values or remained unchanged, whereas the number of background proteins was largely reduced. Noteworthy, the anti-Kv1.1A cross-reactive neurochondrin was not affected by the increase in Ic (Fig. 5D, green bars).
Taken together, relative quantification of protein amounts based on TopCorr-PVs appears well suited for establishing thresholds to discriminate specific protein-protein interactions in AP-MS; it also provides important information on selectivity and efficiency of Abs, as well as on the stability of protein interactions, which may be useful for adjusting the experimental conditions in AP-based approaches.
DISCUSSION
As a central achievement, the present study sets forth a novel procedure for label-free quantification of protein amounts that is based on the PVs of subsets of protein-specific peptides and that displays a dynamic range of up to 4 orders of magnitude. Application of the procedure to complex AP samples showed its suitability for quantitative characterization of Ab properties and AP conditions, as well as for the establishment of thresholds discriminating specifically purified and co-purified proteins from background.
MS Data and Label-free Quantification of Protein Amounts
We first investigated the shortcomings of current label-free quantification methods. Analysis of diluted standard protein digests on some of the most popular high resolution LC-MS/MS instruments, LTQ-FT, LTQ-FT Ultra, and LTQ-Orbitrap XL, resulted in two major findings: the acquisition of spectral counts was highly dependent on the MS setup, sample amount, and complexity, as well as on individual protein properties reflecting the statistical nature of MS/MS fragmentation, precursor competition and intrinsic limitations. In line with the technical properties, MS/MS saturation was most prominent on the LTQ-FT instruments, reaching a maximum at sample loads corresponding to less than 300 ng of protein. Despite its higher fragmentation capacity, the LTQ-Orbitrap XL also showed nonlinear behavior of spectral counts for individual proteins that could not be corrected for by normalization (as, for example, attempted by the exponentially modified protein abundance index). This could not be improved by using other MS/MS spectra-based measures (like Mascot scores, number of fragmentations per peptide) or normalization methods (data not shown). We therefore concluded that MS/MS data are generally not suitable for label-free quantitative evaluation of APs.
In marked contrast, LC-MS signal intensities, determined as m/z time integral PVs of individual peptide species, were rather proportional to the loaded sample amount on all three MS setups, in line with previous studies (14, 23, 29). However, we found that only a small fraction of peptides was linear over the investigated dynamic range, similar to the findings by Liu et al. (23). The majority of peptide signals showed rapid and nonproportional intensity loss at lower sample loads that could be explained neither by poor ionization efficiency nor by errors in the msInspect software used for PV determination. This effect was highly reproducible and largely independent from the type of MS instrument but seemed to be linked to physicochemical properties of individual peptide molecules, most likely leading to absorptive losses along the nano-LC pipeline. Independent of the underlying cause, this qualitative heterogeneity had two major consequences for PV-based quantification: (i) more accurate quantification may be obtained by using a small subset of “high quality” peptides instead of means or medians of the total set, and (ii) differences in relative abundance may be overestimated by classical PV-based methods, especially when exceeding a factor of 10.
We therefore developed a procedure for relative quantification of proteins that first determines the m/z signal intensity for each peptide of a given protein and subsequently selects the two to six most consistent peptides by pair-wise correlation ranking of their PVs across all of the data sets (23). These selected peptides were finally used to calculate ratios, the median of which defines the relative quantity (rPV) in a pair or group of samples. To reduce the influence of remaining assignment errors and LC-MS noise, at least two peptide ratios and a total PV of 100000 intensity units were used for reliable quantification. This method not only eliminated peptides with disproportional behavior but also reduced variations introduced by false-positive or false-negative assignment of LC-MS signals by the software. Application of this method to spiked samples simulating large abundance differences of a particular set of proteins in a complex background showed that relative quantification was linear over more than 3 orders of magnitude. Apart from some limitations intrinsic to very small proteins, sensitivity and reproducibility were reasonable down to 0.1–1 fmol. The ratio distributions of proteins quantified in complex samples mixed from different amounts of membrane fractions from E. coli and rat brain suggested average relative quantification errors ranging from a factor of ∼1.5 for small (1–10-fold) to 3.5 for large (∼1000-fold) abundance differences (Fig. 4C). Although the performance of the new procedure allowed for in-depth evaluation of AP-MS samples, several steps offer room for further improvement. Correct identification and assignment of m/z signals appeared to be most critical. Of the five different software tools tested (see “Experimental Procedures”), all showed rather high assignment error rates (>10%) despite optimization of parameters. Furthermore, noise processing, gain control, and other settings in the acquisition software Xcalibur may be optimized for better detection of low intensity peptide ion signals. Moreover, the addition of protein standards and application of PV slope calibration factors (Figs. 2C and 3A) may help to further reduce systematic errors and variations between individual LC-MS runs.
Application of the Newly Developed Procedure to AP-MS
The newly developed quantification procedure was applied to resolve crucial issues of functional proteomics, as exemplified by APs of the neuronal potassium channel Kv1.1 (26, 28). Relative quantification of its core subunits in APs not only revealed the substantial (>10-fold) differences in purification efficiency of the respective Abs but also verified their selectivity for Kv1α isoforms.
When applied to relative quantification of proteins co-purified in APs from wild type source material versus IgG and Kv1.1 knockout controls, the extended linearity of the quantification procedure revealed the full range of target protein enrichment of specific versus background or cross-reactive proteins (up to a factor of 100000). The resulting distributions of relative protein amounts depicted as two-dimensional log/log scatter plots (Fig. 5B) were well reproducible (supplemental Fig. S4) and showed several interesting features of the respective Ab: proteins fell into categories (quadrants) of specifically co-enriched, cross-reactive, and different types of background-related proteins; the formation of distinct clusters rather than continuous distributions suggests that proteins have been captured not independently from each other but as defined assemblies. Although distribution patterns and derived specificity thresholds may vary with different Abs, targets, source membranes, and AP conditions, the large number of nonspecific proteins compared with candidate Kv1.1-interactors is rather typical (11, 16, 19). This emphasizes the need for both IgG and knockout controls to obtain true positive AP-MS results. Furthermore, the benefits of the TopCorr-PV method were directly assessed in a head to-head comparison with a commonly used quantification method using the means of all PV ratios, backed up by biochemical experiments (supplemental Fig. S3B). In addition, the quantification procedure may also be useful to optimize AP conditions (Fig. 5, C and D). At this end, MS-based quantification provided detailed and unbiased information on the specific or nonspecific enrichment of more than a hundred proteins captured by anti-Kv1.1A under different ionic strength conditions. The protein distribution profiles confirmed that optimum signal-to-noise was indeed reached at slightly increased physiological ionic strength (CL-80).
It should be emphasized that despite elaborate quantitative controls, the determined specificity thresholds and quantitative relationships among purified proteins in APs neither reflect the in vivo situation nor automatically define a physiological interactome. AP-MS results are likely biased by several factors, including selecting or disrupting properties of the Ab, the actual physiological protein expression levels, and intrinsic differences in their interaction stability and binding promiscuity. As an example, LGI1, a well known interaction partner of Kv1.1 with functional and pathophysiological implications (25, 26, 30) failed the rPV(IgG) threshold (Fig. 5B) because of its binding to IgG background proteins. Thus, reliable broad dynamic range quantification of proteins and stringent negative controls may be regarded as mandatory, but the establishment of a full Kv1.1-associated proteome would require more high quality Abs and complementing experiments beyond the scope of this study.
Footnotes
* The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. This work was supported by grants of the Deutsche Forschungsgemeinschaft to B.F. (SFB 746/TP16, SFB780/A3, Fa 332/5–3).
 This article contains supplemental Figs. S1–S4.
This article contains supplemental Figs. S1–S4.
1 The abbreviations used are:
- AP
- affinity purification
- Ab
- antibody
- IgG
- immunoglobulin G
- Kv
- voltage-gated potassium channel
- LGI1
- leucine-rich glioma-inactivated protein 1
- PV
- peak volume
- rPV
- ratio of PVs
- TIC
- total ion current
- XIC
- extracted ion current (signal intensity of an m/z species over time).
REFERENCES
- 1. Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 2. Schulte U., Müller C. S., Fakler B. (2011) Ion channels and their molecular environments: Glimpses and insights from functional proteomics. Semin. Cell Dev. Biol. 22, 132–144 [DOI] [PubMed] [Google Scholar]
- 3. Vermeulen M., Hubner N. C., Mann M. (2008) High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Curr Opin Biotechnol 19, 331–337 [DOI] [PubMed] [Google Scholar]
- 4. Schulte U. (2008) Protein-protein interactions and subunit composition of ion channels. CNS Neurol. Disord. Drug Targets 7, 172–186 [DOI] [PubMed] [Google Scholar]
- 5. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 6. Bildl W., Strassmaier T., Thurm H., Andersen J., Eble S., Oliver D., Knipper M., Mann M., Schulte U., Adelman J. P., Fakler B. (2004) Protein kinase CK2 is coassembled with small conductance Ca(2+)-activated K+ channels and regulates channel gating. Neuron 43, 847–858 [DOI] [PubMed] [Google Scholar]
- 7. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 8. Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 9. Karp N. A., Huber W., Sadowski P. G., Charles P. D., Hester S. V., Lilley K. S. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteomics 9, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 11. Berkefeld H., Sailer C. A., Bildl W., Rohde V., Thumfart J. O., Eble S., Klugbauer N., Reisinger E., Bischofberger J., Oliver D., Knaus H. G., Schulte U., Fakler B. (2006) BKCa-Cav channel complexes mediate rapid and localized Ca2+-activated K+ signaling. Science 314, 615–620 [DOI] [PubMed] [Google Scholar]
- 12. Higgs R. E., Knierman M. D., Gelfanova V., Butler J. P., Hale J. E. (2005) Comprehensive label-free method for the relative quantification of proteins from biological samples. J. Proteome Res. 4, 1442–1450 [DOI] [PubMed] [Google Scholar]
- 13. Mueller L. N., Brusniak M. Y., Mani D. R., Aebersold R. (2008) An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 7, 51–61 [DOI] [PubMed] [Google Scholar]
- 14. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 15. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 16. Marionneau C., LeDuc R. D., Rohrs H. W., Link A. J., Townsend R. R., Nerbonne J. M. (2009) Proteomic analyses of native brain K(V)4.2 channel complexes. Channels 3, 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schwenk J., Harmel N., Zolles G., Bildl W., Kulik A., Heimrich B., Chisaka O., Jonas P., Schulte U., Fakler B., Klöcker N. (2009) Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 18. Schwenk J., Metz M., Zolles G., Turecek R., Fritzius T., Bildl W., Tarusawa E., Kulik A., Unger A., Ivankova K., Seddik R., Tiao J. Y., Rajalu M., Trojanova J., Rohde V., Gassmann M., Schulte U., Fakler B., Bettler B. (2010) Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature 465, 231–235 [DOI] [PubMed] [Google Scholar]
- 19. Müller C. S., Haupt A., Bildl W., Schindler J., Knaus H. G., Meissner M., Rammner B., Striessnig J., Flockerzi V., Fakler B., Schulte U. (2010) Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc. Natl. Acad. Sci. U.S.A. 107, 14950–14957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daniel T. O., Tremble P. M., Frackelton A. R., Jr., Williams L. T. (1985) Purification of the platelet-derived growth factor receptor by using an anti-phosphotyrosine antibody. Proc. Natl. Acad. Sci. U.S.A. 82, 2684–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Müller-Newen G., Köhne C., Keul R., Hemmann U., Müller-Esterl W., Wijdenes J., Brakenhoff J. P., Hart M. H., Heinrich P. C. (1996) Purification and characterization of the soluble interleukin-6 receptor from human plasma and identification of an isoform generated through alternative splicing. Eur. J. Biochem. 236, 837–842 [DOI] [PubMed] [Google Scholar]
- 22. Turck C. W., Falick A. M., Kowalak J. A., Lane W. S., Lilley K. S., Phinney B. S., Weintraub S. T., Witkowska H. E., Yates N. A. (2007) The Association of Biomolecular Resource Facilities Proteomics Research Group 2006 study: Relative protein quantitation. Mol. Cell. Proteomics 6, 1291–1298 [DOI] [PubMed] [Google Scholar]
- 23. Liu K., Zhang J., Wang J., Zhao L., Peng X., Jia W., Ying W., Zhu Y., Xie H., He F., Qian X. (2009) Relationship between sample loading amount and peptide identification and its effects on quantitative proteomics. Anal. Chem. 81, 1307–1314 [DOI] [PubMed] [Google Scholar]
- 24. Pongs O., Schwarz J. R. (2010) Ancillary subunits associated with voltage-dependent K+ channels. Physiol. Rev. 90, 755–796 [DOI] [PubMed] [Google Scholar]
- 25. Irani S. R., Alexander S., Waters P., Kleopa K. A., Pettingill P., Zuliani L., Peles E., Buckley C., Lang B., Vincent A. (2010) Antibodies to Kv1 potassium channel-complex proteins leucine-rich, glioma inactivated 1 protein and contactin-associated protein-2 in limbic encephalitis, Morvan's syndrome and acquired neuromyotonia. Brain 133, 2734–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schulte U., Thumfart J. O., Klöcker N., Sailer C. A., Bildl W., Biniossek M., Dehn D., Deller T., Eble S., Abbass K., Wangler T., Knaus H. G., Fakler B. (2006) The epilepsy-linked Lgi1 protein assembles into presynaptic Kv1 channels and inhibits inactivation by Kvbeta1. Neuron 49, 697–706 [DOI] [PubMed] [Google Scholar]
- 27. Scott V. E., Muniz Z. M., Sewing S., Lichtinghagen R., Parcej D. N., Pongs O., Dolly J. O. (1994) Antibodies specific for distinct Kv subunits unveil a heterooligomeric basis for subtypes of alpha-dendrotoxin-sensitive K+ channels in bovine brain. Biochemistry 33, 1617–1623 [DOI] [PubMed] [Google Scholar]
- 28. Wang F. C., Parcej D. N., Dolly J. O. (1999) Alpha subunit compositions of Kv1.1-containing K+ channel subtypes fractionated from rat brain using dendrotoxins. Eur. J. Biochem. 263, 230–237 [DOI] [PubMed] [Google Scholar]
- 29. Wang G., Wu W. W., Zeng W., Chou C. L., Shen R. F. (2006) Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J. Proteome Res. 5, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 30. Zhou Y. D., Lee S., Jin Z., Wright M., Smith S. E., Anderson M. P. (2009) Arrested maturation of excitatory synapses in autosomal dominant lateral temporal lobe epilepsy. Nat. Med. 15, 1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sailer C. A., Hu H., Kaufmann W. A., Trieb M., Schwarzer C., Storm J. F., Knaus H. G. (2002) Regional differences in distribution and functional expression of small-conductance Ca2+-activated K+ channels in rat brain. J. Neurosci. 22, 9698–9707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pandey A., Mann M. (2000) Proteomics to study genes and genomes. Nature 405, 837–846 [DOI] [PubMed] [Google Scholar]
- 33. Zolles G., Wenzel D., Bildl W., Schulte U., Hofmann A., Müller C. S., Thumfart J. O., Vlachos A., Deller T., Pfeifer A., Fleischmann B. K., Roeper J., Fakler B., Klöcker N. (2009) Association with the auxiliary subunit PEX5R/Trip8b controls responsiveness of HCN channels to cAMP and adrenergic stimulation. Neuron 62, 814–825 [DOI] [PubMed] [Google Scholar]