Abstract
Development of new biomarkers needs to be significantly accelerated to improve diagnostic, prognostic, and toxicity monitoring as well as therapeutic follow-up. Biomarker evaluation is the main bottleneck in this development process. Selected Reaction Monitoring (SRM) combined with stable isotope dilution has emerged as a promising option to speed this step, particularly because of its multiplexing capacities. However, analytical variabilities because of upstream sample handling or incomplete trypsin digestion still need to be resolved. In 2007, we developed the PSAQ™ method (Protein Standard Absolute Quantification), which uses full-length isotope-labeled protein standards to quantify target proteins. In the present study we used clinically validated cardiovascular biomarkers (LDH-B, CKMB, myoglobin, and troponin I) to demonstrate that the combination of PSAQ and SRM (PSAQ-SRM) allows highly accurate biomarker quantification in serum samples. A multiplex PSAQ-SRM assay was used to quantify these biomarkers in clinical samples from myocardial infarction patients. Good correlation between PSAQ-SRM and ELISA assay results was found and demonstrated the consistency between these analytical approaches. Thus, PSAQ-SRM has the capacity to improve both accuracy and reproducibility in protein analysis. This will be a major contribution to efficient biomarker development strategies.
Introduction of new diagnostic assays in the clinical setting requires an operating pipeline to efficiently translate putative biomarkers into validated biomarkers. Despite the discovery platforms' capacity to generate well populated lists of candidate biomarkers, very few proteins reach the patient bedside as fully fledged “FDA-approved” biomarkers. This is largely because of divergences between analytical needs and performances of the techniques available for candidate biomarker evaluation (1, 2).
Candidate biomarker evaluation is a major process of the biomarker pipeline, positioned downstream of the biomarker discovery phase and necessary before clinical validation. Candidate evaluation aims to select, among hundreds of putative biomarkers, those of clinical relevance. Evaluation phase combines two steps which respectively consist in: (1) confirming a difference between physiological and pathological concentrations in biofluids (the so-called “qualification phase”) and (2) assessing the specificity of candidate biomarkers (the so-called “verification phase”) (1). Currently, because of its high throughput and high sensitivity, quantitative ELISA is the preferred assay format for studies evaluating biomarkers. However, as most candidates are likely to fail as relevant biomarkers, developing ELISA tests (with high quality antibodies) for all candidates is a financial burden for the diagnostics industry (3).
Thus, there exists an urgent need to develop analytical methods capable of reliable candidate evaluation, at high throughput and reasonable cost. Selected Reaction Monitoring (SRM)1 mass spectrometry combined with stable isotope dilution (SID-SRM) has shown promise as a solution to this technological hurdle (4, 5). MS analysis in SRM mode offers the unique possibility to specifically and simultaneously monitor the signatures of hundreds of target peptides generated by trypsin digestion of proteins. Combined with isotope-labeled quantification standards (6), SRM can provide quantitative data for each protein targeted (5).
Recently, in an effort to demonstrate the potential of SID-SRM for candidate biomarker evaluation, a multilaboratory study was set up to assess its analytical performances and potential transferability (7). Exogenous proteins, seven in all, were added to unfractionated plasma samples. The spiked samples were analyzed by eight independent laboratories using SRM and isotope-labeled peptides as standards. The results obtained clearly demonstrated the capacity of SID-SRM to specifically and precisely quantify protein biomarkers in plasma. However, the results also revealed that the protein digestion rate was highly variable between laboratories. This variability had a significant effect on peptide recovery and on the accuracy of protein quantification. As suggested by the authors, this type of bias could be avoided if properly folded isotope-labeled protein standards were used as quantification standards (7, 8).
In 2007, we developed the PSAQ™ (Protein Standard Absolute Quantification) method, which uses full-length isotope-labeled proteins as internal standards for absolute quantitative MS analysis. We demonstrated that, in contrast with peptide standards, adding isotope-labeled proteins before sample digestion enables accurate protein quantification, even for proteins resistant to trypsin digestion (9, 10). In addition, we, and others, have shown that this type of protein standard (“PSAQ standard”) also corrects for protein losses that may occur during sample handling prior to trypsin digestion and liquid chromatography (LC)-MS analysis (11–17). This latter feature is a particular advantage for MS analysis of blood biomarkers. Indeed, as plasma/serum are highly complex matrices and display a huge dynamic range, sample prefractionation must be performed to detect low-abundance protein biomarkers (4).
In this study, we have tested a combination of the PSAQ strategy with SRM (PSAQ-SRM) for quantification of cardiovascular biomarkers in serum samples. Selected biomarkers include LDH-B, CKMB, myoglobin, and troponin I. For some of these validated biomarkers, a comparison of PSAQ-SRM data and ELISA results was performed on samples from patients having suffered myocardial infarction.
EXPERIMENTAL PROCEDURES
Biomarkers and Clinical Samples
Human LDH-B, creatine kinase MB heterodimer (CKMB), and myoglobin were purchased from Applichem (Darmstadt, Germany). Human troponin I and healthy human serum were obtained from Sigma-Aldrich (Saint Quentin Fallavier, France). Serum samples from five patients were provided by the Plateforme de Ressources Biologiques (Groupe Hospitalier Henri Mondor, Créteil, France). Five patients undergoing primary percutaneous coronary intervention (PCI) for ST-elevation myocardial infarction were included in the study. The protocol was approved by the hospital's institutional review board and all patients provided written informed consent for participation. Blood samples were taken at hospital admission and at regular intervals following PCI. Samples were collected in nontreated tubes (BD Biosciences, le Pont de Claix, France). All blood samples were centrifuged at 2200 × g for 10 min to obtain serum supernatants. These were immediately aliquoted and frozen at −80 °C. Additional blood samples were analyzed at the clinical chemistry laboratory for standard cardiac biomarker evaluation (Total CK activity, troponin I, and myoglobin) as described below. In this study, only serum samples collected at hospital admission and at 3 or 8 days after PCI were analyzed using the PSAQ-SRM approach.
Production of Full-length Stable-Isotope-Labeled Proteins (PSAQ Standards)
PSAQ standards were synthesized as previously described (9). Briefly, LDH-B, creatine kinase B chain (CKB), creatine kinase M chain (CKM), myoglobin, and troponin I genes were amplified by PCR using a cardiac cDNA library as template (Biochain Institute, Hayward, CA) (see supplemental Table S1 for primer sequences and PCR reaction conditions). Genes were cloned into the pIVEX 2.4d expression vector (5 Prime, Hamburg, Germany) using the In-Fusion™ PCR cloning system (Clontech, Saint Germain en Laye, France). The pIVEX 2.4d vector provides a N-terminal hexahistidine purification tag. Plasmids were cloned into XL1-Blue cells (Agilent Technologies, Massy, France), purified and sequenced (Cogenics, Meylan, France). Cell-free protein expression and isotope-labeling was performed using the RTS 500 Proteomaster E. coli HY kit (5 Prime) in the presence of [13C6, 15N2] l-lysine and [13C6, 15N4] l-arginine (Eurisotop, Saint-Aubin, France). PSAQ standards were purified on a nickel affinity column (Ni Sepharose 6 Fast Flow resin, GE Healthcare, Orsay, France) using an imidazole gradient. PSAQ standards were checked for purity on SDS-PAGE using Coomassie staining (>95% purity). N-terminal hexahistidine purification tags were not removed as they are not expected to significantly modify PSAQ biochemical properties. Isotope-labeled proteins were quantified by amino acid analysis (MScan SA, Plan les Ouates, Switzerland). Isotope incorporation was verified by LC-MS and LC-SRM analysis and was found to be greater than 99% (see supplemental Fig. S1).
Depletion of Serum Samples
Serum samples (14 μl) from healthy donors and patients were spiked with defined amounts of PSAQ standards. To set up a titration experiment, samples from healthy donors were spiked with defined amounts of exogenous, unlabeled LDH-B, CKMB, myoglobin, or troponin I (see supplemental LC-SRM data). Serum samples were depleted of the six most abundant proteins using the human Multiple Affinity Removal Spin cartridge (MARS 6) (Agilent Technologies) according to the manufacturer's instructions. The flow-through was concentrated to 15 μl using a 5000 Da cutoff ultrafiltration device (Vivascience, Hannover, Germany). Laemmli buffer (10 μl) was added before SDS-PAGE analysis.
Troponin I Antibody Biotinylation
Troponin I antibody (Santa Cruz Biotechnology, Heidelberg, Germany) was dialyzed against phosphate-buffered saline (PBS) buffer. The antibody was then incubated with NHS-PEG4-Biotin reagent (Pierce/Thermo Fischer Scientific, Brebières, France) for 3 h at room temperature (NHS-PEG4-Biotin/antibody ratio: 20/1). After biotinylation, the antibody was once again dialyzed in PBS buffer. Biotinylation was checked by dot-blot analysis with neutravidin-HRP on a polyvinylidene difluoride membrane.
Immunoenrichment of Serum Samples
Before immunoenrichment, serum samples (1 ml) were spiked with defined quantities of troponin I PSAQ standard. To generate the titration curve, a range of concentrations of exogenous unlabeled troponin I was added to healthy donor serum samples (see supplemental LC-SRM data). Biotinylated antibody (1 μg) was added to the samples before overnight incubation at 4 °C on a rotating wheel. Dynabeads M-280 Streptavidin (Invitrogen, Cergy Pontoise, France) were added (15 μl Dynabeads per sample) and samples were incubated for a further 4 h at 4 °C with rotation. Supernatants were eliminated and beads were washed three times with PBS/0.1%Tween, then with PBS and finally with water. Captured proteins were eluted from the beads with 0.5% formic acid. Eluates were dried by vacuum centrifugation and resuspended in Laemmli buffer before SDS-PAGE analysis.
SDS-PAGE Analysis and Trypsin Digestion
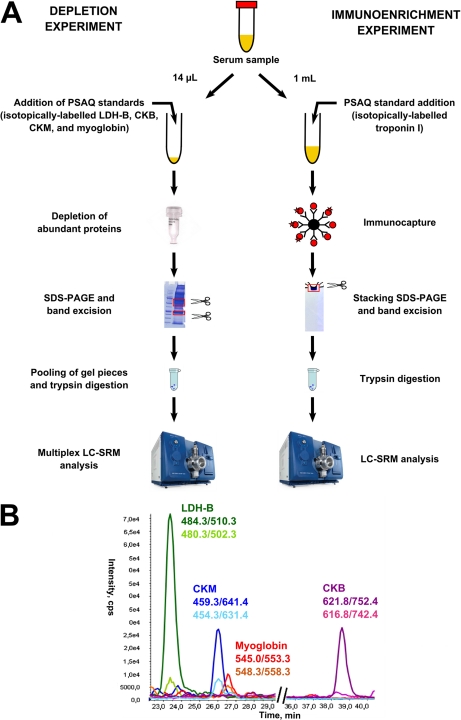
After protein depletion or immunocapture prefractionation, samples were resuspended in Laemmli buffer and loaded onto precast NuPAGE Novex Bis-Tris 4–12% acrylamide gradient gels (Invitrogen). After migration, gels were fixed and stained with Coomassie staining solution (Bio-Rad, Marnes la Coquette, France). Gel regions containing targeted biomarkers were excised and cut into small pieces (see supplemental Figs. 2A, 3A and 4A). For multiplex detection of biomarkers in depleted serum samples, gel pieces obtained from two excised regions (15 to 30 kDa and 35 to 45 kDa regions) were pooled before in-gel digestion (see Fig. 1A and supplemental Fig. 6A). For immunoenriched samples, SDS-PAGE was performed in the “stacking” mode and samples were entirely excised from the gel (see Fig. 1A, supplemental Figs. 5A and 6B). Gel pieces were destained by alternating washing cycles in NH4HCO3 25 mm and NH4HCO3 25 mm (50%)/ACN (50%) for 15 min. After destaining, gel pieces were dried by vacuum centrifugation and oxidized with a 7% H2O2 solution for 15 min (18). Gel pieces were washed with pure water and dehydrated in 100%acetonitrile (ACN) before overnight trypsin (Promega, Madison, WI) digestion at a protein/enzyme ratio of 1:20 (w/w) in 25 mm NH4HCO3 at 37 °C. Tryptic peptides were extracted from the gel in three successive steps (15 min each), using 50% ACN, 5% formic acid, and 100% ACN extraction solutions. After drying by vacuum centrifugation, tryptic peptides were resuspended in 2% ACN and 0.2% formic acid before LC-SRM analysis. Most samples were reconstituted in 10 μl to allow a 6 μl pick-up injection on the nanoLC-chromatography system. However, samples containing higher amounts of biomarkers (highest points of titration curves) were resuspended in a larger volume (20 to 50 μl).
Fig. 1.
Sample preparation workflows for the quantification of cardiovascular biomarkers in serum. A, Sample preparation workflow using either depletion of abundant proteins or troponin I immunoenrichment before SDS-PAGE, in-gel trypsin digestion and LC-SRM analysis. B, Multiplex LC-SRM detection of LDH-B, CKB chain, CKM chain and myoglobin in a healthy donor serum sample after depletion of abundant proteins. For better clarity, a single SRM transition from one proteotypic peptide (labeled and unlabeled versions) is shown for each biomarker.
LC-SRM Analysis
SRM transitions were selected based on proteotypic peptide LC-MS/MS spectra (see supplemental LC-MS/MS data) and were experimentally validated by LC-SRM analysis of trypsin-digests containing each biomarker and its corresponding PSAQ standard. LC-SRM analyses were performed on a 5500 Q-Trap hybrid triple quadrupole/linear ion trap mass spectrometer (400 to 1250 m/z range) equipped with a Nano III electrospray ion source and operating with Analyst software (version 1.5, Applied Biosystems/MDS Sciex, Les Ulis, France). The instrument was coupled to an Ultimate 3000 nanoLC-chromatography system (Dionex, Voisins Le Bretonneux, France). Chromatography was performed using a two-solvent system combining solvent A (2%ACN/0.1%formic acid) and solvent B (80%ACN/0.08%formic acid). First, samples (6 μl pick-up injections) were concentrated for 3 min on a 300-μm x 5-mm PepMap C18 precolumn (Dionex) at 20 μl/min flow rate. Peptide digests were then separated onto a 75-μm x 150-mm C18 column (Dionex) at a flow rate of 300 nl/min and using a 60-min gradient from 10% B to 40% B in 30 min and from 40% to 90% in 5 min. Data were acquired in a positive ion mode with an ion spray voltage of 2200 Volts, curtain gas at 20 p.s.i, a nebulizer gas at 12 p.s.i and an interface heater temperature of 150 °C. Collision exit, declustering and entrance potentials were set to 15, 75 and 12 Volts, respectively. Collision energy values were calculated using linear equations based on the unlabeled peptide precursor m/z ratios:
For doubly charged precursors: CE = 0.05 m/z + 5 (Volts)
For triply-charge precursors: CE = 0.05 m/z + 4 (Volts)
Collision energy was identical for both labeled and unlabeled versions of each peptide (Table I). At least 2 SRM transitions per peptide were monitored. These were acquired at unit resolution both in Q1 and Q3 quadrupoles with a dwell time set at 50 ms and a cycle time set at 4.9 s (multiplex detection in depleted serum samples) or 2.1 s (troponin I detection in immunoenriched serum samples).
Table I. Peptide sequences and SRM transitions. Peptides from PSAQ standards are mentioned with the C-terminal isotope-labelled amino acid in bold. Methionine dioxidation (ox2) or cystein trioxidation (ox3) modification states are indicated.
| Biomarker | UniProt accession number | Peptide sequence | SRM transitions |
Collision energy (Volts) | |
|---|---|---|---|---|---|
| Q1 m/z | Q3 m/z | ||||
| LDH-B | P07195 | IVVVTAGVR | 457.3 | 503.3 | 27.9 |
| 457.3 | 602.4 | 27.9 | |||
| 457.3 | 701.4 | 27.9 | |||
| IVVVTAGVR | 462.3 | 513.3 | 27.9 | ||
| 462.3 | 612.4 | 27.9 | |||
| 462.3 | 711.4 | 27.9 | |||
| GLTSVINQK | 480.3 | 502.3 | 29.0 | ||
| 480.3 | 601.4 | 29.0 | |||
| 480.3 | 688.4 | 29.0 | |||
| GLTSVINQK | 484.3 | 510.3 | 29.0 | ||
| 484.3 | 609.4 | 29.0 | |||
| 484.3 | 696.4 | 29.0 | |||
| SLADELALVDVLEDK | 815.4 | 504.3 | 45.8 | ||
| 815.4 | 718.4 | 45.8 | |||
| 815.4 | 1001.6 | 45.8 | |||
| 815.4 | 1114.6 | 45.8 | |||
| 544.0 | 504.3 | 31.2 | |||
| 544.0 | 718.4 | 31.2 | |||
| 544.0 | 817.4 | 31.2 | |||
| SLADELALVDVLEDK | 819.4 | 512.3 | 45.8 | ||
| 819.4 | 726.4 | 45.8 | |||
| 819.4 | 1009.6 | 45.8 | |||
| 819.4 | 1122.6 | 45.8 | |||
| 546.6 | 512.3 | 31.2 | |||
| 546.6 | 726.4 | 31.2 | |||
| 546.6 | 825.4 | 31.2 | |||
| LIAPVAEEEATVPNNK | 848.0 | 472.3 | 47.4 | ||
| 848.0 | 1201.6 | 47.4 | |||
| LIAPVAEEEATVPNNK | 852.0 | 480.3 | 47.4 | ||
| 852.0 | 1209.6 | 47.4 | |||
| Total LDH: | P00338 | VIGSGC(ox3)NLDSAR | 620.3 | 333.2 | 36.0 |
| LDH-A, LDH-B and LDH-C | P07195 | 620.3 | 448.2 | 36.0 | |
| P07864 | 620.3 | 675.3 | 36.0 | ||
| VIGSGC(ox3)NLDSAR | 625.3 | 343.2 | 36.0 | ||
| 625.3 | 458.2 | 36.0 | |||
| 625.3 | 685.3 | 36.0 | |||
| Creatine kinase B | P12277 | DLFDPIIEDR | 616.8 | 742.4 | 35.8 |
| 616.8 | 857.4 | 35.8 | |||
| DLFDPIIEDR | 621.8 | 752.4 | 35.8 | ||
| 621.8 | 867.4 | 35.8 | |||
| VLTPELYAELR | 652.4 | 990.5 | 37.6 | ||
| 652.4 | 1091.6 | 37.6 | |||
| VLTPELYAELR | 657.4 | 1000.5 | 37.6 | ||
| 657.4 | 1101.6 | 37.6 | |||
| Creatine kinase M | P06732 | FEEILTR | 454.3 | 502.3 | 27.7 |
| 454.3 | 631.4 | 27.7 | |||
| FEEILTR | 459.3 | 512.3 | 27.7 | ||
| 459.3 | 641.4 | 27.7 | |||
| ELFDPIISDR | 602.8 | 700.4 | 35.1 | ||
| 602.8 | 962.5 | 35.1 | |||
| ELFDPIISDR | 607.8 | 710.4 | 35.1 | ||
| 607.8 | 972.5 | 35.1 | |||
| GGDDLDPNYVLSSR | 754.4 | 935.5 | 42.7 | ||
| 754.4 | 1050.5 | 42.7 | |||
| GGDDLDPNYVLSSR | 759.4 | 945.5 | 42.7 | ||
| 759.4 | 1060.5 | 42.7 | |||
| Myoglobin | P02144 | VEADIPGHGQEVLIR | 545.0 | 553.3 | 31.2 |
| 545.0 | 702.9 | 31.2 | |||
| VEADIPGHGQEVLIR | 548.3 | 558.3 | 31.2 | ||
| 548.3 | 707.9 | 31.2 | |||
| Troponin I | P19429 | TLLLQIAK | 450.3 | 685.5 | 27.5 |
| 450.3 | 572.4 | 27.5 | |||
| TLLLQIAK | 454.3 | 693.5 | 27.5 | ||
| 454.3 | 580.4 | 27.5 | |||
| NITEIADLTQK | 623.3 | 675.4 | 36.2 | ||
| 623.3 | 1018.5 | 36.2 | |||
| NITEIADLTQK | 627.3 | 683.4 | 36.2 | ||
| 627.3 | 1026.5 | 36.2 | |||
| ISADAM(ox2)M(ox2)QALLGAR | 756.4 | 303.2 | 42.8 | ||
| 756.4 | 416.3 | 42.8 | |||
| 756.4 | 529.4 | 42.8 | |||
| ISADAM(ox2)M(ox2)QALLGAR | 761.4 | 313.2 | 42.8 | ||
| 761.4 | 426.3 | 42.8 | |||
| 761.4 | 539.4 | 42.8 | |||
LC-SRM Data Analysis
Data analysis was performed using MultiQuant software (version 1.1 Applied Biosystems/MDS Sciex). Unlabeled/labeled peak area ratios were calculated for each SRM transition after careful verification of coelution profiles. Ratios obtained from the different SRM transitions were used to calculate the corresponding average peptide ratio. Then, ratios obtained for the different proteotypic peptides were combined to calculate the protein ratio and determine biomarker concentration in serum (see supplemental LC-SRM data and supplemental Figs. S2 to S5). From the 787 calculated peak area ratios, 11 (1.4%) outlier values were excluded from analysis (see supplemental LC-SRM data). Most of these outliers were related to matrix interferences impairing labeled or unlabeled peptide transition signals.
LLOQ Determination
LLOQ corresponds to the lowest concentration of an analyte that can be determined with acceptable precision and accuracy. Various approaches for determining LLOQ can be used, including signal-to-noise analysis, statistical analysis based on blank sample variance determination (19) and least-squares linear regression analysis (20). In this study, blank samples were not available (non-spiked serum samples contained endogenous levels of biomarkers). Consequently, LLOQ was determined according to the Food and Drug Administration criteria described in the guidelines for bioanalytical method validation (www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances). LLOQ was established as the lowest concentration of the titration curve that was measured with precision (CV) inferior to 20% and accuracy comprised between 80 and 120%.
ELISA and Enzymatic Assays
Myoglobin concentration in patient serum samples was determined using the Human Myoglobin ELISA Kit (Alpco Diagnostics, Salem, NH) according to the manufacturer's instructions. To investigate troponin I concentration in patients' samples, the Access AccuTnI Troponin I Assay (Beckman Coulter, Roissy, France), which is not a highly sensitive troponin I assay, was used as described by the manufacturer. Total CK activity was measured by spectrophotometry using a COBAS system (Hoffman La Roche, Basel, Switzerland) at the local clinical chemistry laboratory. All assay parameters are presented in supplemental Table S2.
RESULTS
Selection of Proteotypic Peptides and SRM Transitions
Once expressed and purified, each PSAQ standard was submitted to SDS-PAGE, in-gel digestion with trypsin and LC-MS/MS analysis (see supplemental LC-MS/MS data). This allowed us to experimentally determine the signature peptides (so-called “proteotypic” peptides) to be monitored for each cardiovascular biomarker targeted. Proteotypic peptides were chosen for their sequence uniqueness, verified by BLAST search, and ease of detection with LC-MS/MS. Peptides containing methionine and cysteine residues were not excluded from the proteotypic peptide panel. Indeed, our group has recently demonstrated that sulfur-containing peptides can be used for quantification if the redox status of the target protein and its PSAQ analog are “equalized” by H2O2 treatment before trypsin digestion (article under preparation, see also supplemental Figs. S2 and S5 which show titration curves obtained with such modified peptides). Based on LC-MS/MS data analysis, a list of putative SRM transitions was established, which included three to six SRM transitions per proteotypic peptide. Then, using prefractionated serum matrix spiked with PSAQ standards and digested with trypsin, we experimentally selected the “best” SRM transitions, i.e. those effectively detected in the matrix and presenting no signs of interference. In the final SRM methods used for titration curves and patient samples, only the “best” SRM transitions (two to four per proteotypic peptide) were retained (Table I). Importantly, for each selected SRM transition, 2 precursor/fragment ions pairs were actually monitored: one for the labeled form of the peptide and one for its unlabeled form. Hence, LDH-B was characterized by four proteotypic peptides. We also monitored peptide VIGSGCNLDSAR, which is shared between the three LDH isoforms (LDH-A, LDH-B and LDH-C). This additional peptide allowed us to measure total LDH content in serum samples. Quantification of myoglobin, which is a small protein (17 kDa), was based on only one proteotypic peptide. Troponin I levels were evaluated using three proteotypic peptides. For CKMB, heterodimer concentrations could not be specifically investigated. However, we could examine the levels of CKB chains (using two proteotypic peptides) and CKM chains (using three proteotypic peptides) independently. Notably, in serum samples, CKB and CKM chains, in addition to making up the CKMB heterodimer, may also form CKMM and CKBB homodimers.
Quantification of Cardiovascular Biomarkers in Depleted Serum Samples
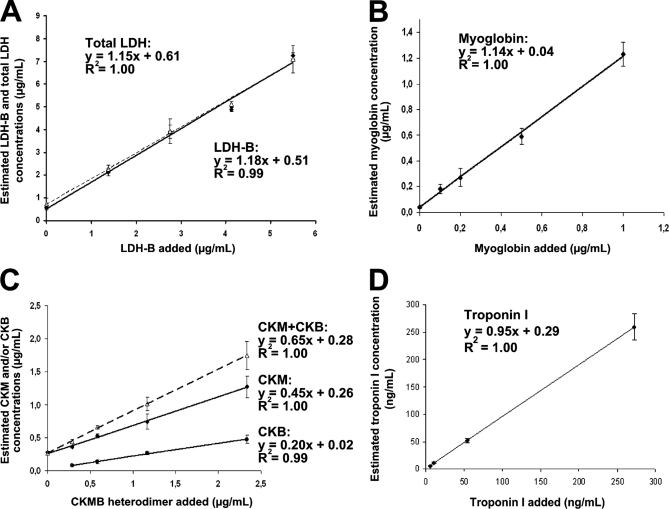
A rapid sample preparation workflow, combining depletion of the six most abundant serum proteins with short SDS-PAGE migration, was developed (Fig. 1A). For each cardiovascular biomarker, a four-point titration experiment was performed by spiking 14 μl serum samples with defined amounts of both unlabeled protein (LDH-B, CKMB, myoglobin, or troponin I) and corresponding PSAQ standard. Zero samples were also constituted. The spiked LDH-B, CKMB, myoglobin, and troponin I quantities were calculated to generate titration curves covering physiological levels to highest pathological concentrations. All titration points were performed in full-process triplicates (see supplemental LC-SRM data). For CKMB heterodimer, we investigated the levels of CKM chains and CKB chains by simultaneously spiking CKM and CKB PSAQ standards. After prefractionation and trypsin digestion, samples were analyzed by LC-SRM. LDH-B, CKM chains and myoglobin could be detected at their physiological levels (i.e. in non-spiked healthy serum samples) estimated at 510 ng/ml, 260 ng/ml and 40 ng/ml, respectively (Fig. 2). For CKB, endogenous levels could not be detected. In healthy donor serum, normal levels of CKB are < 3 ng/ml. Titration curves for LDH-B, CKM and myoglobin were linear for concentrations ranging from endogenous levels to the highest concentrations detected in serum samples. In contrast, troponin I was undetectable in these samples (concentration range: 0.1 ng/ml to 272.5 ng/ml). Based on these titration curves, we determined the analytical performances of the PSAQ-SRM assay for each biomarker targeted (Table II). Ideally, blank serum should have been used to determine LLOQ. However, for CKMB heterodimer, it was difficult to codeplete CKMB, CKBB and CKMM homodimers, which also contain CKB and CKM chains. Thus, we decided to determine LLOQ in healthy donor serum. For each biomarker, LLOQ was deduced from the average titration curve obtained from all proteotypic peptides (Fig. 2) and according to the FDA recommendations (see the “Experimental procedures” section). Despite extensive sample prefractionation and trypsin digestion, excellent quantification accuracy, as indicated by the slope of the average titration curve, was found for both LDH-B and myoglobin (slope value of 1.18 and 1.14, respectively) (Figs. 2A and 2B). Titration of CKMB heterodimer gave unexpected results (Fig. 2C), with slopes of 0.2 (CKB) and 0.45 (CKM), rather than the expected 0.5 for both proteins. This indicates that: (1) commercial CKMB added into serum samples was underestimated and (2) CKM and CKB chains were not added in the same stoeichiometry. To test the latter hypothesis, we investigated “pure” CKMB heterodimer by mixing 900 ng of commercial CKMB with 450 ng of CKB PSAQ standard and 450 ng of CKM PSAQ standard. After in-gel trypsin digestion, the mix was analyzed by LC-SRM which revealed a 2/1 CKM/CKB stoichiometry. This indicates that the commercial CKMB heterodimer contained excess CKM chains, possibly originating from a contaminating CKMM homodimeric form. Commercial CKMB is purified from myocardium containing excess CKMM (75%), thus contamination of CKMB preparations would not be surprising. The underestimation of CKMB in serum may stem from CKB and CKM PSAQ standards and CKMB heterodimer behaving differently during prefractionation and/or digestion. This hypothesis was supported by native-PAGE analysis of CKB and CKM PSAQ standards. CKB PSAQ standard was found to be monomeric whereas CKM PSAQ standard was structured as a CKMM homodimeric (data not shown).
Fig. 2.
Titration of cardiovascular biomarkers in serum samples. A, LDH-B titration curve. This curve was generated using four proteotypic peptides. Titration curves for each proteotypic peptide and related SRM transitions are presented in supplemental Fig. S2. B, Myoglobin titration curve. These results were obtained using one proteotypic peptide. Two SRM transitions were monitored and the corresponding data are presented in supplemental Fig. S3. C, CKM chain, CKB chain and CKMB titration curves. CKMB dimer was artificially spiked into samples. CKB and CKM chain concentrations were estimated independently (from 2 and 3 proteotypic peptides, respectively). The CKM+CKB titration curve represents a combination of the CKB and CKM titration curves. Further details on the different proteotypic peptides monitored and SRM transitions considered are available in supplemental Fig. S4. D, Troponin I titration curve based on three proteotypic peptides. Titration curves for each proteotypic peptide and related SRM transitions are presented in supplemental Fig. S5. For all titration curves, error bars represent standard deviations obtained from 3 full-process replicates.
Table II. Analytical performances of PSAQ-SRM quantifying cardiovascular biomarkers in serum samples.
| Biomarker | Sample prefractionation method | Serum volume | LLOQa (ng/ml) | Range of tested concentrations (ng/ml) | Linearity (R2) | Accuracy (slope value) | Precision at LLOQ (CV in %) | |
|---|---|---|---|---|---|---|---|---|
| LDH-B | Depletion + SDS-PAGE | 14 μl | 510 | 510 (endogenous) → 5500 | 0.99 | 1.18 | 8 | |
| CKB chains | Depletion + SDS-PAGE | 14 μl | ND | 20 (endogenous) → 1125 | 0.99 | 0.20 | 0.65 (CKMB) | ND* |
| CKM chains | Depletion + SDS-PAGE | 14 μl | ND | 260 (endogenous) → 1125 | 1 | 0.45 | ND* | |
| Myoglobin | Depletion + SDS-PAGE | 14 μl | 500 | 40 (endogenous) → 1000 | 1 | 1.14 | 8 | |
| Troponin I | Immunoenrichment + SDS-PAGE | 0.5 to 1 ml | 5.5 | 0.29 (endogenous) → 272.5 | 1 | 0.95 | 3 | |
a LLOQ was established as the lowest concentration of the average titration curve that was measured with precision (CV) inferior to 20% and accuracy comprised between 80 and 120%.
* CKMB LLOQ could not be determined as quantification accuracy was lower than 80% (65%).
Quantification of Troponin I in Immunoenriched Serum Samples
Generally, LC-SRM detection of serum proteins present below 100 ng/ml is difficult when using depletion as unique prefractionation method (4). However, we hypothesized that the combination of depletion and SDS-PAGE might allow troponin I detection, particularly for samples with concentrations in the range of 100 ng/ml. Indeed, myoglobin could be detected at just 40 ng/ml using this procedure. However, troponin I could not be detected, even at the highest concentration tested (272.5 ng/ml). Most likely, this is because of its interaction with abundant proteins retained on the depletion cartridge (19). Therefore, a prefractionation method based on immunocapture coupled to SDS-PAGE was developed to validate the PSAQ-SRM approach for this specific biomarker (Fig. 1A). To perform titration experiments, healthy serum samples (1 ml) were spiked with defined amounts of troponin I and its corresponding PSAQ standard. An immunoenrichment protocol was optimized using a biotinylated antitroponin I antibody and streptavidin coated beads. Because of the strong biotin-streptavidin interaction, antitroponin I antibody was retained on streptavidin beads during elution, further improving sample decomplexification. However, as Laemmli buffer was used to elute troponin I, direct trypsin digestion was not possible. Therefore, samples were submitted to “stacking” SDS-PAGE before in-gel digestion (supplemental Fig. S5A). After trypsin digestion and peptide extraction samples were analyzed using LC-SRM analysis. With this serum prefractionation method, troponin I could be detected at 500 pg/ml of serum, which is slightly higher than physiological concentrations (≈ 350 pg/ml). However, sensitive and accurate troponin I quantification was possible over the pathological concentration range (5.5 to 272.5 ng/ml) (Fig. 2D, supplemental Fig. S5). PSAQ-SRM assay analytical performances after troponin I immunoenrichment are presented in Table II.
Interestingly, the troponin I proteotypic peptide ISADAMMQALLAGR includes serine 149 which was previously reported to be phosphorylatable (21). In this study, all quantification results obtained from this peptide were consistent with those obtained from the two other proteotypic peptides (see supplemental Fig. S5 and supplemental LC-SRM data). Therefore, we can hypothesize that: (1) serine 149 was primarily not phosphorylated or (2) serine 149 phosphorylation was removed as troponin I was released in blood flow.
Multiplexed PSAQ-SRM Analysis of Clinical Samples and Correlation with ELISA/Enzymatic Assay Results
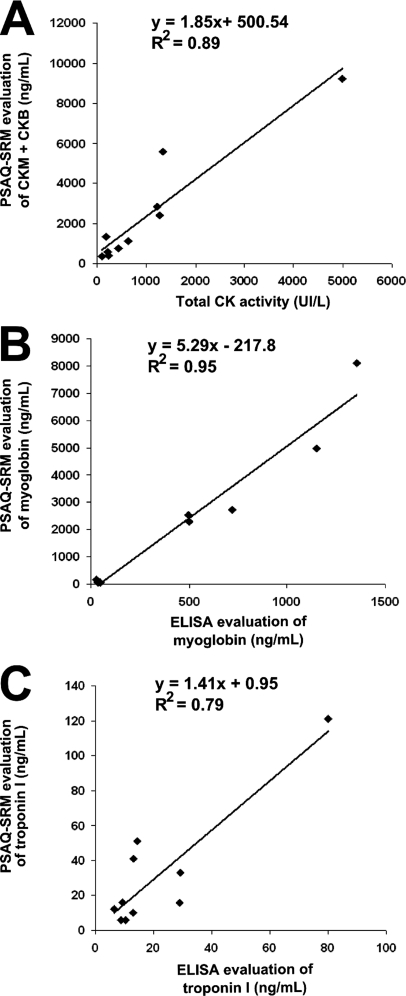
Once the prefractionation and quantification processes had been characterized for each biomarker, multiplexed LC-SRM detection was assessed on healthy donor serum samples. After spiking the samples with defined quantities of PSAQ standards, prefractionation and SDS-PAGE, gel bands were cut, pooled and proteins were digested with trypsin (Fig. 1A). LDH-B, myoglobin, CKM and CKB could be simultaneously detected in depleted serum samples without difficulty and without time-scheduled acquisition (Fig. 1B). We then applied this multiplex PSAQ-SRM method to the analysis of serum samples from patients with myocardial infarction. Serum samples from five patients were collected at two time-points: (1) immediately after hospital admission and (2) 3 to 8 days after PCI. All samples were analyzed using the PSAQ-SRM method after depletion (14 μl) or troponin I immunoenrichment (1 ml). LDH-B, myoglobin, CKM and CKB chains and troponin I were all detected in these samples (Table III, supplemental Figs. S6 and S7). For each patient, biomarker concentration changes between the two collection time-points were consistent with blood-release kinetics i.e. an early increase in troponin I, myoglobin, and CKMB (day 0) and a delayed LDH-B augmentation (days 3 or 8). As myoglobin, CKMB and troponin I are routinely used clinically to confirm myocardial injury, correlation between ELISA or enzymatic assays and the PSAQ-SRM approach could be assessed for these patient samples (Table III and Fig. 3). Total CK enzyme activity and CKB and CKM concentrations estimated by PSAQ-SRM correlated well, with a R2 value of 0.89 (Fig. 3A). Excellent correlation between ELISA and PSAQ-SRM results (R2 = 0.95) were observed for myoglobin. Surprisingly however, the slope of the correlation curve was equal to 5.29, suggesting that PSAQ-SRM systematically quantifies five times more myoglobin than ELISA (Fig. 3B). Following this result, we compared the commercial myoglobin ELISA standard directly to our PSAQ standard by mixing them in equal amounts. After trypsin digestion and LC-SRM analysis of the 1/1 mix, the ELISA standard was found to be 5.39 times less concentrated than the PSAQ standard (see supplemental Fig. S8). From these results, we can surmise that either the concentration of the myoglobin ELISA standard was overestimated by the supplier or that it had been degraded during storage. For troponin I quantification, a correlation coefficient (R2) of 0.79 was found between ELISA and PSAQ-SRM with a correlation curve slope of 1.41 (Fig. 3C). Possibly, this lesser correlation originates from troponin I interactions with serum proteins. By masking epitopes, such interactions could differentially influence ELISA detection (based on two monoclonal antibodies) and PSAQ-SRM immunocapture (based on one monoclonal antibody).
Table III. PSAQ-SRM and ELISA quantification of LDH-B, CKB, CKM, myoglobin and troponin I in patient serum. Biomarker concentrations were determined using at least 1 proteotypic peptide and 2 SRM transitions except for values indicated with:
* 1 proteotypic peptide detected with 1 SRM transition
** 2 proteotypic peptides, each detected with 1 SRM transition
See supplemental Figure 7 which shows the corresponding SRM transition chromatograms.
| Patient, day after hospital admission | PSAQ-SRM |
ELISA or enzymatic assay |
||||||
|---|---|---|---|---|---|---|---|---|
| Depletion |
Immuno-enrichment |
Total CK activity (UI/L) | Myoglobin (ng/ml) | Troponin I (ng/ml) | ||||
| LDH-B (μg/ml) | CKB (ng/ml) | CKM (ng/ml) | Myoglobin (ng/ml) | Troponin I (ng/ml) | ||||
| Patient 1, day 0 | 3.21 | 80** | 9160 | 8120 | 121 | 4983 | 1353 | 80 |
| Patient 1, day 3 | 4.32 | 30* | 730 | 60* | 16 | 439 | 50 | 29 |
| Patient 2, day 0 | 1.07 | 100* | 2320 | 2530* | 41 | 1269 | 496 | 13.3 |
| Patient 2, day 3 | 2.70 | 50* | 1280 | 170 | 12 | 188 | 28 | 6.6 |
| Patient 3, day 0 | 1.93 | 150 | 5430 | 4975 | 51 | 1338 | 1149 | 14.6 |
| Patient 3, day 8 | 2.10 | 120* | 230 | 90 | 6 | 107 | 36 | 10.3 |
| Patient 4, day 0 | 1.03 | 70* | 1050 | 2735 | 16 | 626 | 719 | 9.3 |
| Patient 4, day 3 | 2.18 | 120* | 470 | 80 | 6 | 218 | 36 | 8.9 |
| Patient 5, day 0 | 1.88 | 110 | 2750 | 2305 | 33 | 1219 | 500 | 29.4 |
| Patient 5, day 3 | 4.19 | 20* | 360 | 45 | 10 | 228 | 38 | 12.9 |
Fig. 3.
PSAQ-SRM quantification of cardiovascular biomarkers in patient samples and correlation with enzymatic or ELISA assays. A, Correlation between total CK enzymatic activity and CKMB concentration, as determined by PSAQ-SRM, in clinical serum samples after depletion of abundant proteins (five patients with myocardial infarction, two collection time-points). B, Correlation between ELISA and PSAQ-SRM results for quantification of myoglobin in clinical serum samples after depletion of abundant proteins (five patients with myocardial infarction, two collection time-points). C, Correlation between results for quantification of troponin I in clinical serum samples by ELISA or in immunoenriched samples by PSAQ-SRM (five patients with myocardial infarction, two collection time-points).
DISCUSSION
In 2006, Rifai and co-workers described the biomarker pipeline and the promise MS held for biomarker research (1). In particular, they highlighted how SID-SRM analysis might solve the technological hurdle of biomarker evaluation. However, application of SID-SRM as part of biomarker development requires key analytical performances to be attained, including specificity, sensitivity and confident quantification. In addition, to surpass ELISA it is crucial to offer multiplexing capabilities and antibody-free prefractionation. Recently, an interlaboratory study assessed the analytical features of a multiplexed SID-SRM assay and demonstrated the accelerated throughput and transferability of this type of analysis. This study, however, highlighted quantification accuracy as a limitation of the method, particularly when sample prefractionation was necessary (7, 8). The goal of our study was to demonstrate that the use of full-length isotopically-labeled proteins used as quantification standards (PSAQ standards) could significantly advance the performances of a SRM-based biomarker evaluation platform.
LDH-B, CKMB, myoglobin, and troponin I were chosen as model biomarkers to assess the performances of the PSAQ-SRM method. Troponin I, CKMB, and myoglobin are currently used in hospital laboratories to rapidly confirm myocardial injury. Measurement of LDH-B levels was abandoned because its release in the blood stream is delayed, reaching its maximal concentration 72 h after myocardial infarction. However, to evaluate the PSAQ-SRM method, the combination of these four biomarkers was of particular interest. First because they belong to different concentration classes, with LDH-B being the most abundant (μg/ml of serum) and troponin I requiring very sensitive assays (below 1 ng/ml in serum). Second, ELISA or enzymatic tests are available for troponin I, myoglobin and CKMB, making a comparison between ELISA and PSAQ-SRM assays possible.
Samples were fractionated using a decomplexification method based on the depletion of abundant proteins combined with SDS-PAGE. Using this straightforward sampling, only 14 μl of serum were necessary to simultaneously quantify 3 biomarkers at their physiological levels. Likely because of the high ion-current potential of its proteotypic peptide (22), myoglobin could be quantified down to 40 ng/ml (equivalent to 33 femtomoles in 14 μl). For troponin I quantification, however, this type of fractionation, applied to tiny volumes of serum, was not sufficient. Not only is troponin I a low-abundance biomarker, but it has also been shown to interact with abundant proteins retained by serum depletion devices (19). Therefore, we developed an immunocapture approach to quantify troponin I below ng/ml concentrations in serum. Measurement of this protein might also have been possible using depletion on larger serum volumes, combined with mild-detergents to improve elution from the depletion cartridge and by replacing SDS-PAGE with chromatographic peptide separation. Indeed, Keshishian and coworkers have recently described an antibody-free sampling approach involving protein depletion, trypsin digestion and SCX peptide separation before LC-SRM analysis (19). With this methodology, they were able to detect troponin T down to 5 ng/ml in 100 μl plasma samples. Another recent study detected 4 ng/ml of PSA in 100 μl serum samples using albumin depletion, trypsin digestion, SPE peptide separation, and LC-SRM analysis (23).
Given that most proteins of clinical interest are present in the ng/ml range in serum or plasma, matrix decomplexification is generally mandatory to successfully detect biomarker candidates (4). In this context, the use of isotope-labeled peptide standards, added at late stages of the analytical workflow, may not provide optimal quantification accuracy and reproducibility (7–9). Ideally, quantification standards should correct for losses occurring during sample decomplexification and compensate for digestion variability. Our results demonstrate that accurate and precise biomarker quantification can be achieved by spiking isotopically labeled proteins (PSAQ standards) into serum samples early in the sample preparation process. Provided that PSAQ standards behave exactly like the target proteins, quantification is accurate, even in cases where recovery may have been affected by extensive fractionation (depletion or immunocapture, and SDS-PAGE). Similarly, PSAQ standards also compensate for variable digestion yields (17). In this study, we also tried to quantify a protein heterodimer, CKMB, using both CKB and CKM standards. Possibly because the standards did not exactly mimic the biochemical behavior of the CKMB heterodimer, quantification was less accurate for this protein complex than for the other proteins studied. Nevertheless, in patient serum samples, increases in CKM and CKB chains measured by PSAQ-SRM were highly consistent with total CK activity results.
Accuracy is not essential for biomarker verification as this step is mainly focused on determining the specificity of biomarker candidates. Because of this, the use of isotope-labeled peptides as quantification standards is relevant and straightforward in this phase (19). However, during the qualification phase, the difference between healthy and pathological samples has to be established. For this, a method that can accurately and precisely quantify biomarkers over both physiological and pathological concentration ranges would be of particular value. In addition to meeting this criterion, as PSAQ standards correct for variable recovery due to differences in sample handling and digestion efficiency, their use should significantly improve interassay and interlaboratory reproducibility.
Because of its selectivity, SRM analysis provides a high detection specificity (5). However, when working with highly complex samples such as serum, this selectivity might not be sufficient to avoid matrix interferences (24). In this context, the use of isotope dilution standards, such as labeled peptides or proteins, which coelute with the target improve the specificity of the analysis (12, 25). Finally, we have shown that PSAQ standards offer the largest coverage for quantification even making it possible to take cystein- and methionin-containing peptides into account after protein oxidation (18).
In our study, multiplexing was limited to the detection of four biomarkers in depleted serum samples. However, technological advances, including scheduled SRM (26, 27) and iSRM (28), have made it possible to monitor hundreds of proteins in a single experiment. Recently, Kuzyk and coworkers used isotope-labeled peptide standards and LC-SRM to quantify 45 proteins in plasma. Thirty-one of these proteins were potential biomarkers of cardiovascular diseases (25). The most frequent question about implementation of the PSAQ method concerns the availability of PSAQ standards. In this study, the cell-free expression and isotope labeling of LDH-B, CKB, CKM, myoglobin, and troponin I PSAQ standards were optimized in less than 2 months. These biomarkers are not post-translationally modified, which facilitates their expression in bacterial lysates. However, phosphorylated or glycosylated PSAQ standards can be produced using specific production systems (11, 13). Certainly, the throughput of PSAQ standard production could be further enhanced by availing of cDNA libraries specifically developed for protein expression (29, 30).
In conclusion, this study clearly demonstrates the relevance of using isotope-labeled protein standards for multiplex and reliable quantification of biomarkers in prefractionated clinical samples. We are currently concentrating our efforts on the generation of isotope-labeled protein libraries to increase the availability of PSAQ standards and widen the use of the PSAQ-SRM analytical strategy. Applying PSAQ-SRM as part of the biomarker development pipeline should help bridge the gap between biomarker discovery and clinical applications.
Acknowledgments
We thank Christel Cabon for preparing patient samples, Marie-Laure Bourhis for logistical help, EDyP Service team for LC-MS/MS analyses, Dr Christophe Masselon for scientific discussions and Dr Maighread Gallagher-Gambarelli for editorial assistance.
Footnotes
* This work was supported by grants from the GRAVIT consortium, the French National Agency for Research (Project ANR-08-EBIO-008) and the 7th Framework Programme of the European Union (Contract no. 262067-PRIME-XS).
 This article contains supplemental Figs. S1 to S8 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S8 and Tables S1 and S2.
1 The abbreviations used are:
- CKB
- Creatine kinase B chain
- CKM
- Creatine kinase M chain
- CKBB
- Creatine kinase BB homodimer
- CKMB
- Creatine kinase MB heterodimer
- CKMM
- Creatine kinase MM homodimer
- LDH-B
- Lactate dehydrogenase B
- PCI
- Primary percutaneous coronary intervention
- SID
- Stable isotope dilution
- SRM
- Selected reaction monitoring
- PSAQ
- Protein standard absolute quantification
- PSAQ™
- for accurate biomarker quantification.
REFERENCES
- 1. Rifai N., Gillette M. A., Carr S. A. (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Makawita S., Diamandis E. P. (2010) The Bottleneck in the Cancer Biomarker Pipeline and Protein Quantification through Mass Spectrometry-Based Approaches: Current Strategies for Candidate Verification. Clin. Chem. 56, 212–222 [DOI] [PubMed] [Google Scholar]
- 3. Paulovich A., Whiteaker J., Hoofnagle A., Wang P. (2008) The interface between biomarker discovery and clinical validation: The tar pit of the protein biomarker pipeline. Proteom. Clin. Appl. 2, 1386–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hüttenhain R., Malmström J., Picotti P., Aebersold R. (2009) Perspectives of targeted mass spectrometry for protein biomarker verification. Curr. Opin. Chem. Biol. 13, 518–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoofnagle A. N. (2010) Quantitative Clinical Proteomics by Liquid Chromatography-Tandem Mass Spectrometry: Assessing the Platform. Clin. Chem. 56, 161–164 [DOI] [PubMed] [Google Scholar]
- 9. Brun V., Dupuis A., Adrait A., Marcellin M., Thomas D., Court M., Vandenesch F., Garin J. (2007) Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol. Cell. Proteomics 6, 2139–2149 [DOI] [PubMed] [Google Scholar]
- 10. Brun V., Masselon C., Garin J., Dupuis A. (2009) Isotope dilution strategies for absolute quantitative proteomics. J. Proteomics 72, 740–749 [DOI] [PubMed] [Google Scholar]
- 11. Ciccimaro E., Hanks S. K., Yu K. H., Blair I. A. (2009) Absolute quantification of phosphorylation on the kinase activation loop of cellular focal adhesion kinase by stable isotope dilution liquid chromatography/mass spectrometry. Anal. Chem. 81, 3304–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanke S., Besir H., Oesterhelt D., Mann M. (2008) Absolute SILAC for accurate quantitation of proteins in complex mixtures down to the attomole level. J. Proteome Res. 7, 1118–1130 [DOI] [PubMed] [Google Scholar]
- 13. Heudi O., Barteau S., Zimmer D., Schmidt J., Bill K., Lehmann N., Bauer C., Kretz O. (2008) Towards absolute quantification of therapeutic monoclonal antibody in serum by LC-MS/MS using isotope-labeled antibody standard and protein cleavage isotope dilution mass spectrometry. Anal. Chem. 80, 4200–4207 [DOI] [PubMed] [Google Scholar]
- 14. Izrael-Tomasevic A., Phu L., Phung Q. T., Lill J. R., Arnott D. (2009) Targeting interferon alpha subtypes in serum: a comparison of analytical approaches to the detection and quantitation of proteins in complex biological matrices. J. Proteome Res. 8, 3132–3140 [DOI] [PubMed] [Google Scholar]
- 15. Janecki D. J., Bemis K. G., Tegeler T. J., Sanghani P. C., Zhai L., Hurley T. D., Bosron W. F., Wang M. (2007) A multiple reaction monitoring method for absolute quantification of the human liver alcohol dehydrogenase ADH1C1 isoenzyme. Anal. Biochem. 369, 18–26 [DOI] [PubMed] [Google Scholar]
- 16. Kumar V., Barnidge D. R., Chen L. S., Twentyman J. M., Cradic K. W., Grebe S. K., Singh R. J. (2010) Quantification of serum 1–84 parathyroid hormone in patients with hyperparathyroidism by immunocapture in situ digestion liquid chromatography-tandem mass spectrometry. Clin. Chem. 56, 306–313 [DOI] [PubMed] [Google Scholar]
- 17. Lesur A., Varesio E., Hopfgartner G. (2010) Accelerated tryptic digestion for the analysis of biopharmaceutical monoclonal antibodies in plasma by liquid chromatography with tandem mass spectrometric detection. J. Chromatogr. A. 1217, 57–64 [DOI] [PubMed] [Google Scholar]
- 18. Jaquinod M., Villiers F., Kieffer-Jaquinod S., Hugouvieux V., Bruley C., Garin J., Bourguignon J. (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics 6, 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keshishian H., Addona T., Burgess M., Mani D. R., Shi X., Kuhn E., Sabatine M. S., Gerszten R. E., Carr S. A. (2009) Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 8, 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanagi M. M., Ling S. L., Nasir Z., Hermawan D., Ibrahim W. A., Abu Naim A. (2009) Comparison of signal-to-noise, blank determination, and linear regression methods for the estimation of detection and quantification limits for volatile organic compounds by gas chromatography. J. AOAC Int. 92, 1833–1838 [PubMed] [Google Scholar]
- 21. Buscemi N., Foster D. B., Neverova I., Van Eyk J. E. (2002) p21-activated kinase increases the calcium sensitivity of rat triton-skinned cardiac muscle fiber bundles via a mechanism potentially involving novel phosphorylation of troponin I. Circ. Res. 91, 509–516 [DOI] [PubMed] [Google Scholar]
- 22. Fusaro V. A., Mani D. R., Mesirov J. P., Carr S. A. (2009) Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 27, 190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fortin T., Salvador A., Charrier J. P., Lenz C., Lacoux X., Morla A., Choquet-Kastylevsky G., Lemoine J. (2009) Clinical quantitation of prostate-specific antigen biomarker in the low nanogram/milliliter range by conventional bore liquid chromatography-tandem mass spectrometry (multiple reaction monitoring) coupling and correlation with ELISA tests. Mol. Cell. Proteomics 8, 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sherman J., McKay M. J., Ashman K., Molloy M. P. (2009) How specific is my SRM? The issue of precursor and product ion redundancy. Proteomics 9, 1120–1123 [DOI] [PubMed] [Google Scholar]
- 25. Kuzyk M. A., Smith D., Yang J., Cross T. J., Jackson A. M., Hardie D. B., Anderson N. L., Borchers C. H. (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics 8, 1860–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stahl-Zeng J., Lange V., Ossola R., Eckhardt K., Krek W., Aebersold R., Domon B. (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 6, 1809–1817 [DOI] [PubMed] [Google Scholar]
- 27. Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 28. Kiyonami R., Schoen A., Prakash A., Peterman S., Zabrouskov V., Picotti P., Aebersold R., Huhmer A., Domon B. Increased selectivity, analytical precision, and throughput in targeted proteomics. Mol. Cell. Proteomics, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goshima N., Kawamura Y., Fukumoto A., Miura A., Honma R., Satoh R., Wakamatsu A., Yamamoto J., Kimura K., Nishikawa T., Andoh T., Iida Y., Ishikawa K., Ito E., Kagawa N., Kaminaga C., Kanehori K., Kawakami B., Kenmochi K., Kimura R., Kobayashi M., Kuroita T., Kuwayama H., Maruyama Y., Matsuo K., Minami K., Mitsubori M., Mori M., Morishita R., Murase A., Nishikawa A., Nishikawa S., Okamoto T., Sakagami N., Sakamoto Y., Sasaki Y., Seki T., Sono S., Sugiyama A., Sumiya T., Takayama T., Takayama Y., Takeda H., Togashi T., Yahata K., Yamada H., Yanagisawa Y., Endo Y., Imamoto F., Kisu Y., Tanaka S., Isogai T., Imai J., Watanabe S., Nomura N. (2008) Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 5, 1011–1017 [DOI] [PubMed] [Google Scholar]
- 30. Holz C., Lueking A., Bovekamp L., Gutjahr C., Bolotina N., Lehrach H., Cahill D. J. (2001) A human cDNA expression library in yeast enriched for open reading frames. Genome Res. 11, 1730–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]