Abstract
Lung cancer is the leading cause of cancer-related death in the world. To explore tumor biomarkers for clinical application, two-dimensional fluorescence difference gel electrophoresis and subsequent MALDI-TOF/TOF mass spectrometry were performed to identify proteins differentially expressed in 12 pairs of lung squamous cell tumors and their corresponding normal tissues. A total of 28 nonredundant proteins were identified with significant alteration in lung tumors. The up-regulation of isocitrate dehydrogenase 1 (IDH1), superoxide dismutase 2, 14-3-3ε, and receptor of activated protein kinase C1 and the down-regulation of peroxiredoxin 2 in tumors were validated by RT-PCR and Western blot analysis in independent 15 pairs of samples. Increased IDH1 expression was further verified by the immunohistochemical study in extended 73 squamous cell carcinoma and 64 adenocarcinoma clinical samples. A correlation between IDH1 expression and poor overall survival of non-small cell lung cancer (NSCLC) patients was observed. Furthermore, ELISA analysis showed that the plasma level of IDH1 was significantly elevated in NSCLC patients compared with benign lung disease patients and healthy individuals. In addition, knockdown of IDH1 by RNA interference suppressed the proliferation of NSCLC cell line and decreased the growth of xenograft tumors in vivo. These observations suggested that IDH1, as a protein promoting tumor growth, could be used as a plasma biomarker for diagnosis and a histochemical biomarker for prognosis prediction of NSCLC.
Lung cancer is the leading cause of cancer-related death in the world because of its high morbidity and mortality. Approximately 1.2 million people are diagnosed with lung cancer all over the world per year. Despite the great progresses in cancer research over the last decades, lung cancer remains at a very low 5-year survival rate: 16% compared with 89% for breast cancer, 65% for colon cancer, and 100% for prostate cancer (1). Although lung cancer comprises only about 15% of new cancer diagnosis, it causes over 30% of all cancer-related deaths. Lung cancer is divided into two major clinicopathological classes: small cell lung cancer (∼15% of all lung cancer) and non-small-cell lung cancer (NSCLC, ∼85%).1 The latter includes three major histological subtypes: squamous cell carcinoma (SCC, 40% of NSCLC), adenocarcinoma (AD, 40% of NSCLC), and large cell carcinoma (10% of NSCLC) (2). NSCLC is commonly treated with surgery, whereas small cell lung cancer usually responds better to chemotherapy and radiotherapy. For NSCLC, curative surgery is efficacious only in those who are diagnosed sufficiently early in the disease process. Unfortunately, more than 70% of the patients are diagnosed only at an advanced stage nowadays, which results in the loss of opportunity for curative surgical resection and poor prognosis. To improve the survival of patients with lung cancer, identifying reliable biomarkers for early diagnosis and prognosis prediction and monitoring treatment response remain urgently needed.
Proteomic analysis, a powerful tool for global evaluation of protein expression, has been widely applied in cancer research. Quantitative protein expression profiling allows efficient identification of accurate and reproducible differential expression values for proteins in multiple biological samples. Comparison of protein expression profiles between tumors and normal tissues and among different tumors may lead to discovery of clinically useful tumor biomarkers, new therapeutic targets, and elucidation of molecular mechanisms of cancers (3, 4). Several previous studies have focused on the application of comparative proteomics in screening differentially expressed proteins in cell line or clinical specimens of lung cancer (5–17). Approximately 300 proteins have been identified through these studies, including oncoproteins, signal transduction proteins, metabolic enzymes, and so on. However, few of them have been analyzed for their correlation with clinicopathological characteristics of lung cancer patients to investigate the value for clinical applications and their function in lung tumorigenesis. So far, none of the molecules identified are implemented in routine clinical use yet, and reliable biomarkers of lung cancer are urgently needed (18).
In present study, we used 2D-DIGE to analyze tumors and paired lung tissues from 12 SCC patients. The spots up-regulated or down-regulated were further analyzed by mass spectrometry, and some proteins of interest were validated in an independent patient cohort by RT-PCR and Western blot. Of them, IDH1 was chosen for further validation in paraffin-embedded tissues and plasmas from NSCLC patients, plus functional characterization. The data presented here indicated that IDH1, as a protein promoting tumor growth, might be a novel plasma biomarker for diagnosis and a histochemical biomarker for prognosis prediction of NSCLC.
EXPERIMENTAL PROCEDURES
Samples
All of the tissue and blood specimens were collected from patients in the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences with informed consent and agreement. None of these patients received antineoplastic therapy prior to surgery. All of the tissue samples were taken by experienced surgeons and examined independently by two experienced pathologists. For 2D-DIGE, 12 pairs of fresh primary lung SCC tumors and their corresponding adjacent normal tissues were obtained during 2007–2008 (Table I). The independent 15 pairs of lung SCC samples were collected for RT-PCR and Western blot. Necrotic tissue was excluded, and normal lung tissues were confirmed to contain no tumor cells by histopathologic evaluation. The samples were snap frozen in liquid nitrogen immediately after resection and stored at −80 °C until use. For immunohistochemistry analysis, 137 paraffin-embedded lung tumors and paired adjacent normal lung tissues were randomly obtained from patients during 1999–2001. For ELISA study, preoperative peripheral blood samples were obtained from 200 NSCLC patients (median age of 60 years with a range of 34–79 years) during 2009–2010 including 100 SCCs and 100 ADs. 55 specimens of healthy individuals (median age of 53 years with a range of 41–64 years) were donated on a voluntary basis. In addition, 50 benign lung disease samples were obtained (median age of 53 years with a range of 21–79 years), including 16 chronic lung inflammation, 7 pulmonary tuberculosis, 22 hamartoma, 2 sclerosing hemangioma and 3 inflammatory myofibroblastic tumor. For all the specimens, clinicopathological information (age, gender, pathology, differentiation, smoking history, and TNM stage) was available. The study was approved by the medical ethics committee of Cancer Institute and Hospital of the Chinese Academy of Medical Sciences.
Table I. Clinical and pathologic information of 12 SCC patients.
M, male; F, female.
| Patients | TNM stage | Differentiation | Gender | Age |
|---|---|---|---|---|
| 1 | T2N0M0 | Middle | F | 63 |
| 2 | T2N0M0 | Middle | M | 66 |
| 3 | T2N0M0 | Low | M | 63 |
| 4 | T2N0M0 | Middle-low | M | 52 |
| 5 | T2N0M0 | Low | M | 57 |
| 6 | T2N0M0 | Low | M | 52 |
| 7 | T2N2M0 | Middle | M | 56 |
| 8 | T2N2M0 | Middle | M | 54 |
| 9 | T2N2M0 | Middle | M | 60 |
| 10 | T2N2M0 | Middle | M | 43 |
| 11 | T2N2M0 | Low | F | 53 |
| 12 | T2N2M0 | Low | M | 50 |
2D-DIGE
Approximately 0.5 g of tissue was grinded into powder in liquid nitrogen with a precooled mortar and pestle. The samples were then homogenized on ice in 1 ml of lysis buffer (7 m urea, 2 m thiourea, 4% CHAPS, 30 mm Tris-HCl, pH 8.5, protease inhibitor mixture) using a glass homogenizer. After sonication on ice for 10 s using an ultrasonic processor, the samples were centrifuged for 30 min (40,000 × g) to remove particulate materials. Protein concentrations were determined in duplicate by the Bradford method (Bio-Rad) and confirmed by SDS-PAGE.
The pH of the protein was adjusted to 8.5 by 50 mm NaOH, and the concentration was adjusted to 5 mg/ml with lysis buffer. Equal amounts of proteins from the 12 pairs of samples were pooled together as the internal standard. Tumor and nontumor counterparts of each patient were randomly labeled with Cy3 or Cy5, whereas internal standards were labeled with Cy2 using 400 pmol of fluorochrome/50 μg of protein. Labeling was performed for 30 min on ice in the dark. The reactions were then quenched by the addition of 1 μl of lysine (10 mm) for 10 min on ice in the dark.
The DIGE experimental design is shown in supplemental Table 1. Fifty-microgram Cy3- and Cy5-labeled samples from each patient were combined before mixing with 50 μg of Cy2-labeled internal standard. Then an equal volume of 2× sample buffer (7 m urea, 2 m thiourea, 4% CHAPS, 1% Bio-Lyte, pH 3–10, 20 mg/ml DTT) was added to the sample, and the total volume was made up to 410 μl with rehydration buffer (7 m urea, 2 m thiourea, 4% CHAPS, 0.5% Bio-Lyte, 10 mg/ml DTT).
The samples were actively rehydrated into 24-cm pH 3–10 IPG strips (Bio-Rad) at 17 °C for 12 h using a Protean IEF cell (Bio-Rad). Isoelectric focusing was performed for a total of 80 kV-h (ramped to 250 V in 30 min, held at 1000 V for 1 h, ramped to 10,000 V in 5 h, and held at 10,000 V for 60 kV-h). The IPG strips were equilibrated in equilibration buffer (6 m urea, 2% SDS, 50 mm Tris-HCl, pH 8.8, 30% glycerol) supplemented with 0.5% DTT for 15 min at room temperature followed by 4.5% iodoacetamide in equilibration buffer for another 15-min incubation at room temperature.
IPG strips were placed on the top of 12% homogeneous polyacrylamide gels that had been precast with low fluorescence glass plates using an Ettan DALT 12-gel caster. The second dimension SDS-PAGE was carried out using the Protean Plus system (Bio-Rad). After two-dimensional electrophoresis, the gels were scanned on the Typhoon 9410 scanner with Ettan DALT gel alignment guides using excitation/emission wavelengths specific for Cy2 (488/520 nm), Cy3 (532/580 nm), and Cy5 (633/670 nm). The intensity was adjusted to ensure that the maximum volume of each image was within 60,000–90,000.
Analysis of 2D-DIGE was done using DeCyder 6.5 software (GE Healthcare) according to the manufacturer's recommendations. Briefly, the DeCyder biological variation analysis module was used to detect spots (the estimated number of spots was 2500) and simultaneously match all 36 protein spot maps from 12 gels. All of the matches were also confirmed manually. The paired t test with false discovery rate correction was used for statistical analysis of the data. Protein spots that were differentially expressed in tumors and normal tissues (ratio ≦ −2 or ratio ≥ 2, p ≤ 0.05) were selected. Only spots altered in the same direction with their average ratio in at least five of the 12 patients were chosen for identification.
In-gel Digestion
Spot picking was carried out with preparative gels. Two-dimensional electrophoresis was performed as described under “2D-DIGE” except that the IPG strips were loaded with 1000 μg of protein and the gels were stained with Coomassie Brilliant Blue. Protein spots of interest were excised and destained with 25 mm ammonium bicarbonate, 50% ACN. The gels were then dried completely by centrifugal lyophilization. In-gel digestion was performed with 0.01 μg/μl trypsin (Promega) in 25 mm ammonium bicarbonate for 15 h at 37 °C. The supernatants were collected, and the tryptic peptides were extracted from the gel sequentially with 5% TFA at 40 °C for 1 h and with 2.5% TFA, 50% ACN at 30 °C for 1 h. The extracts were pooled and dried completely by centrifugal lyophilization.
Protein Identification
Peptide mixtures were redissolved in 0.5% TFA, and 1 μl of peptide solution was mixed with 1 μl of matrix (4-hydroxy-α-cyanocinnamic acid in 30% ACN, 0.1% TFA) before spotting on the target plate. MALDI-TOF mass spectrometry and tandem TOF/TOF mass spectrometry were carried out on a 4800 Proteomics Analyzer (Applied Biosystems). Peptide mass maps were acquired in positive reflection mode, averaging 1500 laser shots/MALDI-TOF spectrum and 3000 shots/TOF/TOF spectrum (the resolution was 20,000). The 4800 calibration mixtures (Applied Biosystems) were used to calibrate the spectrum to a mass tolerance within 0.1 Da. Parent mass peaks with a mass range of 600–4000 Da and minimum signal to noise ratio of 15 were picked out for tandem TOF/TOF analysis. Combined mass and mass/mass spectra were used to interrogate human sequences in the IPI human database v3.23 (which contains 66,619 protein entries) using the MASCOT database search algorithms (version 2.1). Searches were performed to allow for carbamidomethylation, oxidation, and a maximum of one missed trypsin cleavage. Peptide tolerance and MS/MS tolerance were both 0.2 Da. All of the automatic data analysis and database searching were fulfilled by the GPS ExplorerTM software (version 3.6; Applied Biosystems). Known contaminant ions (keratin) were excluded. The confident identification had a statistically significant (p ≤ 0.05) protein score (based on combined mass and mass/mass spectra). Redundancy of proteins that appeared in the database under different names and accession numbers was eliminated. Spots in which more than one protein was identified were excluded.
Semi-quantitative RT-PCR
Total RNA was isolated from frozen tissues with TRIzol method (Invitrogen) following the manufacturer's instructions. The first strand cDNA was synthesized from 2 μg of total RNA using RevertAid first strand cDNA synthesis kit (Fermentas). For semi-quantitative PCR analysis, cDNA was amplified by Taq DNA polymerase (Takara). Human 18 S rRNA gene was used as an internal control. The primer sequences and the expected sizes of PCR products were listed in supplemental Table 3. RT-PCR was performed with conditions as follows: denaturation at 94 °C for 2 min; then amplification for 28 cycles at 94 °C for 0.5 min, annealing at 60 °C for 0.5 min, and extension at 72 °C for 0.5 min; and then a terminal elongation step at 72 °C for 10 min and a final holding stage at 4 °C.
Western Blot
Proteins from paired tumors and normal lung tissues or Anip973 cells were extracted and separated by SDS-PAGE and transferred to nitrocellulose membranes (Millipore). These blots were incubated for 1 h at room temperature in TBS-Tween 20 containing 5% skim milk. Primary antibodies used were anti-IDH1 polyclonal antibody (diluted 1:1000; Abcam), anti-SOD2 monoclonal antibody (diluted 1:1000; Epitomics), anti-14-3-3ε polyclonal antibody (diluted 1:2500; Abcam), anti-receptor of activated protein kinase C 1 (RACK1) monoclonal antibody (diluted 1:2000; BD Biosciences), anti-peroxiredoxin 2 (PRDX2) monoclonal antibody (diluted 1:1000; Epitomics), and anti-β-actin monoclonal antibody (diluted 1:5000; ProteinTech). The blots were labeled with peroxidase-conjugated secondary antibody to rabbit or mouse IgG (KPL) and visualized by ECL reagents (Millipore).
Immunohistochemistry Staining
Tissue microarray containing 137 paired NSCLC tumors and corresponding adjacent normal lung tissues was using to examine IDH1 expression. Routine immunohistochemical SP staining was carried out according to the manufacturer's protocol with anti-IDH1 antibody (diluted 1:100; Abcam). The microscopic analysis of the slides was independently performed by two experienced pathologists in a blind fashion without knowledge of the information of patients. The intensity of cell immunostaining was scored as 0, 1, 2, and 3, and the percentage of positive cells was assigned as following categories: 0 (0–5%), 1 (6–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%). The final scores of the sections were recorded as − (negative), + (weakly positive), ++ (positive), and +++ (strongly positive); the scores were based on the intensity and the number of cells staining.
Plasma Biomarker Detection
Plasma samples were prepared by collecting blood in EDTA tubes. The samples were centrifuged at 3,000 rpm for 20 min immediately following collection, and the supernatants were removed and stored in aliquots at −80 °C until use. Plasma levels of IDH1 were measured by ELISA using a commercially available kit (Uscn Life Science) according to the manufacturer's recommendations. Plasma levels of CYFRA21-1, CEA, and CA125 were measured by electrochemiluminescence immunoassay (Roche Applied Science). Plasma levels of SCC-antigen were measured by chemiluminescent assays (CMIA; Abbott Laboratories).
Cell Culture
The six human NSCLC cell lines included Anip973 (lung AD), A549 (lung carcinoma), H2170 (lung SCC), NCI-H322 (bronchioalveolar carcinoma), H460 (lung large cell carcinoma), and H520 (lung SCC). All of the NSCLC cells and HBE cells (HPV transformed normal bronchial epithelium cell line) were grown in monolayer in appropriate medium supplemented with 10% FBS and maintained at 37 °C in humidified air with 5% CO2.
Plasmid Constructions and Cell Transfection
Knockdown of the IDH1 (GenBankTM accession number NM_005896.2) in Anip973 cells was performed by stable transfection of a shRNA-expressing vector. The pGPU6/GFP/Neo-sh-IDH1-1 (sh-IDH1-1, target sequence: 5′-TAACTTTGAAGAAGGTGGTGG-3′) and pGPU6/GFP/Neo-sh-IDH1-2 (sh-IDH1-2, target sequence: 5′-GGTATGAGCATAGGCTCATCG-3′) vectors were constructed and identified by GenePharma Co. (Shanghai, China). The pGPU6/GFP/Neo-sh-NC (sh-NC), which targets a sequence not found in the human, mouse, or rat genome databases (5′-GTTCTCCGAACGTGTCACGT-3′), was used as negative control.
The sh-IDH1-1, sh-IDH1-2 and sh-NC were transfected into Anip973 cells, respectively, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. G418 (Amresco Inc.) at a concentration of 0.8 mg/ml was used to select transfected cells. After selection for 2 weeks, the pools were analyzed by real time PCR and Western blot to detect IDH1 expression level.
Real time Quantitative PCR
To determine the expression of IDH1 gene in Anip973 cells transfected with IDH1 RNA interference vector or control vector, total RNA was isolated with the TRIzol method (Invitrogen). The first strand cDNA was synthesized from 1 μg of total RNA using reagents from Fermentas. Real time PCR (Applied Biosystems 7300 real time PCR system) was performed using custom SYBR assays (Takara) following the manufacturer's instructions. Human 18 S rRNA gene was used as an internal control. The same primers were used for amplifying IDH1 and 18 S RNA in real time PCR as used in RT-PCR described above. All PCRs, including no-template controls, were performed in triplicate.
CCK-8 Assay
In vitro proliferation of the transfected cells was measured using CCK-8 assay. Anip973 cells stably transfected with sh-IDH1-1, sh-IDH1-2, or sh-NC vector were plated at a density of 2,000 cells/well onto 96-well plates. For 7 days, cell viability was measured using CCK-8 (Dojindo Laboratories, Japan) and quantified by SoftMax® Pro 5 program (Molecular Devices) every day.
In Vivo Tumorigenesis
Anip973 cells stably transfected with sh-IDH1-1 or sh-NC vector were suspended in PBS and then injected subcutaneously in the right flank of athymic nude mice (2 × 106 cells/mouse, eight mice/group). Tumor growth curve was plotted by means of tumor volumes monitored at indicated times. Tumor volume was calculated according to the following formula: length × (width)2/2. The mice were killed at day 28, and the xenograft tumors were dissected and weighed.
Statistical Analysis
The differences between tumors and paired normal lung tissues were evaluated by Wilcoxon matched pairs test, whereas the differences between plasma sample groups were evaluated by the Mann-Whitney U test using GraphPad Prism version 5 for Windows. Survival curves were generated according to the Kaplan-Meier method using SPSS 13.0 software, and the statistical analysis was performed by log rank test. Multivariate analysis was evaluated by Cox proportional hazard models. Statistical significance was defined as p ≤ 0.05.
RESULTS
Differentially Expressed Proteins in Lung SCC Tumors and Paired Normal Lung Tissues
To screen differentially expressed proteins, tumors and their adjacent normal tissues from lung SCC patients were analyzed by 2D-DIGE. Twelve paired samples labeled with Cy3 or Cy5 were run in 12 gels along with a pooled standard labeled with Cy2, and the images were analyzed by Decyder 6.5 software. Among 2173 matched protein spots, 72 were up-regulated (Ratiotumor/normal ≥ 2, p ≤ 0.05; paired t test with false discovery rate correction), and 112 were down-regulated (Ratiotumor/normal ≥ −2, p ≤ 0.05) in tumors versus normal tissues.
Identification of Differentially Expressed Proteins
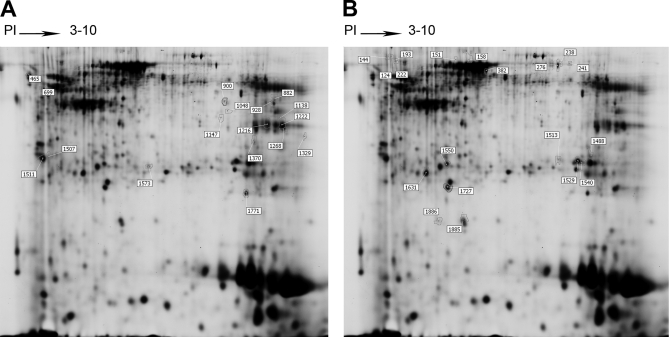
53 differentially expressed protein spots showing sufficient intensity, clear separation from surrounding spots, and consistent spot shape and size were excised and subsequently in-gel digested by trypsin and then successfully analyzed by MALDI-TOF/TOF. The nine spots identified as known contaminants (keratin 1, 2, and 10) and the eight spots with multiple proteins identification were excluded from further analysis. Among those remaining 36 spots (17 up-regulated spots and 19 down-regulated spots shown in Fig. 1, A and B, respectively), 17 up-regulated and 11 down-regulated non-redundant proteins in tumor samples were identified. The details of these proteins are listed in supplemental Tables 2a and 2b.
Fig. 1.
Representative two-dimensional gel image of the Cy2-labeled proteins that comprise the internal standard. The 36 identified protein spots are marked with master numbers and displayed. A, identified protein spots up-regulated in tumors. B, identified protein spots down-regulated in tumors. The information for the proteins corresponding to the spot numbers is listed in supplemental Table 2a and 2b.
Validating Expression of Certain Candidate Proteins by RT-PCR and Western Blot
Subsequently, we chose IDH1, SOD2, 14-3-3ε, RACK1, and PRDX2 to be further validated by RT-PCR and Western blot, which were reported to be involved in some important pathways in tumor development and progression. Among them, IDH1, SOD2, 14-3-3ε, and RACK1 were up-regulated in tumors compared with corresponding normal lung tissues with an average ratio of 3.08, 2.16, 2.53, and 2.34, respectively, by 2D-DIGE analysis. PRDX2 was identified as a tumor down-regulated protein with an average ratio of −2.11 (supplemental Fig. 1A). The mass spectra of these five differentially expressed proteins were shown in supplemental Fig. 1B.
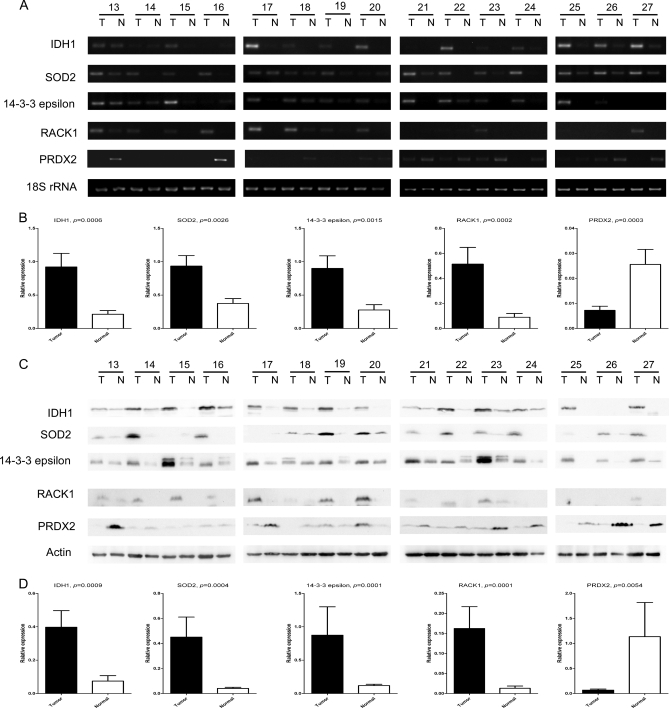
RT-PCR and Western blot were performed in independent 15 pairs of samples. RT-PCR showed that mRNA levels of IDH1, SOD2, 14-3-3ε, and RACK1 were significantly higher in tumors than those in paired normal lung tissues, whereas that of PRDX2 was lower in tumors (IDH1, 4.3-fold, p = 0.0006; SOD2, 2.5-fold, p = 0.0026; 14-3-3ε, 3.3-fold, p = 0.0015; RACK1, 5.7-fold, p = 0.0002; PRDX2, −3.5-fold, p = 0.0003; Wilcoxon matched pairs test) (Fig. 2, A and B). Western blot showed that the expression pattern of these five genes in protein level was identical with that in mRNA level. The abundance of IDH1, SOD2, 14-3-3ε, and RACK1 protein increased markedly in most of 15 tumors compared with their paired normal tissues, and PRDX2 proteins decreased in most tumors (IDH1, 5.3-fold, p = 0.0009; SOD2, 11.1-fold, p = 0.0004; 14-3-3ε, 7.4-fold, p = 0,0001; RACK1, 12.5-fold, p = 0.0001; PRDX2, −16.7-fold, p = 0.0054; Wilcoxon matched pairs test) (Fig. 2, C and D). These results were consistent with the observations in 2D-DIGE. The variation of the expression of these proteins may have been due to the heterogeneity of the samples.
Fig. 2.
Validation of differential expression of IDH1, SOD2, 14-3-3ε, RACK1, and PRDX2 in paired lung SCC tissues by semi-quantitative RT-PCR and Western blot. Semi-quantitative RT-PCR (A) and Western blot (C) were performed in 15 independent pairs of lung SCC tumors and corresponding normal tissues. 18 S RNA and β-actin were used as internal controls. The agarose gel images (B) and Western blot images (D) were quantified by densitometric scanning, and a Wilcoxon matched pairs test was used after the intensity values were normalized against those of controls.
Up-regulation of IDH1 in Tumors by Immunohistochemistry
Redox reaction plays an important role in carcinogenesis and cancer therapy (19, 20). Three proteins (IDH1, SOD2, and PRDX2) involved in cellular redox regulation were identified and validated in this study. To the best of our knowledge, this is the first evidence that IDH1 is up-regulated in tumors over paired normal tissues, although IDH1 was widely studied because of its cancer-related mutation in Arg-132 recently. Immunohistochemistry staining in clinical samples from extended NSCLC patients were performed to further validate the elevated IDH1 in tumors and evaluate its relationship with the clinicopathological factors of NSCLC.
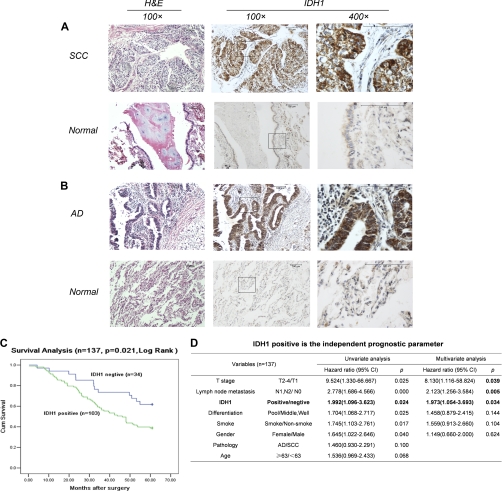
The expression of IDH1 was examined in formalin-fixed and paraffin-embedded tumor tissues and corresponding normal tissues from extended 73 SCC and 64 AD patients by immunohistochemistry using antibody against human IDH1. As shown in Fig. 3 (A and B), the expression of IDH1 protein was located in the cytoplasm of tumor cells of SCC and AD. For SCC, the expression of IDH1 was significantly higher in tumor tissues with 72.6% positive staining, compared with paired normal lung tissues with only 8.2% positive staining (chi-squared test, p < 0.001) (Fig. 3A and Table II). Similarly to SCC, positive expression of IDH1 was detected in 78.1% of AD tumors and only 7.9% of corresponding normal lung tissues (chi-squared test, p < 0.001) (Fig. 3B and Table II). No significant difference of IDH1 expression was found between tumors of SCC and AD (chi-squared test, p = 0.849). The IDH1 expression was significantly higher in this panel of 137 NSCLC tumors with 75.2% positive staining, compared with corresponding normal lung tissues, with only 8.1% positive staining (chi-squared test, p < 0.001) (Table II).
Fig. 3.
Immunohistochemical analysis of IDH1 expression in NSCLC tumors and paired normal lung tissues. A and B, staining against IDH1 showed an obvious increase of IDH1 expression in tumors of both squamous cell carcinoma (A) and adenocarcinoma (B) compared with corresponding normal lung tissues. IDH1 proteins were strongly stained in the cytoplasm of tumor cells but weakly or negatively expressed in normal respiratory bronchiolar cells and alveolar cells. C, Kaplan-Meier curves showed that IDH1 expression was related to decreased overall survival. A log rank test was used. D, multivariate analysis using the Cox proportional hazard model indicated that IDH1-positive staining as well as T stage and lymph node metastasis are independent prognostic factors.
Table II. Immunohistochemistry results for IDH1 in lung cancer patients.
The scores of the tissue spots based on the intensity and the number of cells staining were recorded as: −, negative; +, weakly positive; ++, positive; and +++, strongly positive. The proportions and numbers of samples with different immunohistochemistry staining results in tumors, and normal tissues are listed in the table.
| Total cases | − | + | ++ | +++ | p valuea | |
|---|---|---|---|---|---|---|
| SCC | ||||||
| Tumor | 73 | 27.4% (20/73) | 27.4% (20/73) | 24.7% (18/73) | 20.5% (15/73) | <0.001 |
| Normal | 73 | 91.8% (67/73) | 6.8% (5/73) | 1.4%(1/73) | 0 | |
| AD | ||||||
| Tumor | 64 | 21.9% (14/64) | 29.7% (19/64) | 23.4%(15/64) | 25.0% (16/64) | <0.001 |
| Normal | 64 | 92.2% (59/64) | 6.3% (4/64) | 1.6% (1/64) | 0 | |
| SCC + AD | ||||||
| Tumor | 137 | 24.8% (34/137) | 28.5% (39/137) | 24.1% (33/137) | 22.6% (31/137) | <0.001 |
| Normal | 137 | 92.0% (126/137) | 6.6% (9/137) | 1.5% (2/137) | 0 |
a The p values of chi-squared test.
IDH1 Is an Unfavorable Prognostic Factor for NSCLC
The correlation between expression of IDH1 and clinicopathological variables was evaluated by chi-squared test. No significant correlation was observed between IDH1 expression and gender, age, smoking habit, family history, TNM stage, lymph node metastasis, tumor size (T stage), pathology, and differentiation (Table III). However, the overall 5-year survival rate is significantly higher in the IDH1-negative group than in the IDH1-positive group (61.8% versus 38.8%, p = 0.020). Kaplan-Meier survival analysis was further performed in 137 NSCLC patients. The survival curves showed that the patients with positive expression of IDH1 had a shorter survival than the patients with negative expression (p = 0.021, log rank test) (Fig. 3C). To determine whether the prognostic value of IDH1 expression was independent of other risk factors associated with the clinical outcome of NSCLC, multivariate analysis was performed using the Cox proportional hazard model. Univariate Cox analysis showed that T stage, lymph node metastasis, IDH1 expression, differentiation, smoking habit, and gender were significantly associated with survival of NSCLC patients. Subsequently, a multivariate Cox proportional hazard regression analysis using all of these variables indicated that IDH1 expression was an independently unfavorable prognostic factor (p = 0.034; risk ratio 1.973; 95% confidence interval, 1.054–3.693) for NSCLC patients along with T stage (p = 0.039; risk ratio 8.130; 95% confidence interval, 1.116–58.824) and lymph node metastasis (p = 0.005; risk ratio 2.123; 95% confidence interval, 1.256–3.584) (Fig. 3D). These results suggested that IDH1 could be used as a histochemical biomarker for prognosis of NSCLC patients.
Table III. Correlation between IDH1 expression and clinicopathological factors of NSCLC patients.
| IDH1 positive (number of cases) | IDH1 negative (number of cases) | p valuea | |
|---|---|---|---|
| Gender | 0.203 | ||
| Male | 70 | 27 | |
| Female | 33 | 7 | |
| Age | 0.801 | ||
| <63 | 48 | 15 | |
| ≥63 | 55 | 19 | |
| Smoking | 0.312 | ||
| Yes | 66 | 25 | |
| No | 37 | 9 | |
| Family history | 0.982 | ||
| Yes | 18 | 6 | |
| No | 85 | 28 | |
| TNM stage | 0.233 | ||
| I | 39 | 19 | |
| II | 29 | 9 | |
| III | 30 | 5 | |
| IV | 5 | 1 | |
| Lymph node metastasis | 0.107 | ||
| N0 | 40 | 20 | |
| N1 | 29 | 9 | |
| N2 | 32 | 4 | |
| N3 | 2 | 1 | |
| T stage | 0.417 | ||
| T1 | 10 | 1 | |
| T2 | 85 | 32 | |
| T3 | 7 | 1 | |
| T4 | 1 | 0 | |
| Pathology | 0.455 | ||
| SCC | 53 | 20 | |
| AD | 50 | 14 | |
| Differentiation | 0.125 | ||
| Well | 5 | 2 | |
| Middle | 71 | 17 | |
| Poor | 27 | 15 | |
| 5-year survival | 0.020 | ||
| Yes | 40 | 21 | |
| No | 63 | 13 |
a The p values of chi-squared test.
IDH1 Is a Potential Plasma Biomarker for Diagnosis of NSCLC
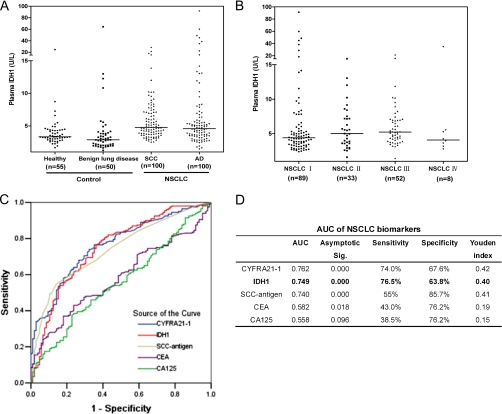
We further investigated the plasma level of IDH1 and its potential value as a plasma biomarker for NSCLC. The plasma levels of IDH1 in 200 NSCLC patients (100 SCCs and 100 ADs), 55 healthy individuals, and 50 benign lung disease patients were assessed by ELSIA. Plasma levels of IDH1 were significantly elevated in the 200 NSCLC patients (median = 4.69 units/liter) in comparison with 55 healthy individuals (median = 3.34 units/liter; p < 0.0001, Mann-Whitney test) or 50 benign lung disease patients (median = 2.87 units/liter; p < 0.0001, Mann-Whitney test) (Fig. 4A). According to histological types of NSCLC, the median plasma levels of IDH1 were 4.77 units/liter in 100 SCC patients and 4.58 units/liter in 100 AD patients; no significant difference between the two histological types was found (p = 0.2168, Mann-Whitney test). High levels of plasma IDH1 were detected even in patients with earlier stage NSCLC, and no significant differences were observed among each stage (p = 0.1869, Kruskal-Wallis test) (Fig. 4B).
Fig. 4.
Plasma level of IDH1 and four conventional tumor biomarkers (CYFRA21-1, SCC-antigen, CEA, and CA125) in NSCLC patients, healthy individuals, and benign lung disease patients. A, distribution of IDH1 plasma level determined by ELISA in SCC patients, AD patients, healthy individuals, and benign lung disease patients. Median values are shown with a horizontal line. Differences were significant between NSCLC patients and healthy individuals/benign lung disease patients (p < 0.0001, respectively, Mann-Whitney test), between SCC patients and healthy individuals/benign lung disease patients (p < 0.0001, respectively), and between AD patients and healthy individuals/benign lung disease patients (p = 0.0002/p < 0.0001). No significant difference between SCC patients and AD patients was observed (p = 0.217). B, distribution of IDH1 in plasmas of patients with various TNM stages of NSCLC. No significant differences were observed among each stage (p = 0.187, Kruskal-Wallis test). C, ROC curves of IDH1, CYFRA21-1, SCC-antigen, CEA, and CA125 in discriminating NSCLC patients from controls (healthy individuals and benign lung disease patients). x axis, 1-specificity; y axis, sensitivity. D, AUC of each biomarker was listed along with the sensitivity and specificity of each biomarker at their optimal cutoff value, which was determined by maximizing the Youden index.
The receiver operating characteristic (ROC) curve of the plasma IDH1 levels of 200 NSCLC patients and 105 controls (55 healthy individuals and 50 benign lung disease patients) was shown in Fig. 4C. The area under the ROC curve (AUC) of IDH1 was 0.749, significantly higher than that of the null hypothesis (true area was 0.5; p = 0.000) (Fig. 4D). To discriminate NSCLC patients from healthy individuals and benign lung disease patients, the optimal cutoff value of IDH1 level in plasma was 3.54 units/liter in this study, a level at which the Youden index is maximized (0.40) with 76.5% sensitivity and 63.8% specificity. In addition to IDH1, four conventional NSCLC tumor markers (CYFRA21-1, SCC-antigen, CEA, and CA125) were also measured, in the same set of plasma samples from cancer patients and control individuals. ROC analysis determined the optimal cutoff values to be 2.03 ng/ml for CYFRA21-1 (sensitivity, 74.0%; specificity, 67.6%; Youden index, 0.42), 0.95 ng/ml for SCC-antigen (sensitivity, 55.0%; specificity, 85.7%; Youden index, 0.41), 2.68 ng/ml for CEA (sensitivity, 43.0%; specificity, 76.2%; Youden index, 0.19), and 14.7 units/ml for CA125 (sensitivity, 38.5%; specificity, 76.2%; Youden index, 0.15). The AUC of IDH1 (0.749) was similar to CYFRA21-1 (0.762) and SCC-antigen (0.740) and larger than CEA (0.582) and CA125 (0.558) (Fig. 4, C and D), suggesting that IDH1 has comparable diagnostic efficacy with CYFRA21-1 and SCC-antigen for NSCLC.
IDH1 Regulates NSCLC Cell Proliferation in Vitro and in Vivo
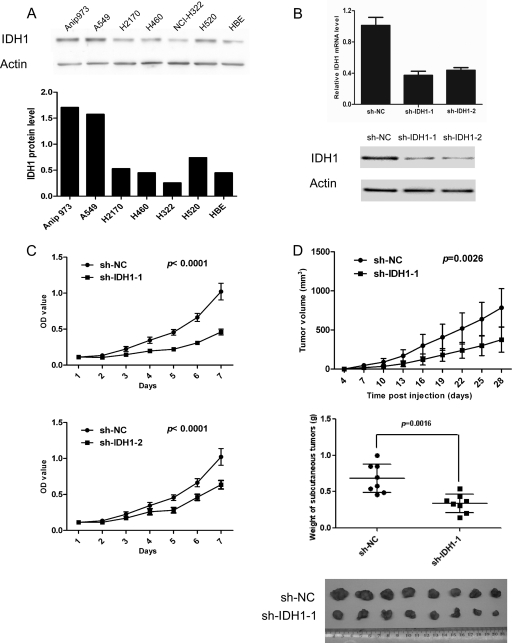
Expression of IDH1 in six NSCLC cell lines (Anip973, A549, H2170, H460, NCI-H322, and H520) was detected by Western blot, with a HPV-transformed normal bronchial epithelium cell line (HBE) as control cell line. Higher IDH1 expression was observed in Anip973, A549, and H520 than in the HBE cell line (Fig. 5A). To further investigate the effect of IDH1 on NSCLC tumor growth, Anip973 was stably transfected with shRNA-expressing vector to knock down the IDH1 expression. Real time PCR and Western blot showed that IDH1 expression was markedly decreased in cells transfected with sh-IDH1-1 or sh-IDH1-2 vector compared with cells transfected with sh-NC vector (Fig. 5B). To investigate the effect of IDH1 knockdown on cell proliferation, growth of cells was assessed by CCK-8 assay. As shown in Fig. 5C, knockdown of IDH1 obviously decreased proliferation of Anip973 cells cultured in vitro. Furthermore, we evaluated the effect of IDH1 on tumor growth in vivo using a xenograft model. Anip973 cells stably transfected with sh-IDH1-1 or sh-NC vector were implanted in nude mice subcutaneously, and tumor size was measured every 3 days. The mice were killed at day 28, and the tumors were dissected and weighed. As shown in Fig. 5D, the cells with IDH1 knockdown resulted in slower growing xenografts as compared with cells with control vector. In 28 days, tumors from IDH1 knockdown cells were on average 50% smaller in weight than tumors from control cells (0.684 g versus 0.339 g, respectively; p = 0.0016, t test) (Fig. 5D). The results indicated that the down-regulation of IDH1 expression decreased the growth of NSCLC tumor cells.
Fig. 5.
Tumor cell proliferation assessed by CCK-8 assays and tumor xenografts. A, expression of IDH1 in six NSCLC cell lines and a normal bronchial epithelium cell line were detected by Western blot (upper panel). β-Actin was used as an internal control. The intensity values of IDH1 expression in various cell lines were normalized against those of controls and shown (lower panel). B, Anip973 cells were stably transfected with two vectors expressing different shRNAs silencing IDH1 (sh-IDH1-1 and sh-IDH1-2) or negative control vector (sh-NC). Knockdown efficiencies of two independent shRNAs were assessed by real time PCR (upper panel) and Western blot (lower panel). C, proliferation of the cells stably transfected with sh-IDH1-1, sh-IDH1-2, and sh-NC were analyzed by CCK-8 assay. The data are expressed as the means ± S.D. from eight replicates in each group. D, cells stably transfected with sh-IDH1-1 or sh-NC were injected subcutaneously in the right flank of athymic nude mice (2 × 106 cells/mouse, eight mice/group). Tumor volume was monitored at indicated times, and tumor growth curves were plotted (top panel). The data are expressed as the means ± S.D. from eight mice in each group. The mice were killed at day 28, and the xenograft tumors were weighed (middle panel) and shown (bottom panel). The data shown are representative of three independent experiments.
DISCUSSION
To identify potential biomarkers of lung cancer using the 2D-DIGE technique coupled with MALDI-TOF/TOF, we analyzed the differential proteome of paired tumors and normal lung tissues from 12 lung SCC patients. A total of 28 differentially expressed proteins were identified in this study. Most of them were involved in metabolism, cytoskeleton, redox regulation, binding and transport, transcription, stress response, and other functions. These differentially expressed proteins are very likely involved in the carcinogenesis of lung cancer and provide hypotheses for mechanism-based studies. Some of them may serve as biomarkers to guide lung cancer diagnosis and therapy. In our study, vimentin, tropomyosin 3, fructose-bisphosphate aldolase A, phosphoglycerate kinase 1, and heat shock protein β-1, were related to the lung cancer, which has been reported in previous proteomic researches (5, 9, 12, 17). For proteomic study, further validations by independent methods were necessary for candidate proteins identified by MS analysis. Therefore, the expression of four up-regulated proteins (IDH1, SOD2, 14-3-3ε, and RACK1) and one down-regulated protein (PRDX2) was validated by RT-PCR and Western blot in an independent patient cohort in our study. Furthermore, IDH1 expression was screened by immunochemistry staining in 137 paired NSCLC tissues and ELISA assay in plasmas from 200 NSCLC patients, 55 healthy individuals, and 50 benign lung disease patients. These results were consistent with 2D-DIGE data, indicating that our proteomic analysis in lung cancer was reliable.
14-3-3 protein is a family of highly conserved regulatory molecules, containing seven isoforms named beta (β), gamma (γ), epsilon (ε), eta (η), tau (τ), sigma (σ), and zeta (ζ) in mammalian cell. Through binding and regulating a wide range of binding partners, 14-3-3 proteins are involved in many cancer-related processes, including cell cycle regulation, metabolism control, apoptosis, and transcription regulation (21). Previous studies revealed that some of 14-3-3 isoforms were related to lung cancer. Isoform sigma is decreased in lung cancer because of DNA hypermethylation, whereas isoform zeta was increased in lung cancer and implicated as a prognostic and therapeutic target (22, 23). 14-3-3ε was also found to be up-regulated in lung cancer in our study, although its significance and molecular mechanism were unknown as yet. More investigations about involvement of all seven 14-3-3 isoforms in lung cancer are needed to discover the relationship between this family and lung tumorigenesis.
RACK1, a homologue of the β-subunit of heterotrimeric G proteins, was shown to be increased in lung SCC tumors in our study. It is regarded as an anchoring protein and activator for several kinases, including PKC (24), PDE4D5 (25), Src (26), and so on. By interacting with and regulating different kinase, it is involved in distinct cancer-related signaling pathways and may play roles in carcinogenesis. Up-regulation of RACK1 was also found in tumors of oral squamous cell cancer (27, 28), melanoma (29), and breast cancer (30) and is a poor prognostic factor for oral squamous cell cancer and breast cancer. Recently, RACK1 showed significantly high and frequent expression in lung AD (77 of 123 samples) but bare expression in lung SCC with only a few positive (3 of 44 samples) in an immunohistochemistry study (31). In our study, more frequent expression of RACK1 protein in lung SCC (7 of 12 samples by 2D-DIGE and 10 of 15 samples by Western blot) was detected. The inconsistent results may have been due to methodological differences and the limited number of samples. Further study containing a larger number of samples will be required to clarify the expression pattern of RACK1 in lung SCC and evaluate its potential as a biomarker.
Redox regulation plays an essential role in multiple cellular pathways related to carcinogenesis (e.g. proliferation, inflammatory responses, apoptosis, and senescence) (32). Elevated reactive oxygen species (ROS) levels were observed in many cultured carcinoma cell lines and solid tumors. To survive increased ROS stress, cancer cell would acquire adaptive mechanisms to counteract the toxic effects of elevated ROS and maintain redox homeostasis. The enhanced antioxidant capability seemed to be a major adaptive mechanism (19, 20). Three proteins related to ROS scavenging were identified and validated in our study. IDH1 and SOD2 were up-regulated, whereas PRDX2 was down-regulated in lung SCC tumors. SOD2 in carcinogenesis has been widely studied but remains ambiguous. As one of the major antioxidant enzymes, it was proved to inhibit growth of tumor cells in cultured cancer cells and experimental animal models (33–36), but was up-regulated in various solid tumors, including lung cancer (37–42). Consistent with previous studies that found the increased SOD2 in lung tumors and in the sera of lung SCC patients (41, 42), SOD2 was observed to be up-regulated in lung SCC tumors compared with corresponding normal lung tissues in our study. IDH1, which catalyzes oxidative decarboxylation of isocitrate into α-ketoglutarate, is one of three enzymes responsible for production of NADPH in cytoplasm (43–45). NADPH is an essential cofactor for regeneration of GSH by glutathione reductase in addition to playing critical roles in maintenance of thioredoxin in its reduced state (46). Both GSH and reduced thioredoxin are important antioxidants for mammalian cells to resist oxidative damage. Therefore, IDH1 plays a role in antioxidant system by producing NADPH. Up-regulation of SOD2 and IDH1 in lung tumors in this study may be an adaptive alteration for tumor cells to antagonize and survive increased oxygen stress. PRDX2, another protein participating in cellular antioxidant defense, was down-regulated in lung SCC tumors in our study. Human PRDXs comprise a family of six enzymes (PRDX 1–6). Previous study showed that PRDXs 1, 2, 3, and 5 predominantly expressed in normal lung tissue, and the isoforms 1 and 3 were overexpressed in lung cancer tissues compared with normal lungs (47). There was no difference in expression of PRDX2 between lung tumors and paired normal lung tissues observed in the study of Park et al. (47). Their report and our findings suggested that PRDX2 was at least not increased in cancerous lung tissues. However, redox alterations in cancer cells are very complex because many factors are involved in the redox regulation and stress response. Our findings in differential expression of these three antioxidation-related proteins provide some clues to further research for understanding the complicated redox regulation of lung cancer.
In the present study, IDH1 was further characterized by immunochemistry staining and ELISA assay in an extended series of patients. Immunohistochemistry staining showed that IDH1 was up-regulated in tumors of more than 70% of NSCLC, and increased IDH1 was correlated to a lower 5-year survival rate. Furthermore, IDH1 was proved to be an independent unfavorable prognostic factor for overall survival of NSCLC patients by multivariate analysis. It is indicated that the IDH1 expression in tumor tissues may be able to act as a histochemical biomarker for prognosis prediction of NSCLC, and the patients with IDH1-positive tumors may need more active follow-up and more postoperative adjuvant therapy. However, the specificity for IDH1 alone is not high enough to predict 5-year survival of NSCLC patients (82% sensitivity and 34% specificity). A combination of IDH1 expression and other prognosis related factors, such as tumor size, lymph node metastasis, differentiation, smoking habit, and gender, will probably make a better prediction for NSCLC prognosis. A study employing a larger sample size is needed to develop a more reliable “prognosis predictor,” which will be calculated by mathematical model using these prognosis-related variables. ELISA analysis revealed that plasma levels of IDH1 in NSCLC patients were significantly higher than those in healthy individuals and benign lung disease patients. The ROC curve is a useful method for evaluating clinical usefulness of a biomarker and for comparing the effectiveness between different biomarkers. A larger AUC generally represents more reliability and better discrimination. Based on our preliminary analysis, the AUC of IDH1 reached 0.749, indicating a comparable diagnostic performance with conventional tumor biomarkers for NSCLC (CYFRA21-1 or SCC-antigen) that are being used in clinical practice. It was worth noting that IDH1 was elevated in an earlier stage of the disease, which might be useful in early detection of NSCLC. However, further validations using a larger set of blood samples from lung cancer patients in various clinical stages and from other cancers are required before clinical application of IDH1 as a plasma biomarker for NSCLC diagnosis.
Up-regulated expression of IDH1 in tumor cells suggests its potential role in tumorigenesis for lung cancer. Recently, in a study performed by Ward et al. (48), knockdown of IDH1 could significantly reduce the proliferative capacity of cultured glioma cancer cell, indicating that IDH1 plays a critical role in survival and proliferation of cancer cell. In addition, overexpression of IDH1 down-regulated the expression of tumor suppressors, p53 and p21, in mammalian cells (49). In the present study, we observed the effects of IDH1 suppression in cultured tumor cells and tumor xenografts. It was noteworthy that silencing of IDH1 by RNA interference resulted in an obviously decreased growth for tumor cells both in vitro and in vivo. These studies suggest that IDH1 may be a key player in the growth of certain cancers.
There is increasing interest in IDH1 recently in cancer biology because of its cancer-related mutation in enzymatic active site, Arg-132 (50). The mutation at Arg-132 of IDH1 results not only in the loss of physiological function to produce α-ketoglutarate and NADPH (51, 52) but also in the gain of new function to produce an oncometabolite, 2-hydroxyglutarate (53). This tumorigenic mutation was observed in more than 70% of WHO grade II and III gliomas and secondary glioblastomas (51), in 8.5% of acute myeloid leukemias (54), and in rare prostate carcinomas and B-acute lymphoblastic leukemia (55). In previous reports from other investigators, a total of 321 lung cancer samples (107 samples by Bleeker, 35 samples by Yan, and 179 samples by Kang) had been examined, but no IDH1 Arg-132 mutation was found in lung cancer (50, 51, 55). We sequenced IDH1 exon 4 in the 12 tumor samples that underwent proteomic analysis in this study, and no Arg-132 mutation was identified (supplemental Fig. 2). Therefore, it may be up-regulated expression and not mutation in Arg-132 that plays a role in IDH1-related tumorigenesis of NSCLC.
In conclusion, we provide global profiling of proteomic alteration of lung SCCs. IDH1, one of the up-regulated proteins in tumors, was further proved as a plasma biomarker for diagnosis and a histochemical biomarker for prognosis prediction of NSCLC. Silencing of IDH1 by RNA interference suppressed the tumor cell growth both in vitro and in vivo, indicating its role in growth of NSCLC tumor. However, more studies are required to determine the applicability of IDH1 in clinical practice.
Footnotes
* The work was supported by Chinese National Natural Science Foundation Grant 81101772; International Science and Technology Corporation and Exchange Project Grant 2010DFB30650 from the Chinese Ministry of Science and Technology; Key Technologies R & D Program Grant 2006BAI02A02; Chinese State Key Projects for Basic Research Grants 2011CB910601, 2011CB910700, 2010CB912700, and 2011CB505304; Chinese State Key Project Specialized for Infectious Diseases Grants 2008ZX10002-016 and 2009ZX10004-103; the National Key Technologies R & D Program for New Drugs Grant 2009ZX09301-002; International Scientific Collaboration Program Grants 2009DFB33070 and 2010DFA31260; and State Key Laboratory of Proteomics Grants SKLP-Y200901 and SKLP-O200901. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental Tables 1–3 and Figs. 1 and 2.
This article contains supplemental Tables 1–3 and Figs. 1 and 2.
1 The abbreviations used are:
- NSCLC
- non-small cell lung cancer
- 2D-DIGE
- two-dimensional fluorescence difference gel electrophoresis
- SCC
- squamous cell carcinoma
- AD
- adenocarcinoma
- IDH1
- isocitrate dehydrogenase 1
- SOD2
- superoxide dismutase 2
- RACK1
- receptor of activated protein kinase C 1
- PRDX2
- peroxiredoxin 2
- ROC
- receiver operation characteristics
- AUC
- area under the curve
- CYFRA21-1
- cytokeratin 19 fragment 21-1
- CEA
- carcinoembryonic antigen
- CA125
- cancer antigen 125
- ROS
- reactive oxygen species.
REFERENCES
- 1. Jemal A., Siegel R., Xu J., Ward E. (2010) Cancer statistics. CA-Cancer J. Clin. 60, 277–300 [DOI] [PubMed] [Google Scholar]
- 2. Hoffman P. C., Mauer A. M., Vokes E. E. (2000) Lung cancer. Lancet. 355, 479–485 [DOI] [PubMed] [Google Scholar]
- 3. Simpson R. J., Bernhard O. K., Greening D. W., Moritz R. L. (2008) Proteomics-driven cancer biomarker discovery: Looking to the future. Curr. Opin. Chem. Biol. 12, 72–77 [DOI] [PubMed] [Google Scholar]
- 4. Cho W. C., Cheng C. H. (2007) Oncoproteomics: Current trends and future perspectives. Exp. Rev. Proteomics 4, 401–410 [DOI] [PubMed] [Google Scholar]
- 5. Li C., Xiao Z., Chen Z., Zhang X., Li J., Wu X., Li X., Yi H., Li M., Zhu G., Liang S. (2006) Proteome analysis of human lung squamous carcinoma. Proteomics 6, 547–558 [DOI] [PubMed] [Google Scholar]
- 6. Huang L. J., Chen S. X., Luo W. J., Jiang H. H., Zhang P. F., Yi H. (2006) Proteomic analysis of secreted proteins of non-small cell lung cancer. Chin. J. Cancer 25, 1361–1367 [PubMed] [Google Scholar]
- 7. Maciel C. M., Junqueira M., Paschoal M. E., Kawamura M. T., Duarte R. L., Carvalho Mda G., Domont G. B. (2005) Differential proteomic serum pattern of low molecular weight proteins expressed by adenocarcinoma lung cancer patients. J. Exp. Ther. Oncol. 5, 31–38 [PubMed] [Google Scholar]
- 8. Li C., Chen Z., Xiao Z., Wu X., Zhan X., Zhang X., Li M., Li J., Feng X., Liang S., Chen P., Xie J. Y. (2003) Comparative proteomics analysis of human lung squamous carcinoma. Biochem. Biophys. Res. Commun. 309, 253–260 [DOI] [PubMed] [Google Scholar]
- 9. Deng B., Ye N., Luo G., Chen X., Wang Y. (2005) Proteomics analysis of stage-specific proteins expressed in human squamous cell lung carcinoma tissues. Cancer Biomark. 1, 279–286 [DOI] [PubMed] [Google Scholar]
- 10. Li C., Zhan X., Li M., Wu X., Li F., Li J., Xiao Z., Chen Z., Feng X., Chen P., Xie J., Liang S. (2003) Proteomic comparison of two-dimensional gel electrophoresis profiles from human lung squamous carcinoma and normal bronchial epithelial tissues. Genomics Proteomics Bioinformatics 1, 58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu X., Xiao Z., Chen Z., Li C., Li J., Feng X., Yi H., Liang S., Chen P. (2004) Differential analysis of two-dimension gel electrophoresis profiles from the normal-metaplasia-dysplasia-carcinoma tissue of human bronchial epithelium. Pathol. Int. 54, 765–773 [DOI] [PubMed] [Google Scholar]
- 12. Chen G., Gharib T. G., Wang H., Huang C. C., Kuick R., Thomas D. G., Shedden K. A., Misek D. E., Taylor J. M., Giordano T. J., Kardia S. L., Iannettoni M. D., Yee J., Hogg P. J., Orringer M. B., Hanash S. M., Beer D. G. (2003) Protein profiles associated with survival in lung adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 100, 13537–13542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen G., Gharib T. G., Huang C. C., Thomas D. G., Shedden K. A., Taylor J. M., Kardia S. L., Misek D. E., Giordano T. J., Iannettoni M. D., Orringer M. B., Hanash S. M., Beer D. G. (2002) Proteomic analysis of lung adenocarcinoma: Identification of a highly expressed set of proteins in tumors. Clin. Cancer Res. 8, 2298–2305 [PubMed] [Google Scholar]
- 14. Li R., Wang H., Bekele B. N., Yin Z., Caraway N. P., Katz R. L., Stass S. A., Jiang F. (2006) Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene 25, 2628–2635 [DOI] [PubMed] [Google Scholar]
- 15. Seike M., Kondo T., Fujii K., Yamada T., Gemma A., Kudoh S., Hirohashi S. (2004) Proteomic signature of human cancer cells. Proteomics 4, 2776–2788 [DOI] [PubMed] [Google Scholar]
- 16. Seike M., Kondo T., Fujii K., Okano T., Yamada T., Matsuno Y., Gemma A., Kudoh S., Hirohashi S. (2005) Proteomic signatures for histological types of lung cancer. Proteomics 5, 2939–2948 [DOI] [PubMed] [Google Scholar]
- 17. Poschmann G., Sitek B., Sipos B., Ulrich A., Wiese S., Stephan C., Warscheid B., Klöppel G., Vander Borght A., Ramaekers F. C., Meyer H. E., Stühler K. (2009) Identification of proteomic differences between squamous cell carcinoma of the lung and bronchial epithelium. Mol. Cell. Proteomics 8, 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bührens R. I., Amelung J. T., Reymond M. A., Beshay M. (2009) Protein expression in human non-small cell lung cancer: A systematic database. Pathobiology 76, 277–285 [DOI] [PubMed] [Google Scholar]
- 19. Trachootham D., Alexandre J., Huang P. (2009) Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. 8, 579–591 [DOI] [PubMed] [Google Scholar]
- 20. Acharya A., Das I., Chandhok D., Saha T. (2010) Redox regulation in cancer: A double-edged sword with therapeutic potential. Oxid. Med. Cel. Longev. 3, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Obsilova V., Silhan J., Boura E., Teisinger J., Obsil T. (2008) 14-3-3 proteins: A family of versatile molecular regulators. Physiol. Res. 3, (57 Suppl.) S11–S21 [DOI] [PubMed] [Google Scholar]
- 22. Osada H., Tatematsu Y., Yatabe Y., Nakagawa T., Konishi H., Harano T., Tezel E., Takada M., Takahashi T. (2002) Frequent and histological type-specific inactivation of 14-3-3sigma in human lung cancers. Oncogene 21, 2418–2424 [DOI] [PubMed] [Google Scholar]
- 23. Fan T., Li R., Todd N. W., Qiu Q., Fang H. B., Wang H., Shen J., Zhao R. Y., Caraway N. P., Katz R. L., Stass S. A., Jiang F. (2007) Up-regulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 67, 7901–7906 [DOI] [PubMed] [Google Scholar]
- 24. Besson A., Wilson T. L., Yong V. W. (2002) The anchoring protein RACK1 links protein kinase Cepsilon to integrin beta chains: Requirements for adhesion and motility. J. Biol. Chem. 277, 22073–22084 [DOI] [PubMed] [Google Scholar]
- 25. Steele M. R., McCahill A., Thompson D. S., MacKenzie C., Isaacs N. W., Houslay M. D., Bolger G. B. (2001) Identification of a surface on the beta-propeller protein RACK1 that interacts with the cAMP-specific phosphodiesterase PDE4D5. Cell. Signal. 13, 507–513 [DOI] [PubMed] [Google Scholar]
- 26. Chang B. Y., Chiang M., Cartwright C. A. (2001) The interaction of Src and RACK1 is enhanced by activation of protein kinase C and tyrosine phosphorylation of RACK1. J. Biol. Chem. 276, 20346–20356 [DOI] [PubMed] [Google Scholar]
- 27. Wang Z., Jiang L., Huang C., Li Z., Chen L., Gou L., Chen P., Tong A., Tang M., Gao F., Shen J., Zhang Y., Bai J., Zhou M., Miao D., Chen Q. (2008) Comparative proteomics approach to screening of potential diagnostic and therapeutic targets for oral squamous cell carcinoma. Mol. Cell. Proteomics 7, 1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Z., Zhang B., Jiang L., Zeng X., Chen Y., Feng X., Guo Y., Chen Q. (2009) RACK1, an excellent predictor for poor clinical outcome in oral squamous carcinoma, similar to Ki67. Eur. J. Cancer 45, 490–496 [DOI] [PubMed] [Google Scholar]
- 29. Egidy G., Julé S., Bossé P., Bernex F., Geffrotin C., Vincent-Naulleau S., Horak V., Sastre-Garau X., Panthier J. J. (2008) Transcription analysis in the MeLiM swine model identifies RACK1 as a potential marker of malignancy for human melanocytic proliferation. Mol. Cancer 7, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cao X. X., Xu J. D., Liu X. L., Xu J. W., Wang W. J., Li Q. Q., Chen Q., Xu Z. D., Liu X. P. (2010) RACK1: A superior independent predictor for poor clinical outcome in breast cancer. Int. J. Cancer 127, 1172–1179 [DOI] [PubMed] [Google Scholar]
- 31. Nagashio R., Sato Y., Matsumoto T., Kageyama T., Satoh Y., Shinichiro R., Masuda N., Goshima N., Jiang S. X., Okayasu I. (2010) Expression of RACK1 is a novel biomarker in pulmonary adenocarcinomas. Lung Cancer 69, 54–59 [DOI] [PubMed] [Google Scholar]
- 32. Wu W. S. (2006) The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. 25, 695–705 [DOI] [PubMed] [Google Scholar]
- 33. Yan T., Oberley L. W., Zhong W., St Clair D. K. (1996) Manganese-containing superoxide dismutase overexpression causes phenotypic reversion in SV40-transformed human lung fibroblasts. Cancer Res. 56, 2864–2871 [PubMed] [Google Scholar]
- 34. Zhong W., Oberley L. W., Oberley T. D., Yan T., Domann F. E., St Clair D. K. (1996) Inhibition of cell growth and sensitization to oxidative damage by overexpression of manganese superoxide dismutase in rat glioma cells. Cell Growth Differ. 7, 1175–1186 [PubMed] [Google Scholar]
- 35. Cullen J. J., Weydert C., Hinkhouse M. M., Ritchie J., Domann F. E., Spitz D., Oberley L. W. (2003) The role of manganese superoxide dismutase in the growth of pancreatic adenocarcinoma. Cancer Res. 63, 1297–1303 [PubMed] [Google Scholar]
- 36. Ough M., Lewis A., Zhang Y., Hinkhouse M. M., Ritchie J. M., Oberley L. W., Cullen J. J. (2004) Inhibition of cell growth by overexpression of manganese superoxide dismutase (MnSOD) in human pancreatic carcinoma. Free Radic. Res. 38, 1223–1233 [DOI] [PubMed] [Google Scholar]
- 37. Izutani R., Asano S., Imano M., Kuroda D., Kato M., Ohyanagi H. (1998) Expression of manganese superoxide dismutase in esophageal and gastric cancers. J. Gastroenterol. 33, 816–822 [DOI] [PubMed] [Google Scholar]
- 38. Kim J. J., Chae S. W., Hur G. C., Cho S. J., Kim M. K., Choi J., Nam S. Y., Kim W. H., Yang H. K., Lee B. L. (2002) Manganese superoxide dismutase expression correlates with a poor prognosis in gastric cancer. Pathobiology 70, 353–360 [DOI] [PubMed] [Google Scholar]
- 39. Janssen A. M., Bosman C. B., Sier C. F., Griffioen G., Kubben F. J., Lamers C. B., van Krieken J. H., van de Velde C. J., Verspaget H. W. (1998) Superoxide dismutases in relation to the overall survival of colorectal cancer patients. Br. J. Cancer 78, 1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haapasalo H., Kyläniemi M., Paunul N., Kinnula V. L., Soini Y. (2003) Expression of antioxidant enzymes in astrocytic brain tumors. Brain Pathol. 13, 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chung-man Ho J., Zheng S., Comhair S. A., Farver C., Erzurum S. C. (2001) Differential expression of manganese superoxide dismutase and catalase in lung cancer. Cancer Res. 61, 8578–8585 [PubMed] [Google Scholar]
- 42. Yang F., Xiao Z. Q., Zhang X. Z., Li C., Zhang P. F., Li M. Y., Chen Y., Zhu G. Q., Sun Y., Liu Y. F., Chen Z. C. (2007) Identification of tumor antigens in human lung squamous carcinoma by serological proteome analysis. J. Proteome Res. 6, 751–758 [DOI] [PubMed] [Google Scholar]
- 43. Des Rosiers C., Di Donato L., Comte B., Laplante A., Marcoux C., David F., Fernandez C. A., Brunengraber H. (1995) Isotopomer analysis of citric acid cycle and gluconeogenesis in rat liver: Reversibility of isocitrate dehydrogenase and involvement of ATP-citrate lyase in gluconeogenesis. J. Biol. Chem. 270, 10027–10036 [DOI] [PubMed] [Google Scholar]
- 44. DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. (2008) The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 57, 11–20 [DOI] [PubMed] [Google Scholar]
- 45. Ronnebaum S. M., Ilkayeva O., Burgess S. C., Joseph J. W., Lu D., Stevens R. D., Becker T. C., Sherry A. D., Newgard C. B., Jensen M. V. (2006) A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J. Biol. Chem. 281, 30593–30602 [DOI] [PubMed] [Google Scholar]
- 46. Holmgren A. (2000) Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid. Redox Signal. 2, 811–820 [DOI] [PubMed] [Google Scholar]
- 47. Park J. H., Kim Y. S., Lee H. L., Shim J. Y., Lee K. S., Oh Y. J., Shin S. S., Choi Y. H., Park K. J., Park R. W., Hwang S. C. (2006) Expression of peroxiredoxin and thioredoxin in human lung cancer and paired normal lung. Respirology 11, 269–275 [DOI] [PubMed] [Google Scholar]
- 48. Ward P. S., Patel J., Wise D. R., Abdel-Wahab O., Bennett B. D., Coller H. A., Cross J. R., Fantin V. R., Hedvat C. V., Perl A. E., Rabinowitz J. D., Carroll M., Su S. M., Sharp K. A., Levine R. L., Thompson C. B. (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kil I. S., Huh T. L., Lee Y. S., Lee Y. M., Park J. W. (2006) Regulation of replicative senescence by NADP+-dependent isocitrate dehydrogenase. Free Radic. Biol. Med. 40, 110–119 [DOI] [PubMed] [Google Scholar]
- 50. Bleeker F. E., Lamba S., Leenstra S., Troost D., Hulsebos T., Vandertop W. P., Frattini M., Molinari F., Knowles M., Cerrato A., Rodolfo M., Scarpa A., Felicioni L., Buttitta F., Malatesta S., Marchetti A., Bardelli A. (2009) IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum. Mutat. 30, 7–11 [DOI] [PubMed] [Google Scholar]
- 51. Yan H., Parsons D. W., Jin G., McLendon R., Rasheed B. A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G. J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K. W., Velculescu V. E., Vogelstein B., Bigner D. D. (2009) IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., Yu W., Li Z., Gong L., Peng Y., Ding J., Lei Q., Guan K. L., Xiong Y. (2009) Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 324, 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dang L., White D. W., Gross S., Bennett B. D., Bittinger M. A., Driggers E. M., Fantin V. R., Jang H. G., Jin S., Keenan M. C., Marks K. M., Prins R. M., Ward P. S., Yen K. E., Liau L. M., Rabinowitz J. D., Cantley L. C., Thompson C. B., Vander Heiden M. G., Su S. M. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mardis E. R., Ding L., Dooling D. J., Larson D. E., McLellan M. D., Chen K., Koboldt D. C., Fulton R. S., Delehaunty K. D., McGrath S. D., Fulton L. A., Locke D. P., Magrini V. J., Abbott R. M., Vickery T. L., Reed J. S., Robinson J. S., Wylie T., Smith S. M., Carmichael L., Eldred J. M., Harris C. C., Walker J., Peck J. B., Du F., Dukes A. F., Sanderson G. E., Brummett A. M., Clark E., McMichael J. F., Meyer R. J., Schindler J. K., Pohl C. S., Wallis J. W., Shi X., Lin L., Schmidt H., Tang Y., Haipek C., Wiechert M. E., Ivy J. V., Kalicki J., Elliott G., Ries R. E., Payton J. E., Westervelt P., Tomasson M. H., Watson M. A., Baty J., Heath S., Shannon W. D., Nagarajan R., Link D. C., Walter M. J., Graubert T. A., DiPersio J. F., Wilson R. K., Ley T. J. (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kang M. R., Kim M. S., Oh J. E., Kim Y. R., Song S. Y., Seo S. I., Lee J. Y., Yoo N. J., Lee S. H. (2009) Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int. J. Cancer 125, 353–355 [DOI] [PubMed] [Google Scholar]