Abstract
CYP3A4, an integral endoplasmic reticulum (ER)-anchored protein, is the major human liver cytochrome P450 enzyme responsible for the disposition of over 50% of clinically relevant drugs. Alterations of its protein turnover can influence drug metabolism, drug-drug interactions, and the bioavailability of chemotherapeutic drugs. Such CYP3A4 turnover occurs via a classical ER-associated degradation (ERAD) process involving ubiquitination by both UBC7/gp78 and UbcH5a/CHIP E2-E3 complexes for 26 S proteasomal targeting. These E3 ligases act sequentially and cooperatively in CYP3A4 ERAD because RNA interference knockdown of each in cultured hepatocytes results in the stabilization of a functionally active enzyme. We have documented that UBC7/gp78-mediated CYP3A4 ubiquitination requires protein phosphorylation by protein kinase (PK) A and PKC and identified three residues (Ser-478, Thr-264, and Ser-420) whose phosphorylation is required for intracellular CYP3A4 ERAD. We document herein that of these, Ser-478 plays a pivotal role in UBC7/gp78-mediated CYP3A4 ubiquitination, which is accelerated and enhanced on its mutation to the phosphomimetic Asp residue but attenuated on its Ala mutation. Intriguingly, CYP3A5, a polymorphically expressed human liver CYP3A4 isoform (containing Asp-478) is ubiquitinated but not degraded to a greater extent than CYP3A4 in HepG2 cells. This suggests that although Ser-478 phosphorylation is essential for UBC7/gp78-mediated CYP3A4 ubiquitination, it is not sufficient for its ERAD. Additionally, we now report that CYP3A4 protein phosphorylation by PKA and/or PKC at sites other than Ser-478, Thr-264, and Ser-420 also enhances UbcH5a/CHIP-mediated ubiquitination. Through proteomic analyses, we identify (i) 12 additional phosphorylation sites that may be involved in CHIP-CYP3A4 interactions and (ii) 8 previously unidentified CYP3A4 ubiquitination sites within spatially associated clusters of Asp/Glu and phosphorylatable Ser/Thr residues that may serve to engage each E2-E3 complex. Collectively, our findings underscore the interplay between protein phosphorylation and ubiquitination in ERAD and, to our knowledge, provide the very first example of gp78 substrate recognition via protein phosphorylation.
Hepatic cytochromes P450 (P450s)1 are endoplasmic reticulum (ER)-anchored hemoproteins involved in the metabolism of numerous endo- and xenobiotics. Of these, CYP3A4 is particularly noteworthy because it comprises 30% of the human liver microsomal P450 complement and is responsible for the metabolism of >50% clinically relevant drugs, as well as hepatotoxins such as aflatoxin B1 (1). In common with other ER-integral P450s, it is a monotopic protein, N-terminally anchored to the ER membrane, with the bulk of its catalytic domain in the cytosol. We have documented that CYP3A4, in common with its CYP3A orthologs, incurs ubiquitin (Ub)-dependent proteasomal degradation (UPD) in a classical ER-associated degradation (ERAD) process (2–4). Indeed, irrespective of any detectable structural lesion(s) in their cytosolic domain, CYPs 3A function as typical ERAD-C substrates (2–8). Accordingly, we have previously shown that both native and structurally/functionally inactivated CYPs 3A including CYP3A4 incur protein phosphorylation, followed by ubiquitination by UBC7/gp78 and UbcH5a/CHIP (C terminus of Hsc70-interacting protein) E2-E3 systems, p97-mediated ER-extraction, and subsequent 26 S proteasomal degradation (8–14).
CYP3A4 is but one of the many mammalian P450s known to be phosphorylated in vivo and in vitro (Ref. 15 and references therein; Ref. 16). These studies have invariably implicated protein kinase (PK) A and PKC as protagonists in this process. Over the years, since the phosphorylation of a P450 was first reported (17–19), it has been ascribed various roles (15, 20–31) including targeting P450s to degradation by a microsomal Ser protease (29–31), cytosolic ATP-dependent protease (24) or as suggested by us, UPD (9–12). However, we were the first to document that such post-translational protein phosphorylation occurs at multiple P450 S/T sites, is synergistic in character and serves to target P450s for UPD/ERAD (10–12). Accordingly, our LC-MS/MS analyses of purified CYP3A4 incubated with intact rat liver cytosol documented the phosphorylation of its CYP3A4 Ser-478, Thr-264, and Ser-420 residues (10, 11). The use of specific and general kinase inhibitors as probes in this system led to the identification of rat liver PKA and PKC as the relevant hepatic kinases (10). This was further confirmed in an in vitro screen of various commercially available kinases,2 enabling us in our subsequent studies to employ purified PKA and PKC as reagents to simplify our proteomic analyses. Furthermore, our studies in yeast and HEK293T cells revealed that phosphorylation of CYP3A4 Ser-478, Thr-264, and Ser-420 residues was relevant to its UPD/ERAD, because simultaneous Ala mutation of all three residues attenuated its turnover (11). Our in vitro studies of UBC7/gp78-mediated CYP3A4 ubiquitination also indicated that Ala mutation of these residues greatly impaired this process (11). However, the relative importance of each kinase and/or each of these phosphorylatable residues to CYP3A4 ubiquitination by UBC7/gp78 remained to be elucidated. Additionally, although we have documented an essential, albeit complementary role of both UBC7/gp78 and UbcH5a/CHIP E2-E3 systems in CYP3A UPD/ERAD (14), it was unclear whether protein phosphorylation was required for UbcH5a/CHIP-mediated CYP3A ubiquitination.
Herein, we provide additional mechanistic insight by examining the role of each kinase and describe the relative importance of CYP3A4 Ser-478 phosphorylation using both in vitro and intracellular assays. We further extend our findings to CYP3A5, a polymorphically expressed adult human liver CYP3A isoform that also happens to be a natural phosphomimetic (Asp-478) mutant. We report our findings of CYP3A4 phosphorylation by either PKA or PKC in vitro, coupled with proteomic analyses of the phosphorylated protein. Our findings reveal that whereas Ser-478 is specifically phosphorylated by PKA, Thr-264 and Ser-420 are phosphorylated by PKC. Moreover, we find that each of these kinases significantly phosphorylates additional, previously unidentified CYP3A4 Ser/Thr residues. Because PKA-dependent protein phosphorylation is generally believed to stem from signaling cascades, the physiological relevance of the observed in vitro CYP3A4 Ser-478 phosphorylation by the PKA catalytic subunit remains equivocal, despite the overwhelming evidence from our own laboratory and that of others that PKA is indeed involved in P450 phosphorylation (10, 17–30). In support of the in vivo relevance of this phosphorylation, we now also provide proteomic evidence that the corresponding residue Ser-479 in the orthologous rat liver CYP3A23 is indeed phosphorylated intracellularly in cultured hepatocytes upon acceleration of its ERAD through 3,5-dicarbethoxy-2,6-dimethyl-4-ethyl-1,4-dihydropyridine (DDEP)-elicited CYP3A suicide inactivation. We provide additional evidence that this DDEP-induced CYP3A ubiquitination and turnover in rat hepatocytes is blocked by treatment with specific inhibitors of either PKA or PKC. Furthermore, we document that the phosphorylation of just Ser-478 greatly accelerates and enhances the extent of CYP3A4 ubiquitination by UBC7/gp78 E2-E3 system in vitro. Accordingly, mutation of Ser-478 to the phosphomimetic Asp residue accelerates this process, whereas its mutation to Ala considerably attenuates the extent of this post-translational modification. Thus, to our knowledge, CYP3A4 and CYP2E1 (12) are the first examples of UBC7/gp78 substrates whose ERAD targeting entails protein phosphorylation. By contrast, even though protein phosphorylation enhanced UbcH5a/CHIP-mediated CYP3A4 ubiquitination, the phosphorylation of Ser-478, Thr-264, and Ser-420 played no apparent role in this process.
We have also assessed the relative relevance of Ser-478, Thr-264, and Ser-420 residues to CYP3A4 UPD/ERAD in human HepG2 cells following expression of adenoviral vectors (AdV) encoding wild type CYP3A4 and CYP3A5 or their site-specific mutants. Our findings described herein underscore a pivotal role of CYP3A4 Ser-478 in its UPD/ERAD. Additionally, using proteomic analyses, we have also identified for the first time, eight CYP3A4 Lys residues (Lys-115, Lys-127, Lys-168, Lys-173, Lys-282, Lys-466, Lys-487, and Lys-492) specifically ubiquitinated in vitro by UBC7/gp78 and/or UbcH5a/CHIP. Inspection of the CYP3A4 structure reveals that these ubiquitinated Lys residues lie within spatially associated, negatively charged Asp/Glu and phosphorylatable Ser/Thr surface clusters. The potential relevance of this finding to CYP3A4 ubiquitination is discussed. We believe that our findings of CYP3A4 as a model target substrate are applicable to other UPD/ERAD targeted ER proteins.
EXPERIMENTAL PROCEDURES
Materials
The sources of most chemicals have been previously reported (8, 11–14). Human cytosolic C-terminal gp78 domain (residues 309–643) (E3) and murine UBC7 (E2) were expressed in Escherichia coli and purified as described (13) from plasmids kindly provided by Dr. A. M. Weissman. CHIP was also expressed in E. coli from a plasmid kindly provided by Dr. C. Patterson and purified as described (13). Recombinant cytochrome b5 (b5) and P450 oxidoreductase (POR) were expressed in E. coli and purified as detailed (32). All other buffers and reagents were of the highest commercial grade.
Expression and Purification of CYP3A4(His)6, CYP3A5(His)6, and Corresponding Site-specific Mutants
Each cDNA encoding a C-terminally His6-tagged human CYP3A4 or CYP3A5 or its corresponding site-specific mutant was incorporated into a pCWori+ vector and expressed in DH5α cells as described (11). Recombinant CYPs 3A were purified to homogeneity by nickel affinity chromatography, followed by detergent exchange by hydroxylapatite chromatography as described previously (11).
Construction and Preparation of CYP3A4/CYP3A5 AdV
Full-length human cDNA of CYP3A4*1 was subjected to site-directed mutagenesis to Ala at Thr-264, Ser-420, and Ser-478 (T264A/S420A/S478A) or at Ser-478 to Asp (S478D) before being subcloned into the Gateway entry vector. Full-length human CYP3A5 cDNA and its K264T mutant were also similarly subcloned. These CYP3A4/CYP3A5 cDNAs were then recombined into an ∼38.2-kb AdV-BGFP-Gateway destination vector that also encoded β-galactosidase and green fluorescent protein. The CYP3A4/CYP3A5 AdV and the AdV-BGFP (vector control) were grown, propagated, and purified, and their titer was determined essentially as detailed (33, 34). CYP3A4/CYP3A5 AdV and the AdV-BGFP (100 multiplicity of infection) were transduced into 80–90% confluent HepG2 cells (≈5 × 106) in 25-cm2 flasks with 10 μg/ml polybrene. The cells were grown for 48 h to allow P450 protein expression, at which time the medium was replaced with a Cys/Met-free medium for 1 h. The cells were pulsed with 35S-EXPRESS (111 μCi/flask) and then 1 h later were chased with excess unlabeled cysteine (1.4 mm) and methionine (10 mm) and harvested at the indicated times over a 6-h period. At the time of chase, some cells were treated with the proteasomal inhibitor MG132 (final concentration, 20 μm) for 6 h. The cells were harvested, and lysates were subjected to CYP3A immunoprecipitation with goat anti-CYP3A IgG (1 mg of lysate protein to 2 mg of IgG) (8). Equivalent aliquots of immunoprecipitated CYPs 3A were subjected to SDS-PAGE on 4–20% gels. The gels were dried and exposed to a PhosphorImager screen for detection and visualized using a Typhoon scanner (8). Other aliquots were used for quantifying 35S-CYP3A radioactivity by scintillation counting.
Structural-Functional Validation of Site-specific CYP3A5K264T Mutant
As previously, the hallmark P450 CO-reduced binding spectra were compared to exclude any structural alteration relative to the wild type (WT) CYP3A5. Functional perturbations/disruptions were excluded by monitoring a CYP3A-selective functional probe: 7-benzyloxy-4-trifluoromethylcoumarin O-debenzylase activity of the recombinant CYPs 3A in CHAPS-solubilized E. coli membranes after functional reconstitution with POR (POR/P450 molar ratios, 4:1) and b5 (b5/P450 molar ratios, 2:1) assayed as detailed (35).
In Vitro Reconstitution Assays of CYP3A Ubiquitination
Purified recombinant human CYP3A4[His]6 WT or mutant was inactivated for 15 min at 37 °C with 1 mm CuOOH, as detailed (11, 12), to accelerate the phosphorylation/ubiquitination reactions. CuOOH-inactivated CYP3A4 WT or mutant (250 pmol) was then incubated in a reconstituted UBC7/gp78 ubiquitination system containing E1 (0.68 μm), human UBC7 (4.25 μm), purified recombinant gp78C (the cytosolic 63-KDa E3 ligase domain; 1 μm), ATP (5 mm), creatine phosphokinase (20 units), creatinine phosphate (20 mm), MgCl2 (12 mm), 32P-Ub (167 μm; prepared as detailed (13)), Hepes buffer (50 mm, pH 7.4, containing 20% (v/v) glycerol), EGTA (0.5 mm), and EDTA (0.5 mm) with or without PKA (0.004 units) and/or PKC (0.004 units) addition in a final volume of 70 μl as described previously (11–13). CHIP-mediated CYP3A4 ubiquitination was monitored in the presence of Hsp70 and Hsp40 in the incubation as described previously (12).
Control reactions were incubated in parallel in the absence of ATP or CuOOH-inactivated CYP3A4 (11–13). Except where otherwise indicated, all of the reactions were incubated at 30 °C for 90 min. At the end of each reaction, N-ethylmaleimide (final concentration, 5 mm) was added followed by 2% w/v SDS (final concentration). CYP3A4 was immunoprecipitated with goat anti-CYP3A4 IgG as detailed (8, 11). Equivalent aliquots (48 μl) of CYP3A4 immunoprecipitates were subjected to SDS-PAGE. The dried gels were exposed to PhosphorImager screens and visualized using a Typhoon scanner as detailed (8, 11–13).
Phosphorylation of Native/CuOOH-inactivated CYP3A4 by PKA/PKC
Purified recombinant CYP3A4 inactivated with CuOOH (CuOOH-inactivated) or without (native CYP3A4) was incubated with PKA or PKC (with a sample incubated without ATP as a control) at 30 °C for 30 min, as described (11).
In Vivo Effects of PKA/PKC Inhibitors on CYP3A4 UPD/ERAD in Cultured Rat Hepatocytes
Rat hepatocytes cultured exactly as described (8) were pretreated with the CYP3A-inducer dexamethasone for 5 days and then subjected to 35S pulse-chase analyses on day 5 (8). Following chase, they were treated with DDEP (100 μm), and 30 min later they were treated with DMSO (vehicle), KT-5720 (5 μm; a specific PKA inhibitor), bisindolylmaleimide III (BisIII; 5 μm; a specific PKC inhibitor), or staurosporine (1 μm; a general PKA/PKC/PKG inhibitor). The cells were harvested at 0, 2, and/or 4 h following DDEP treatment. CYPs 3A were immunoprecipitated as described above, and aliquots were subjected to radioactive scintillation counting, as well as SDS-PAGE/PhosphorImager/Typhoon scanning analyses as described above.
MS Analysis and Protein/Peptide Identification
Protein samples were reduced, alkylated, and subjected to SDS-PAGE separation as described (11, 12). Protein bands were excised from the gel, followed by in-gel digestion with lysyl endopeptidase C (Lys-C) as detailed previously (11, 12). The peptides were analyzed by LC-MS/MS analyses on LTQ-FT or LTQ Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA), equipped with a Waters NanoAcquity LC system (Milford, MA), exactly as described previously (12). To verify the accuracy of their assignments, some of the low abundance modified peptides were reanalyzed using the LTQ Velos Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA), because of its high mass resolution and greater sensitivity for detection of high energy dissociation (HCD) product ions. The MS/MS data were searched against the SwissProt database using the in-house Protein Prospector search engine (version 5.0.0beta6) (36, 37), with a concatenated database consisting of normal and randomized decoy databases (38, 39). For database searches, variable phosphorylations on serine and threonine were allowed. The phosphopeptide identification and the site assignment were manually verified by inspection of the raw MS/MS spectra. False discovery rates for phosphorylation and ubiquitination assays were estimated to be 1 and 5%, corresponding to the maximum expectation values of 0.01 and 0.05, respectively, and a minimum peptide score of 15 was used in both assays. The retention times of modified peptides provided additional evidence to support the modified peptide identification especially for multisite phosphorylated peptides. Mass tolerance for parent/precursor ions was 20 ppm, and mass tolerances of 0.5 Da for CID ions detected in ion trap and 20 ppm for HCD ions detected in Orbitrap were respectively employed. A maximum of two missed and/or nonspecific cleavages were permitted.
Estimation of PKA/PKC-catalyzed CYP3A4 Phosphorylation Stoichiometry
CYP3A4 phosphorylation stoichiometry was estimated by the application of a label-free semi-quantitative analysis as described (12). Protein/peptide identification, the phosphorylation site assignment, as well as information on the relative stoichiometry were obtained from two LC-MS/MS experiments with nonphosphorylated (control) and phosphorylated (sample) preparations (40, 41). Briefly, two samples were prepared as described under the CYP3A4 phosphorylation assay (11). One with ATP added represented the phosphorylated sample and the other without ATP inclusion provided the control sample. Both the phosphorylated and control samples were freshly prepared and analyzed by LC-MS/MS individually. The MS/MS scan ion peak lists from the two LC-MS/MS raw data were generated with the in-house PAVA program (version, July 28, 2009), and searched against the SwissProt database (June 5, 2010 with a concatenated database consisting of normal and randomized decoy databases; 1,035,604/1,035,604 entries searched) with the in-house Protein Prospector search engine (version 5.0.0beta6) to identify phosphorylated and nonphosphorylated peptides. The information of m/z, z, and retention time of the nonphosphorylated/phosphorylated peptide obtained from the database search of MS/MS scans was used to extract ion chromatographic peak area values from the MS level scans. Although the phosphorylated peptide ion should not be observed in LC-MS spectra of control sample (without ATP), LC peak information of the phosphorylated peptide ion and the corresponding nonphosphorylated/intact peptide can be obtained from the raw data of the phosphorylated (with ATP) sample. Two strategies were used for the semi-quantitative label-free analysis based on extracted ion chromatograms to estimate the phosphorylation stoichiometry. First, the ratio of the extracted ion chromatographic peak areas of phosphorylated peptide over that of the corresponding nonphosphorylated peptide is directly calculated with the assumption that their ionization efficiencies are similar. Peptides with a miscleavage site or different charge but containing the same phosphorylation site need to be combined for estimation. Alternatively, the depletion of the nonphosphorylated peptide as determined from the chromatographic peak areas was used to estimate the relative quantity of the corresponding phosphorylated peptide. This strategy works well if the nonphosphorylated peptide is detected with good signal to noise ratios in both the control and phosphorylated P450 samples, and the phosphorylation stoichiometry is high (≈>20%), but it does not work well in the case of a low stoichiometry level of phosphorylation or multiple phosphorylation sites in the same peptide such as Ser-116 and Ser-119 in the same CYP3A4 phosphorylated Ser-116–Lys-127 peptide. In the case of low stoichiometry, the intensity of the phosphorylated peptide was used directly. Because the ionization efficiency of the phosphopeptide is typically lower than that of its corresponding nonphosphorylated peptide, its estimation can be significantly under-represented. Thus, in the case of low stoichiometry, the method may indeed estimate smaller values because the difference in the ionization efficiency between phosphorylated and nonphosphorylated peptides is not known.
Identification of CYP3A4 Ubiquitination Sites
Purified human liver CYP3A4 (500 pmol) was subjected to in vitro ubiquitination for 90 min with either the complete UBC7/gp78 or UbcH5a/CHIP system as detailed above. The reaction was terminated with a 2% w/v SDS (final concentration). After boiling the mixture, the protein was reduced with DTT (25 mm) and then placed in the dark and alkylated with iodoacetamide (75 mm) at room temperature for an hour. This mixture was then subjected to immunoaffinity capture by disuccinimidyl suberate cross-linked goat anti-CYP3A4 IgG-protein A-Sepharose beads (≈500 μg of goat anti-CYP3A4 IgG; 200 μl) (12). This step removed any unreacted iodoacetamide, thereby effectively excluding any subsequent artifactual iodoacetamide-induced 2-acetaminoacetamidation of Lys residues that because of its striking mass similarity (114.0429 Da) can confound the mass spectrometric detection of truly ubiquitinated Lys residues (42).
The ubiquitinated CYP3A4 protein was eluted with SDS-PAGE loading buffer (80 μl) as described (12). The eluate was subjected to 4–15% SDS-PAGE analysis followed by Coomassie Blue gel staining. Gel pieces corresponding to a molecular mass range of 65–250 kDa were excised, finely chopped, destained, and dehydrated with acetonitrile. The dried gel pieces were then rehydrated with 50 mm ammonium bicarbonate containing Lys-C/trypsin in an ≈1:20 (w/w) ratio of each protease to CYP3A4 and digested at 37 °C for at least 12 h. The extracted peptides were concentrated, desalted, and then subjected to LC-MS/MS analysis as described (12). Ubiquitination was identified by use of a two-stage database search.
In the first stage, a default setting for modifications was used (fixed modification = carbmidomethylation; variable modifications = acetylation on protein N terminus, glutamine on peptide N-terminal to glutamic acid, methionine loss from protein N terminus, methionine loss from protein N terminus and acetylation, methionine oxidation) to search against mammalian sequences in the Swiss-Prot (version, July 07, 2009) database (471,472 entries). In the second stage, a restricted database was constructed consisting of CYP3A4 and in vitro reaction components, which were identified by the first search. The second search used mass modification for ubiquitination (Gly-Gly and Leu-Arg-Gly-Gly on lysine) in addition to the default PTM (i.e. phosphoserine and phosphothreonine modifications) setting against the restricted database. Peptides with expectation values of 0.05 or less were accepted. Phosphorylated/ubiquitinated peptides were confirmed by manual inspection of the raw MS/MS spectra (see supplemental data). MS/MS spectra were inspected for the diagnostic loss of GG- or KGG-associated mass (114.043 or 242.1 Da, respectively) that not only verified the Ub-derived protein modification but also served to identify the precise CYP3A4 Lys residue ubiquitinated (43). Mass tolerance for parent/precursor ions of 20 ppm and for fragment ions of 0.5 Da for Orbitrap/FT and 0.1 Da for Velos were employed. A maximum of two missed and/or nonspecific cleavages were permitted.
RESULTS
Identification of Human Liver CYP3A4 Residues Phosphorylated by PKA through Proteomic Analyses
Sites phosphorylated in the native and CuOOH-inactivated CYP3A4 after their incubation with either PKA or PKC were identified by LC-MS/MS analyses. Ion peaks were selected for MS/MS analyses based on a so-called dynamic exclusion scheme as previously described (12). As previously, fragmentation spectra were inspected for the characteristic phosphorylation signature stemming from the elemental loss of phosphoric acid (98 Da) from the parent peptides (11, 12). The manually inspected MS/MS spectra of each phosphorylated peptide indicating the target site are appended as evidence (supplemental materials). The stoichiometry of phosphorylation at those sites was estimated using a label-free quantification method, as described (“Experimental Procedures”).
In vitro PKA-incubation of purified recombinant native CYP3A4 resulted in the detectable phosphorylation of four Ser residues (Ser-116, Ser-119, Ser-134, and Ser-478), as monitored by LC-MS/MS analyses. Their phosphorylation extents (i.e. % of the total phosphorylatable residue in the protein) as monitored by semi-quantitative analyses were found to be 0.2 ± 0.05, 0.01 ± 0.004, 0.04 ± 0.01, and 0.32 ± 0.07%, respectively, and thus low. Upon CuOOH-mediated CYP3A4 inactivation, this extent was appreciably increased for Ser-478 (1.29 ± 0.12%) and Ser-119 (0.42 ± 0.06%). In addition, Ser-259 was found to be detectably phosphorylated (0.49 ± 0.05%) upon CuOOH-mediated inactivation (Tables I and II and supplemental Fig. S1, top left panel).
Table I. List of identified PKA-catalyzed CYP3A4 phosphorylation sites.
For experimental details, see “Experimental Procedures.” CYP3A4 peptides were derived from Lys-C digest. Annotated spectra are provided in the supplemental materials. RT, peptide retention time; Expect val, expectation value; ox, oxidation; p, phosphorylation.
| No. | Site | Peptide sequence | m/z | z | Mass error (ppm) | RT (min) | Score | Expect val | NetPhos score |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Ser-116 | pSAISIAEDEEWK | 729.31 | 2 | 8.1 | 29.59 | 22.5 | 1.4e-4 | 0.011 |
| 2 | Ser-119 | SAIpSIAEDEEWK | 729.31 | 2 | 2.7 | 37.74 | 35.3 | 9.0e-6 | 0.993 |
| 3 | Ser-134 | RLRSLLpSPTFTSGK | 821.94 | 2 | 7.4 | 28.61 | 19.9 | 4.1e-5 | 0.932 |
| 4 | Ser-259 | RM(ox)KEpSRLEDTQK | 808.87 | 2 | −4.6 | 21.98 | 18.9 | 3.3e-4 | 0.927 |
| 5 | Ser-478 | ETQIPLKLpSLGGLQPEKPVVLK | 860.83 | 3 | 4.7 | 40.17 | 23.6 | 3.8e-4 | 0.232 |
Table II. PKA-mediated phosphorylation: semi-quantitative comparison of native and CuOOH-inactivated CYP3A4.
The values are the means ± S.D. of three individual determinations. Control samples had no detectable (—) phosphorylated peptides.
| No. | Residue | Control | Native 3A4 (%) | CuOOH-3A4 (%) |
|---|---|---|---|---|
| 1 | Ser-116 | — | 0.20 ± 0.05 | 0.27 ± 0.08 |
| 2 | Ser-119 | — | 0.01 ± 0.004 | 0.42 ± 0.06 |
| 3 | Ser-134 | — | 0.04 ± 0.01 | 0.06 ± 0.03 |
| 4 | Ser-259 | — | — | 0.49 ± 0.05 |
| 5 | Ser-478 | — | 0.32 ± 0.07 | 1.29 ± 0.12 |
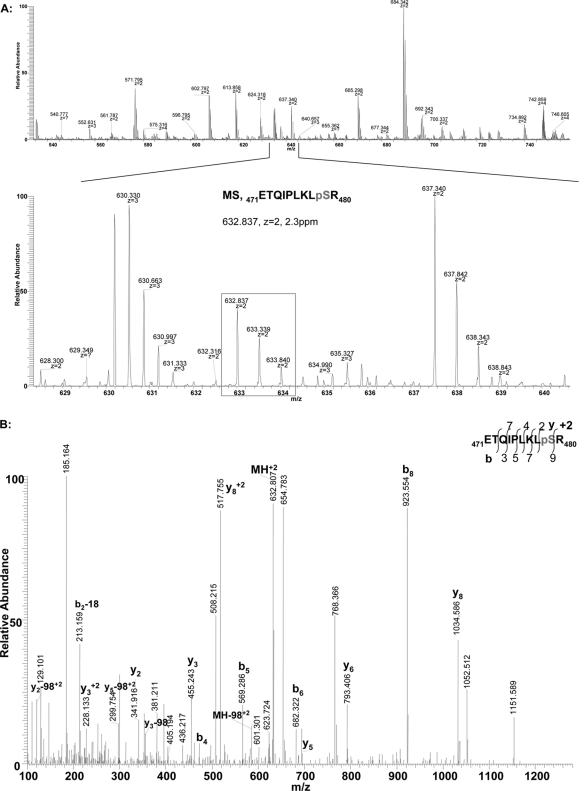
To verify that this in vitro Ser-478 phosphorylation by the PKA catalytic subunit is physiologically relevant, we examined whether this residue was phosphorylated in the orthologous dexamethasone-inducible CYPs 3A (3A23/3A2) in cultured rat hepatocytes treated with DDEP to stimulate UPD/ERAD. Immunoaffinity capture of rat hepatic CYPs 3A followed by Lys-C digestion and LC-MS/MS analyses revealed that Ser-479, the residue corresponding to CYP3A4 Ser-478, was indeed phosphorylated intracellularly during its DDEP-elicited ERAD.3 The corresponding MS and the annotated MS/MS spectra of the phosphorylated Ser-479-containing CYP3A23 peptide are documented (Fig. 1).
Fig. 1.
Proteomic evidence for the intracellular phosphorylation of CYP3A23 Ser-479 in cultured rat hepatocytes. A, LC-MS spectrum of ETQIPLKLpSR, a CYP3A23 phosphorylated peptide (residues 471–480; containing Ser-479 corresponding to CYP3A4 Ser-478) derived from CYP3A23 following its immunoaffinity capture from rat hepatocyte cultures treated with DDEP for 2 h. B, its manually annotated LC-MS/MS spectrum.
Identification of Human Liver CYP3A4 Residues Phosphorylated by PKC by Proteomic Analyses
Similar incubation of native CYP3A4 with PKC, and subsequent LC-MS/MS analyses revealed detectable PKC-mediated phosphorylation of multiple CYP3A4 residues (Thr-92, Ser-100, Thr-103, Ser-131, Ser-134, Thr-136, Ser-139, Thr-264, Thr-284, and Ser-420). Of these, Thr-264 and Thr-284 are to be underscored as two residues phosphorylated to a very high extent (24.2 ± 1.26 and 40.1 ± 0.56%, respectively) in the native protein, whereas the phosphorylation of the other residues was modest, ranging from 0.07 ± 0.01 to 2.08 ± 0.24%. The manually inspected MS/MS spectra are appended as evidence (supplemental data). Thus, with the exception of Ser-134, PKC phosphorylates a different set of CYP3A4 residues than PKA.
CuOOH-mediated CYP3A4 inactivation also considerably increased the PKC-mediated phosphorylation extent of several residues that were significantly (Thr-284), modestly (Ser-131, Ser-134, Ser-139, and Ser-420), poorly (Thr-92 and Thr-136), or not at all (Ser-116, Ser-119, Ser-259, and Ser-398) detectably phosphorylated in the native protein (Tables III and IV and supplemental Fig. S1, bottom left panel). However, the phosphorylation extent of Thr-264, a CYP3A4 residue significantly modified in the native protein, was not further increased upon CuOOH inactivation.
Table III. List of identified PKC-catalyzed CYP3A4 phosphorylation sites.
For experimental details, see “Experimental Procedures.” CYP3A4 peptides were derived from Lys-C digest. Annotated spectra are provided in the supplemental materials. cm, carbamidomethylation; ox, oxidation; p, phosphorylation; RT, peptide retention time; Expect val, expectation value.
| No. | Site | Peptide sequence | m/z | z | Mass error (ppm) | RT (min) | Score | Expect val | NetPhos score |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Thr-92 | VWGFYDGQQPVLAITDPDMIKpTVLVK | 1005.18 | 3 | 3.6 | 41.66 | 44.8 | 3.5e-5 | 0.414 |
| 2 | Ser-100 | EC(cm)YpSVFTNRRPFGPVGFMK | 791.36 | 3 | 7.3 | 31.61 | 25.1 | 2.5e-2 | 0.027 |
| 3 | Thr-103 | EC(cm)YSVFpTNRRPFGPVGFM(ox)K | 796.69 | 3 | 7.9 | 29.12 | 25.1 | 3.4e-4 | 0.737 |
| 4 | Ser-116 | pSAISIAEDEEWK | 729.31 | 2 | −1.0 | 32.77 | 37.7 | 3.0e-6 | 0.011 |
| 5 | Ser-119 | SAIpSIAEDEEWK | 729.31 | 2 | 6.1 | 34.58 | 34.6 | 9.0e-6 | 0.993 |
| 6 | Ser-131 | RLRpSLLSPTFTSGK | 548.29 | 3 | 3.4 | 26.72 | 36.4 | 1.5e-4 | 0.156 |
| 7 | Ser-134 | RLRSLLpSPTFTSGK | 821.94 | 2 | 7.4 | 26.73 | 22.7 | 7.6e-4 | 0.932 |
| 8 | Thr-136 | RLRSLLSPpTFTSGK | 821.94 | 2 | 6.6 | 26.71 | 23.7 | 0.096 | 0.006 |
| 9 | Ser-139 | RLRSLLSPTFTpSGK | 548.29 | 3 | 4.3 | 26.73 | 18.6 | 7.6e-4 | 0.486 |
| 10 | Ser-259 | RM(ox)KEpSRLEDTQK | 808.88 | 2 | −4.6 | 16.55 | 18.9 | 0.011 | 0.927 |
| 11 | Thr-264 | ESRLEDpTQK | 593.26 | 2 | 3.9 | 5.42 | 15.8 | 3.1e-4 | 0.919 |
| 12 | Thr-284 | HRVDFLQLMIDSQNSKEpTESHK | 681.32 | 4 | 1.4 | 29.12 | 39.3 | 6.0e-3 | 0.133 |
| 13 | Ser-398 | GVVVMIPpSYALHRDPK | 621.32 | 3 | 2.2 | 34.93 | 26.5 | 9.8e-5 | 0.005 |
| 14 | Ser-420 | FLPERFpSKK | 616.31 | 2 | 3.7 | 18.02 | 20.3 | 1.4e-4 | 0.996 |
| FLPERFpSK | 552.26 | 2 | 2.8 | 26.06 | 23.2 | 0.011 |
Table IV. PKC-mediated phosphorylation: semi-quantitative comparison of native and CuOOH-inactivated CYP3A4.
The values are the means ± S.D. of three individual determinations. Control samples had no detectable (—) phosphorylated peptides.
| No. | Residue | Control | Native 3A4 (%) | CuOOH-3A4 (%) |
|---|---|---|---|---|
| 1 | Thr-92 | — | 0.07 ± 0.01 | 0.15 ± 0.07 |
| 2 | Ser-100 | — | 2.08 ± 0.24 | 2.28 ± 0.46 |
| 3 | Thr-103 | — | 1.30 ± 0.23 | 2.22 ± 0.53 |
| 4 | Ser-116 | — | — | 0.28 ± 0.09 |
| 5 | Ser-119 | — | — | 0.25 ± 0.08 |
| 6 | Ser-131 | — | 0.98 ± 0.23 | 10.5 ± 1.28 |
| 7 | Ser-134 | — | 0.53 ± 0.12 | 2.72 ± 0.16 |
| 8 | Thr-136 | — | 0.31 ± 0.12 | 2.21 ± 0.18 |
| 9 | Ser-139 | — | 0.97 ± 0.15 | 3.69 ± 0.34 |
| 10 | Ser-259 | — | — | 10.1 ± 1.20 |
| 11 | Thr-264 | — | 24.2 ± 1.26 | 23.2 ± 1.59 |
| 12 | Thr-284 | — | 40.1 ± 0.56 | 52.9 ± 2.34 |
| 13 | Ser-398 | — | — | 1.89 ± 0.27 |
| 14 | Ser-420 | — | 1.26 ± 0.06 | 14.1 ± 0.28 |
The Effects of PKA and/or PKC on in Vitro UBC7/gp78-mediated CYP3A4 Ubiquitination, and the Role of Ser-478 Phosphorylation in This Process
We have previously shown that both UbcH5a/CHIP and UBC7/gp78 E2-E3 complexes catalyze CYP3A ubiquitination in vitro and in cultured rat hepatocytes or human HepG2 cells (13, 14). We have also shown that PKA- and PKC-mediated phosphorylation of Ser-478, Thr-264, and Ser-420 residues was important for the UBC7/gp78-mediated ubiquitination of CuOOH-inactivated CYP3A4, because simultaneous Ala mutation of these residues dramatically inhibited this process (11). Indeed, we confirmed that CYP3A4 was appreciably ubiquitinated upon immunoprecipitation/SDS-PAGE/PhosphorImager/Typhoon scanning analyses from a 90-min incubation of an UBC7/gp78-reconstituted ubiquitination system (Fig. 2A). Inclusion of either PKA or PKC in the incubation further enhanced the extent of this ubiquitination, although PKA was relatively more robust than PKC in eliciting this enhancement (Fig. 2A). Nevertheless, the inclusion of both PKA/PKC in the incubation further increased the extent of this CYP3A4 ubiquitination (Fig. 2A),4 consistent with our previous findings (11). These results are also consistent with PKA and PKC each phosphorylating a different set of residues.
Fig. 2.
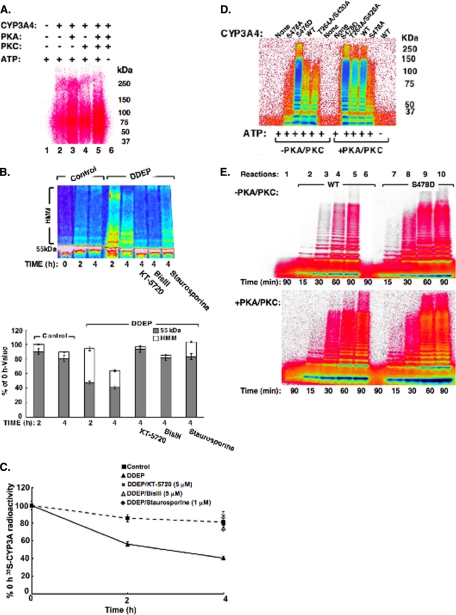
Effects of PKA and/or PKC on CYP3A ubiquitination and/or turnover. A, ubiquitination of CYP3A4 in an in vitro reconstituted UBC7/gp78 system. PhosphorImager analyses of UBC7/gp78-mediated ubiquitination of CuOOH-inactivated CYP3A4 in the presence or absence of PKA and/or PKC or in the presence or absence of ATP in the incubation. CYP3A4 was immunoprecipitated from the incubation mixture at 90 min of incubation and subjected to SDS-PAGE analyses. For details of the complete CYP3A4 ubiquitination analyses, see “Experimental Procedures.” Data from a typical experiment are depicted. Lane 1 corresponds to the complete reaction mixture with ATP, but no CYP3A4, PKA, or PKC. Lanes 2–5 correspond to the complete ATP-supplemented CYP3A4 ubiquitination system in the presence of no kinase (2), either PKA (3) or PKC (4) or both kinases (5). Lane 6 corresponds to the complete reaction mixture with CYP3A4 and PKA/PKC, but no ATP. Each experiment was conducted in its entirety at the least three separate times. B, CYP3A ubiquitination and turnover in cultured rat hepatocytes treated with PKA and/or PKC inhibitors. Typhoon scanning analyses of a typical gel. The cells were subjected to 35S pulse-chase analyses. At time 0 of chase, the cells were treated with DDEP (solid line) or DMSO (vehicle; dashed line). Solid line represents hepatocytes treated with DDEP alone; dashed line represents corresponding control untreated hepatocytes incubated in parallel without any DDEP and/or kinase inhibitor. Thirty min later, they were treated with KT-5720 (5 μm), BisIII (5 μm), staurosporine (1 μm), or DMSO (vehicle). At 0, 2, and/or 4 h following DDEP, 35S-CYP3A was immunoprecipitated from cell lysates, and aliquots were subjected to SDS-PAGE and PhosphorImager analyses as described (top panel). The color wheel intensity code is as follows: white > magenta > red > orange > yellow > green > light blue > dark blue > black. The corresponding quantitation (mean ± S.D.) of the relative intensities of the 55-kDa 35S-CYP3A parent species and corresponding HMM ubiquitinated 65–250-kDa species from three individual experiments (one of which is depicted on top) is shown at the bottom. C, effects of PKA and/or PKC inhibitors on DDEP-induced 35S-CYP3A turnover. Equivalent aliquots of 35S-CYP3A immunoprecipitates from cells treated in B above were also subjected to scintillation counting. The values are the means ± S.D. of three individual experiments. The time course of DDEP (solid line) or DMSO (vehicle; dashed line) treatment on 35S-CYP3A content over 0–4 h of treatment is shown. The corresponding effects of KT-5720 (5 μm; ×), BisIII (5 μm; open triangle), staurosporine (1 μm; solid diamond), and DMSO (vehicle; solid square) on 35S-CYP3A content at 4 h following DDEP are shown. D, ubiquitination of CYP3A4 or mutants in an in vitro reconstituted UBC7/gp78 system. Shown are the results of analyses of CYP3A4WT or its Ser-478, Thr-264, and Ser-420 mutants incubated for 90 min in a functionally reconstituted UBC7/gp78 ubiquitination system similar to those in A, except that aliquots of incubation mixtures were directly subjected to SDS-PAGE analyses without CYP3A4 immunoprecipitation. E, time course of UBC7/gp78-mediated ubiquitination of CuOOH-inactivated CYP3A4 or its S478D mutant in the presence or absence of PKA and/or PKC. Lane 1 corresponds to the complete reaction mixture incubated for 90 min with ATP, but without CYP3A4, PKA, or PKC (top panel) or without CYP3A4 (bottom panel). Lane 6 corresponds to the complete incubation mixture without ATP. Aliquots of the incubated mixtures were directly subjected to SDS-PAGE analyses without CYP3A4 immunoprecipitation. The color wheel intensity code is as follows: black > dark blue > light blue > green > yellow > orange > red > magenta > white.
An intracellular role for both kinases was also evident from our studies of DDEP-elicited CYP3A ubiquitination and turnover in cultured rat hepatocytes following a 4-h treatment with KT-5720 (a specific PKA inhibitor), BisIII (a specific PKC inhibitor), or staurosporine (a general PKA/PKC/PKG inhibitor). As in the case of CYP2E1 (12), each of these inhibitors blocked the time-dependent CYP3A ubiquitination (appearance of the high molecular mass (HMM) species; Fig. 2B), as well as degradation (loss of the parent 55-kDa species; Fig. 2, B and C), relative to the corresponding 4-h control levels. This indicated that both kinases are required for CYP3A4 UPD/ERAD.
We have previously reported that Ser-478, Thr-264, and Ser-420 were relevant for both UBC7/gp78-mediated CYP3A4 ubiquitination in vitro and CYP3A4 turnover in S. cerevisiae and HEK293T cells (11). Of these residues, Ser-478 was found to be the most dominant determinant of CYP3A4 turnover in Saccharomyces cerevisiae (11). We therefore sought to further explore its involvement, if any, in UBC7/gp78-mediated CYP3A4 ubiquitination as follows. Recombinant purified CYP3A4 mutants with Ser-478 mutated to either Ala (S478A) or to the phosphomimetic residue Asp (S478D) were evaluated as substrates relative to the WT enzyme or a CYP3A4T264A/S420A mutant in an in vitro functionally reconstituted UBC7/gp78 ubiquitination system, in the presence or absence of both PKA/PKC (Fig. 2D). In the absence of PKA/PKC in the incubation, the ubiquitination of the CYP3A4WT enzyme and that of its T264A/S420A mutant (both of which contain Ser-478) were comparable, whereas that of the S478A mutant was much weaker (Fig. 2D). By contrast, the ubiquitination of the phosphomimetic S478D mutant was dramatically enhanced even in the absence of PKA/PKC (Fig. 2D). Inclusion of PKA/PKC in the incubation greatly increased the ubiquitination of both CYP3A4 WT and its T264A/S420A mutant (Fig. 2D). However, in the presence of both kinases, the ubiquitination of the WT enzyme was nominally greater than that of the T264A/S420A mutant. Furthermore, the ubiquitination ladders of these P450s were extended to higher molecular mass levels, indicating that protein phosphorylation may have enhanced the efficiency and/or extent of their UBC7/gp78-mediated ubiquitination (Fig. 2D). The overall ubiquitination of the S478D mutant, which was robustly ubiquitinated in the absence of PKA/PKC, was only further modestly increased by the inclusion of both kinases (Fig. 2D). Inclusion of PKA/PKC in the incubation increased the otherwise sluggish ubiquitination of CYP3A4S478A to a small extent, thereby underscoring a minor role for other phosphorylatable residues in the UBC7/gp78-mediated CYP3A4 ubiquitination. Together these findings reveal a salient role for CYP3A4 Ser-478 in its UBC7/gp78-mediated ubiquitination.
This is further strengthened by the time course of UBC7/gp78-mediated ubiquitination in the absence of PKA/PKC, which revealed that the ubiquitination of CYP3A4S478D mutant is both accelerated and enhanced (extension to higher molecular mass species) relative to that of the WT enzyme (Fig. 2E). Inclusion of PKA/PKC in the incubation further enhanced this process for both proteins (Fig. 2E). Thus, despite the dominant role of Ser-478 phosphorylation, other CYP3A4 phosphorylatable residues must also be involved in the observed synergistic acceleration/enhancement of its UBC7/gp78-mediated ubiquitination (Fig. 2E).
The Effects of PKA and/or PKC on in Vitro UbcH5a/CHIP-mediated CYP3A4 Ubiquitination, and a Role for Phosphorylation of Residues Other Than Ser-478, Thr-264, and/or Ser-420 in This Process
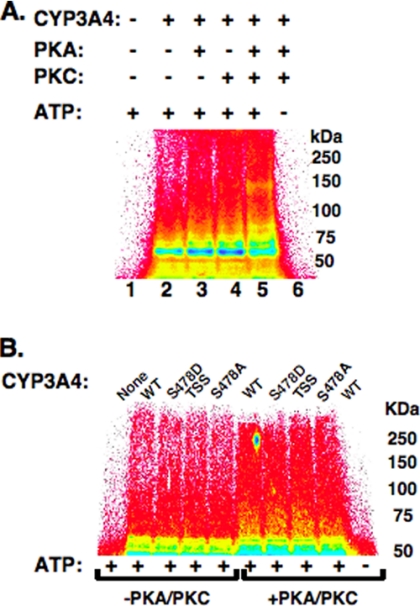
We have previously reported that CYP3A UPD/ERAD involves the concerted function of both gp78 and CHIP E3 ligases (13, 14). Given the role of PKA- and PKC-mediated phosphorylation in UBC7/gp78-mediated CYP3A4 ubiquitination, we investigated whether each of these kinases similarly influenced UbcH5a/CHIP-mediated CYP3A4 ubiquitination. Indeed, the inclusion of either PKA or PKC in the incubation greatly increased CYP3A4 ubiquitination by this E2-E3 system, whereas inclusion of both kinases further enhanced this process, as documented by the upward shift of the HMM species (Fig. 3A). However, no relative differences were observed in either the presence or the absence of both kinases between the ubiquitination extents of the CYP3A4 WT and its S478D, S478A, or CYP3A4S478A/T264A/S420A mutants (Fig. 3B). These findings indicate that although protein phosphorylation by each kinase clearly enhances UbcH5a/CHIP-mediated CYP3A4 ubiquitination, this enhancement must involve residues other than Ser-478, Thr-264, and Ser-420. Given the involvement of both PKA and PKC in CYP3A4 ubiquitination by both the E2-E3 systems, all of the subsequent studies were conducted in their presence.
Fig. 3.
Effects of PKA and/or PKC on in vitro reconstituted UbcH5a/CHIP-mediated CYP3A ubiquitination. A, ubiquitination of CYP3A4 in an in vitro reconstituted UbcH5a/CHIP system PhosphorImager analyses of UbcH5a/CHIP-mediated ubiquitination of CuOOH-inactivated CYP3A4 in the presence or absence of PKA and/or PKC or in the presence or absence of ATP in the incubation. CYP3A4 was immunoprecipitated from the incubation mixture at 90 min of incubation and subjected to SDS-PAGE analyses. For details of the complete CYP3A4 ubiquitination analyses, see “Experimental Procedures.” The data from a typical experiment are depicted. Lane 1 corresponds to the complete reaction mixture with ATP, but no CYP3A4, PKA, or PKC. Lanes 2–5 correspond to the complete ATP-supplemented CYP3A4 ubiquitination system in the presence of no kinase (2), either PKA (3) or PKC (4) or both kinases (5). Lane 6 corresponds to the complete reaction mixture with CYP3A4 and PKA/PKC, but no ATP. Each experiment was conducted in its entirety at the least three separate times. B, ubiquitination of CYP3A4 or mutants in an in vitro reconstituted UbcH5a/CHIP system. Analyses of CYP3A4WT or its Ser-478, Thr-264, and Ser-420 mutants incubated for 90 min in a functionally reconstituted UbcH5a/CHIP ubiquitination system similar to that in A, and CYP3A4 immunoprecipitates were directly subjected to SDS-PAGE and Typhoon scanning analyses. Data from a typical experiment are depicted. Lane 1 corresponds to the complete reaction mixture with ATP, but no CYP3A4, PKA, or PKC. Lanes 2–4 correspond to the complete ATP-supplemented ubiquitination system containing CYP3A4 or one its Ser/Thr mutants in the absence of both PKA and PKC. Lanes 5–9 are the corresponding incubations carried out in the presence of both kinases. Lane 10 corresponds to the complete reaction mixture with CYP3A4 and PKA/PKC, but no ATP. Each experiment was conducted in its entirety at least three separate times. The color wheel intensity code is as follows: white > magenta > red > orange > yellow > green > light blue > dark blue > black.
Identification of Human Liver CYP3A4 Residues Ubiquitinated by UBC7/gp78 or UbcH5a/CHIP by Proteomic Analyses
As previously, LC-MS/MS analyses of the tryptic/Lys-C digests of ubiquitinated CYP3A4 were employed to detect the corresponding mass signatures of Ub-derived tags (-GG76 or -LRGG76) through the associated 114.04- or 383.23-Da mass increase of the modified peptide relative to that of the unmodified parent peptide. Subsequent MS/MS fragmentation of this modified peptide resulted in a characteristic loss of KGG- or KGGRL-associated mass (242.1 or 383.23 Da, respectively) that not only verified the Ub-derived protein modification but also served to identify the precise Lys residue ubiquitinated in the protein of interest. We used this approach to identify several CYP3A4 residues ubiquitinated by UBC7/gp78 (Table V) or UbcH5a/CHIP in the presence of PKA/PKC (Table VI).
Table V. List of identified UBC7/gp78-catalyzed CYP3A4 ubiquitination sites.
For experimental details, see “Experimental Procedures.” CYP3A4 peptides were derived from Lys-C/tryptic digests. ox, oxidation; p, phosphorylation; Expect val, expectation value.
| No | Site | Peptide sequence | m/z | z | Mass error (ppm) | Score | Expect val. |
|---|---|---|---|---|---|---|---|
| 1 | Lys-115 | RPFGPVGFMK(GlyGly)SAISIAEDEEWKa | 870.104 | 3 | 4.6 | 23.2 | 7.6e-3 |
| RPFGPVGFMK(GlyGly)SAIpSIAEDEEWK | 896.770 | 3 | 15.8 | 22.0 | 5.0e-2 | ||
| RPFGPVGFM(ox)K(GlyGly)SAIpSIAEDEEWK | 902.091 | 3 | 4.4 | 16.7 | 2.4e-2 | ||
| 2 | Lys-168 | EAETGK(GlyGly)PVTLKa | 643.855 | 2 | 5.8 | 22.6 | 2.5e-3 |
| REAETGK(LeuArgGlyGly)PVTLKa | 571.333 | 3 | 2.0 | 17.1 | 9.4e-3 | ||
| 3 | Lys-282 | VDFLQLMIDSQNSK(GlyGly)ETESHKa | 821.745 | 3 | 15.3 | 20.7 | 1.0e-2 |
| VDFLQLM(ox)IDSQNSK(GlyGly)ETESHK | 827.004 | 3 | 12.0 | 20.3 | 2.1e-2 | ||
| VDFLQLM(ox)IDSQNSK(LeuArgGlyGly)ETESHK | 550.483 | 5 | 8.4 | 17.8 | 3.5e-2 | ||
| 4 | Lys-492 | LSLGGLLQPEKPVVLK(GlyGly)VESR | 759.430 | 3 | −6.0 | 16.1 | 3.3e-2 |
a Manually annotated spectra provided in the supplemental materials.
Table VI. List of identified UbcH5a/CHIP-catalyzed CYP3A4 ubiquitination sites.
For experimental details, see “Experimental Procedures.” CYP3A4 peptides were derived from Lys-C/tryptic digests. cm, carbamidomethylation; p, phosphorylation.
| No | Site | Peptide sequence | m/z | z | Mass error (ppm) | Score | Expect val |
|---|---|---|---|---|---|---|---|
| 1 | Lys-127 | SAISIAEDEEWK(GlyGly)Ra | 549.939 | 3 | 4.4 | 20.5 | 6.7e-3 |
| 2 | Lys-168 | EAETGK(GlyGly)PVTLK | 643.856 | 2 | 7.5 | 33.6 | 2.3e-6 |
| REAETGK(LeuArgGlyGly)PVTLK | 428.749 | 4 | −3.0 | 25.6 | 1.5e-3 | ||
| 3 | Lys-173 | EAEpTGKPVTLK(LeuArgGlyGly)DVFGAYSMDVITSTSFGVNIDSLNNPQDPFVENpTK | 1380.420 | 4 | 10 | 23.2 | 0.043 |
| EAETGKPVpTLK(LeuArgGlyGly)DVFGAYSMDVITSTSFGVNIDpSLNNPQDPFVENTK | 789.240 | 7 | 8.9 | 16.1 | 0.050 | ||
| 3 | Lys-466 | VLQNFSFK(GlyGly)PC(cm)Ka | 494.592 | 3 | 2.0 | 32.0 | 3.9e-7 |
| 4 | Lys-487 | EpTQIPLKLpSLGGLLQPEK(GG)PVVLKa | 925.506 | 3 | 5.8 | 23.6 | 8.0e-3 |
| LSLGGLLQPEK(GlyGly)PVVLKVESR | 1138.653 | 2 | 15.8 | 15.8 | 1.0e-2 | ||
| 5 | Lys-492 | LSLGGLLQPEKPVVLK(GlyGly)VESRa | 569.830 | 4 | −15.6 | 15.7 | 1.5e-2 |
a Manually annotated spectra provided in the supplemental materials.
Several CYP3A4 peptides containing detectably ubiquitinated Lys residues (Lys-115, Lys-168, Lys-282, and Lys-492) were isolated from the UBC7/gp78 system (Table V). Similar analyses of UbcH5a/CHIP-ubiquitinated CYP3A4 peptides identified Lys-127, Lys-168, Lys-173, Lys-466, Lys-487, and Lys-492 as target sites (Table VI). Seven of these sites could be verified via LC-MS/MS analyses to release the diagnostic KGG and/or KGGRL tag (Tables V and VI and supplemental data). These findings reveal that although each E2-E3 system ubiquitinates unique Lys residues, their target sites are not mutually exclusive.5 Our LC-MS/MS analyses also enabled us to determine that CYP3A4 polyubiquitination by UBC7/gp78 entailed largely a Lys-48-Ub-Ub linkage (supplemental Fig. S2), whereas that by UbcH5a/CHIP also included Lys-11- and Lys-63-Ub-Ub linkages (data not shown), as previously reported with CYP2E1 (12).
Structural-Functional Validation of CYP3A4 and CYP3A5 Site-specific Mutants
We have previously verified through bona fide structural/functional probes i.e. CO-reduced binding spectra and testosterone 6β-hydroxylase activity (a relatively selective CYP3A functional marker) that mutation of CYP3A4 Thr-264, Ser-478, and/or Ser-420, individually or in combination to Ala, or of CYP3A4 Ser-478 to Asp has no appreciable effect on either its structure or function following functional reconstitution of the recombinant mutant proteins with POR and b5 (11, 44). In this present study, we similarly verified that the structure (CO-reduced binding spectra) and function of CYP3A5 and CYP3A5K264T were comparable. The 7-benzyloxy-4-trifluoromethylcoumarin O-debenzylase activities (6.21 ± 0.25 and 5.94 ± 0.25 pmol of 7-hydroxy-4-trifluoromethylcoumarin formed/pmol P450/min, respectively) of CYP3A5 and CYP3A5K264T in CHAPS-solubilized E. coli membranes, although as expected considerably lower than that of CYP3A4 (11.4 ± 0.12 pmol of 7-hydroxy-4-trifluoromethylcoumarin formed/pmol P450/min), were found to be comparable. Collectively, these findings revealed that this mutation did not appreciably alter CYP3A5 structure or function.
Intracellular Role of Ser-478 Phosphorylation in CYP3A4 UPD/ERAD
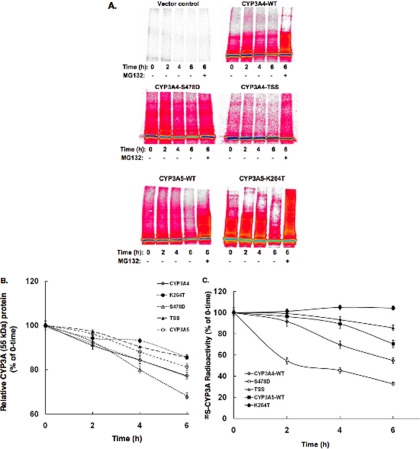
To probe whether CYP3A4 Ser-478 was relevant to its physiological turnover, we carried out 35S pulse-chase analyses of CYP3A4WT, CYP3A4S478D, CYP3A4S478A/T264A/S420A, and CYP3A5WT, expressed along with a control AdV in human HepG2 cells (Fig. 4A). PhosphorImager analyses of immunoprecipitated 35S-CYP3A confirmed our previous finding in HEK293T cells (11) that the turnover of CYP3A4S478A/T264A/S420A was significantly slower than that of CYP3A4WT. Our current findings also revealed that both CYP3A4S478D and CYP3A5WT were indeed ubiquitinated to a greater extent than CYP3A4, as judged by the 35S-CYP3A HMM species ranging from 65 to 250 kDa (Fig. 4A). This ubiquitination profile was further enhanced on proteasomal inhibition by MG132 treatment, consistent with an UPD process. However, whereas the turnover of CYP3A4S478D was accelerated, that of CYP3A5WT, despite its enhanced ubiquitination, was slower than that of CYP3A4WT (Fig. 4B).
Fig. 4.
Pulse-chase analyses of AdV expressed CYP3A4WT, CYP3A5WT, and mutant proteins in HepG2 cells. For details see “Experimental Procedures.” The experiments were conducted in triplicate for each P450 protein. A, a prototypical PhosphorImager scan is shown for each CYP3A protein. Some cells were also treated in parallel with MG132 for 6 h. TSS stands for the CYP3A4T264A/S420A/S478A mutant. The color wheel intensity code is as follows: black > dark blue > light blue > green > yellow > orange > red > magenta > white. B, the time-dependent loss of each 35S-labeled parent CYP3A protein following pulse-chase by densitometric quantification of the corresponding 55-kDa band. The values are the means ± S.D. of three individual HepG2 cultures, each individually immunoprecipitated. The calculated half-life of each parent CYP3A protein was as follows: CYP3A4WT (13.2 h), CYP3A5WT (15.9 h), CYP3A4S478D (9.5 h), TSS (20.3 h), and CYP3A5K264T (22.7 h). C, the time-dependent loss of each 35S-labeled CYP3A protein (parent + ubiquitinated HMM species) following pulse-chase as determined by scintillation counting of aliquots of 35S-CYP3A immunoprecipitates. The values are the means ± S.D. of three individual HepG2 cultures. The 100% (mean ± S.D.) values for 0-h 35S-CYP3A immunoprecipitates were 74943 ± 2780 cpm.
CYP3A5WT exhibits 85% sequence identity to CYP3A4WT (1). Among its divergent residues, CYP3A5 contains Asp-478 and Lys-264 instead of Ser-478 and Thr-264, two residues whose phosphorylation along with that of Ser-420 is relevant for CYP3A4 ERAD (Ref. 11 and Fig. 4A). To determine whether the lack of Thr-264 in CYP3A5 affects its turnover, the CYP3A5K264T mutant that contains both Thr-264 and Asp-478 residues was also included in the relative 35S-CYP3A turnover analysis. CYP3A5 ubiquitination was clearly further enhanced by the K264T mutation, as revealed by the HMM 35S-CYP3A species. This is consistent with the relative importance of both Asp-478 and Thr-264 residues to CYP3A ubiquitination. However, despite its enhanced ubiquitination relative to that of CYP3A4WT or CYP3A5WT, the CYP3A5K264T mutant was not degraded any faster than CYP3A4WT and was even proteolytically more stable than CYP3A5WT (Fig. 4B). The observed enhanced stabilization of the ubiquitinated 35S-CYP3A5K264T HMM species upon MG132 treatment indicated that its degradation was also UPD-dependent.
DISCUSSION
Proteomic analyses of in vitro phosphorylated CYP3A4 revealed that five of its residues (Ser-116, Ser-119, Ser-134, Ser-259, and Ser-478) are phosphorylated by PKA, and 14 residues (Thr-92, Ser-100, Thr-103, Ser-116, Ser-119, Ser-131, Ser-134, Thr-136, Ser-139, Ser-259, Thr-264, Thr-84, Ser-398, and Ser-420) by PKC. This is the first identification of CYP3A4 phosphorylation sites other than Ser-134 (16) and Thr-264, Ser-420, and Ser-478 (10, 11), which were previously identified by LC-MS/MS analyses.6 Some of these sites (Ser-116, Ser-119, Ser-259, and Ser-398) are detectably phosphorylated only after CuOOH-elicited inactivation of the enzyme, which may structurally unravel the P450 protein and render these sites PKA/PKC-accessible. However, structural analyses reveal that the specific residues phosphorylated in the native CYP3A4 by either PKA or PKC are not only different but are also located in distinct topological domains.
We have previously shown that CYP3A4 phosphorylation at all three (Ser-478, Thr-264, and Ser-420) sites greatly enhances and accelerates its UBC7/gp78-mediated ubiquitination, whereas the simultaneous Ala mutation of these three residues greatly reduces it (11). Our current findings single out Ser-478 as the most important of these residues for UBC7/gp78-mediated CYP3A4 ubiquitination, despite the low extent of its PKA-mediated phosphorylation. Although such an in vitro CYP3A4 Ser-478 phosphorylation by the PKA catalytic subunit may be deemed physiologically irrelevant, it is noteworthy that the corresponding CYP3A23 Ser-479 residue is also phosphorylated intracellularly to a significant extent in cultured rat hepatocytes. Furthermore, a specific PKA inhibitor (KT-5720) significantly (p ≤ 0.01) decreased both CYP3A ubiquitination and degradation (Fig. 2, B and C). Thus, if not PKA itself, then a liver cytosolic PKA-like kinase(s) (other than PKC) is clearly involved. Nevertheless, we emphasize that such a low extent (1.29 ± 0.12%) of CYP3A4 Ser-478 phosphorylation is entirely consistent with the comparably low extent of P450 protein ubiquitination and its relatively slow pace. Accordingly, only 1–3% of P450s are ubiquitinated at any given time (data not shown), and thus detection of ubiquitinated P450 species requires 32P-labeled Ub and/or sensitive immunoprecipitation/immunoblotting analyses. Equally noteworthy in this context is that the particular site of P450 phosphorylation rather than its overall extent may be the relevant determinant of P450 UPD. For instance, we have reported that CYP2E1S129 located in a well defined RRFS129 PKA recognition motif is phosphorylated by >98% in the native enzyme (12). However, despite this large extent of phosphorylation, its site-directed mutation to the nonphosphorylatable Ala or Gly residue fails to stabilize the enzyme by inhibiting its proteolytic degradation, even though each mutant is just as functionally active as the parent CYP2E1 (45). Further support for this concept is provided by our findings that Ser-420, a residue phosphorylated to a much greater extent (14.1 ± 0.28%) than Ser-478 in the CuOOH-inactivated CYP3A4 (Table IV), contributes only minimally to CYP3A4 UPD relative to Ser-478 (11).
The relevance of Ser-478 phosphorylation to UBC7/gp78-mediated CYP3A4 ubiquitination is further underscored by our finding that the mutation of CYP3A4 Ser-478 to the phosphomimetic Asp residue greatly accelerated/enhanced this process. By contrast, its Ala mutation significantly reduced this modification, whereas the ALA mutation of both Thr-264 and Ser-420 in the absence of the kinases had very little effect relative to that of CYP3A4WT. However, in the presence of both kinases, the ubiquitination of the WT enzyme was noticeably greater than that of the T264A/S420A mutant, thereby revealing an appreciable influence of Thr-264 and Ser-420 phosphorylation on this process. It is to be noted that whereas PKA phosphorylates Ser-478, PKC phosphorylates both Thr-264 and Ser-420 residues. Thus, these modifications may be complementary and account for the observed PKA/PKC synergistic enhancement of UBC7/gp78-mediated CYP3A4 ubiquitination. Furthermore, we found that CYP3A5, a natural S478D mutant, is constitutively ubiquitinated to a greater extent than CYP3A4 in HepG2 cells (Fig. 4A). This CYP3A5 ubiquitination extent is further augmented by its K264T mutation, thereby revealing the relative importance to this process of a phosphorylatable residue at this site.
Our comprehensive proteomic analyses of PKA/PKC-mediated CYP3A4 phosphorylation have identified several additional residues (Thr-92, Ser-100, Thr-103, Ser-116, Ser-119, Ser-131, Ser-134, Thr-136, Ser-139, and Thr-284), some phosphorylated to a very significant extent in the native protein and others (Ser-259 and Ser-398) following CuOOH-mediated CYP3A4 inactivation (Tables II and IV). The role of these newly identified phosphorylation sites, if any, is unknown. However, our analyses of UbcH5a/CHIP-mediated CYP3A4 ubiquitination reveal that although this reaction was also enhanced by its PKA/PKC-mediated phosphorylation (Fig. 3A), Ser-478, Thr-264, and Ser-420 can be excluded as relevant sites in this process (Fig. 3B). Thus, it is conceivable that one or more of these additional residues may similarly promote CYP3A4 interactions with CHIP and/or Hsp70, its co-chaperone partner. Furthermore, such an enhancement by CYP3A4 protein phosphorylation may reconcile the discrepancy in our previous findings of the very feeble role of UbcH5a/CHIP in CYP3A4 ubiquitination observed in vitro in the absence of any added PKA/PKC (13) and the essential role of CHIP in CYP3A4 ERAD established in cultured hepatocytes through RNA interference (14).
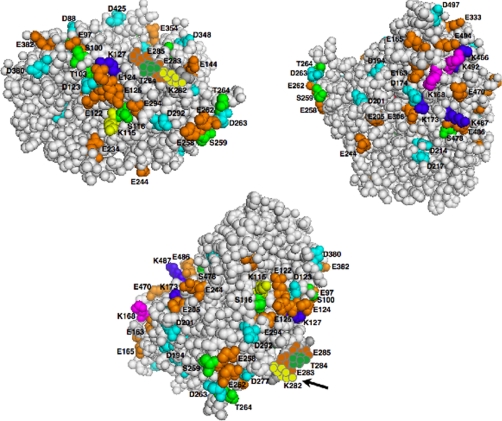
Inspection of the CYP3A4 structure reveals that the phosphorylated Ser/Thr residues in this protein reside on its surface within a cluster of acidic (Asp/Glu) residues (Fig. 5). Thus, Ser/Thr phosphorylation would further enhance the negatively charged character of this domain by imparting additional negative charges within this cluster. Accordingly, we propose that such acidic patches may indeed be important for ERAD substrate recognition by corresponding basic residues/domains in the E3 Ub ligases and/or E2 Ubcs. Thus, Ser/Thr phosphorylation by filling in the missing gaps in these negatively charged clusters could control the timing of CYP3A4 recognition by UBC7/gp78 and/or UbcH5a/CHIP and thus its turnover.
Fig. 5.
PyMol depiction of CYP3A4 Lys residues ubiquitinated by UBC7/gp78 and/or UbcH5a/CHIP. Three different CYP3A4 structural orientations (59) are shown with UBC7/gp78-ubiquitinated Lys residues in yellow, UbcH5a/CHIP-ubiquitinated Lys residues in blue, and those ubiquitinated by both systems in magenta. These ubiquitinated Lys residues lie within spatially associated surface clusters of phosphorylated Ser/Thr residues in green, Glu residues in orange, and Asp residues in cyan. The arrow indicates the disordered Lys-282–Glu-283–Thr-284–Glu-285 region not featured in the CYP3A4 crystal structure (59) within which UBC7/gp78-ubiquitinated Lys-282 resides. Note that Thr-284 is highly phosphorylated in the native protein and even more so following CuOOH inactivation.
Multisite protein phosphorylation within discrete conserved primary sequence motifs known as phosphodegrons is a common feature of target substrate recognition by the SCF (SKP1-CUL1-F-Box protein complex) E3 Ub-ligases (46–48). On the other hand, CHIP substrate recognition is known to involve “distributed phosphodegrons,” consisting of phosphorylated residues dispersed over the entire protein sequence (49–51). Such phosphodegrons apparently function cooperatively in a highly specific phosphorylation- and sequence-dependent manner in CHIP-mediated substrate recruitment (49–51). Thus, the observed multisite protein phosphorylation within spatially associated negatively charged surface clusters in CYP3A4 (discussed above) and in CYP2E1 (12) could represent distributed surface phosphodegrons for the recognition of phosphorylated P450s as CHIP target substrates.
By contrast, gp78 substrate recognition is mechanistically quite versatile, apparently involving different gp78 topological domains and/or various determinants (52). However, to our knowledge, none of the extant gp78 substrate recognition mechanisms has been shown to involve substrate protein phosphorylation (52–58). Thus, we believe the observed striking enhancement of gp78-mediated ubiquitination by P450 protein phosphorylation at a single residue (Ser-478) introduces a novel dimension to the mechanistic diversity of gp78 substrate recognition. We find it intriguing that Ser-478 is apparently strategically situated within the CYP3A4 C-terminal loop (residues 475–485) (59), a region on the enzyme surface that is thought to exhibit “pronounced structural plasticity” (60). Molecular dynamics simulation reveals that phosphorylation or mutation of Ser-478 is unlikely to have a large conformational effect on the protein, because this C-terminal loop is relatively mobile (supplemental Fig. S4). This evidence, in addition to our findings that Ser-478 mutants are fully functional (11), suggests that mutating this residue would not result in significant conformational changes at the protein-wide level. CYP3A4 structural analyses reveal that Ser-478 forms a polar surface patch with Gln-484 and Glu-486 (supplemental Fig. S4). S478A would reduce the hydrophilicity and size of this patch relative to the WT enzyme, thus resulting in a less surface exposed residue at position 478. By contrast, S478D (and by inference, the Ser-478-phosphorylated CYP3A4) has increased hydrophilicity and size relative to the WT enzyme, and thus this mutant would likely exhibit a larger polar surface patch and a more surface-exposed residue at position 478. Thus, it is not surprising that the interactions between the S478A mutant and UBC7/gp78 would be weaker, thereby accounting for its weaker ubiquitination profile. On the other hand, Ser-478 phosphorylation would strengthen these interactions, thereby enabling the UBC7/gp78 ubiquitination of Lys residues (i.e. Lys-492) in its vicinity.
We find it compelling that in addition to phosphorylatable residues, two consecutive acidic residues (Glu and Asp) at positions −2 and −1 of an ubiquitinatable Lys residue (position 0) are found in yeast integral membrane proteins (61). Furthermore, lysines in KEEE or KNEEEE motifs of HMGCoA (3-hydroxy-3-methylglutaryl coenzyme A) reductase are also found to be preferentially ubiquitinated by UBC7/gp78 (62). Intriguingly, UBC7/gp78-ubiquitinated CYP3A4 Lys-115 resides within a spatially associated surface cluster of acidic/phosphorylatable residues (Fig. 5). CYP3A4 Lys-282 present in a “disordered” region not featured in the crystal structure (59) is flanked by Ser-281 and Glu-283–Thr-284-Glu-285–Ser-286 and is also in the very close vicinity of another negatively charged cluster, Glu-258–Ser-259–Glu-262–Asp-263–Thr-264 (Fig. 5). It is noteworthy that CYP3A4 Ser-259, Thr-264, and Thr-284 within these clusters are very highly phosphorylated by PKC, which would increase the negative charge of their respective clusters (Table IV). Similar analyses of every gp78- and CHIP-ubiquitinated CYP3A4 and CYP2E1 (12) Lys residue reveal its location within or in close proximity of an Glu/Asp/Ser/Thr surface cluster. Thus, it is conceivable that these spatially associated negatively charged clusters represent “conformational phosphodegrons” that are important for P450 recognition by UBC7/gp78 and UbcH5a/CHIP E2/E3 ubiquitination complexes.
Our findings described above reveal that CYP3A4 is phosphorylated at multiple sites by PKA and PKC. Of these sites, Ser-478 is clearly important for its UBC7/gp78-mediated ubiquitination. However, pulse-chase analyses of CYP3A5, a CYP3A4 isoform that contains the phosphomimetic Asp-478 residue, and of its K264T mutant revealed that although these proteins are indeed ubiquitinated intracellularly to a much greater extent than CYP3A4WT, this is insufficient for their faster turnover via ERAD (Fig. 4). Thus, although our finding with the CYP3A4T264A/S420A/S478A mutant indicates that the phosphorylation of these residues is clearly essential for CYP3A4 ERAD, additional determinants must exist and remain to be elucidated. They may include (i) CYP3A4 structural motifs/“degrons” or phosphodegrons not present in CYP3A5 and/or (ii) sufficiently high catalytic turnover for significant futile catalytic uncoupling and consequent oxidative/structural damage to mark the protein for UPD/ERAD. Nevertheless, the detection of significant constitutive ubiquitination of CYP3A5 under physiological conditions we find noteworthy. Such a CYP3A5 post-translational modification may further contribute to its functional sluggishness in vivo and may be therapeutically relevant in individuals that polymorphically express CYP3A5 as their dominant hepatic drug-metabolizing CYP3A enzyme (63, 64). Together, our findings reveal that post-translational multisite phosphorylation may enhance the susceptibility of a P450 protein for ubiquitination by both gp78 and CHIP E3 ligases and may thus control clinically relevant hepatic P450 content.
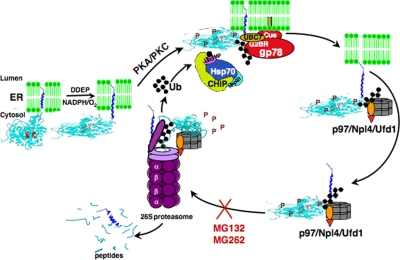
Collectively, our findings above together with our previous reports (5–14) led us to propose a mechanistic scheme for CYP3A ERAD/UPD (Fig. 6), wherein the P450 protein requires to be first phosphorylated by both PKA (or a PKA-like kinase) and PKC on a distinct set of residues, for its subsequent recognition by UbcH5a/CHIP/Hsp70 and UBC7/gp78 E2-E3 ubiquitination complexes, following which the ubiquitinated protein is extracted out of the ER by the p97/Ufd1/Npl4 AAA ATPase complex and delivered to the 26 S proteasome for degradation.
Fig. 6.
Mechanistic scheme for CYP3A ERAD/UPD.
Acknowledgments
We sincerely thank Dr. A. M. Weissman (NCI, National Institutes of Health) for the UBC7 and gp78C expression plasmids, Dr. Cam Patterson (University of North Carolina) for the CHIP expression plasmid, Dr. L. Waskell (University of Michigan) for the b5 expression plasmid, and Dr. C. B. Kasper (University of Wisconsin) for the POR expression plasmid. We gratefully acknowledge St. Jude Children's Research Hospital Vector Core Facility (Memphis, TN) for the AdV production. We also thank Ahalya Sriskandarajan for technical assistance in some studies and Chris Her (UCSF Liver Center Cell and Tissue Biology Core Facility) for rat hepatocyte isolation.
Footnotes
* These studies were supported by National Institutes of Health Grants GM44037 (to M. A. C.), DK26506 (to M. A. C.), NCRRRR 01614 (to A. L. B.), and GM09441 (to E. S.). The UCSF Liver Center Core on Cell and Tissue Biology was supported by National Institutes of Health NIDDK Center Grant P30DK26743. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental materials.
This article contains supplemental materials.
2 Y. Q. Wang and M. A. Correia, unpublished observations.
3 In addition to Ser-479, our proteomic analyses of Lys-C/tryptic digests from cultured rat hepatocytes have uncovered several phosphorylated sites in the orthologous rat liver CYPs 3A (Thr-136, Thr-138, Ser-263, and Ser-427) that are in the same structural regions as the corresponding phosphorylated Ser/Thr residues of CYP3A4 (Tables I–IV).
4 Our findings (supplemental Fig. S3) exclude the involvement of any intrinsic PKA/PKC-mediated activation of the ubiquitination machinery and/or its individual components (E1, E2, and/or E3) in the observed PKA/PKC-mediated enhancement of CYP3A4 ubiquitination, thus further substantiating a role for target substrate phosphorylation.
5 Indeed, our preliminary proteomic analyses reveal that Lys-168, an ubiquitination target of both UBC7/gp78 and UbcH5a/CHIP in vitro, is also found to be ubiquitinated in the orthologous rat liver CYPs 3A in cultured rat hepatocytes.
6 PKA has been previously reported to phosphorylate Ser-393 of CYP3A23 in vitro (30). However, the corresponding CYP3A4 residue is a Trp.
1 The abbreviations used are:
- P450s or CYPs
- cytochromes P450
- b5
- cytochrome b5
- CuOOH
- cumene hydroperoxide
- DDEP
- 3,5-dicarbethoxy-2,6-trimethyl-4-ethyl-1,4-dihydropyridine
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- HMM
- high molecular mass
- Lys-C
- lysylendopeptidase C
- PKA
- cAMP-dependent protein kinase A
- PKC
- protein kinase C
- POR
- NADPH-P450 oxidoreductase
- Ub
- ubiquitin
- UBC
- genes for Ub-conjugating enzymes
- UPD
- Ub-dependent proteasomal degradation
- AdV
- adenoviral vector(s)
- WT
- wild type
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- BisIII
- bisindolylmaleimide III.
REFERENCES
- 1. Guengerich F. P. (2005) Human cytochrome P450 enzymes in Cytochrome P450: Structure, Mechanism and Biochemistry (Ortiz de Montellano P., ed) pp. 377–530, Kluwer-Academic/Plenum Press, New York [Google Scholar]
- 2. Correia M. A. (2003) Hepatic cytochrome P450 degradation: Mechanistic diversity of the cellular sanitation brigade. Drug Metab. Rev. 35, 107–143 [DOI] [PubMed] [Google Scholar]
- 3. Correia M. A., Sadeghi S., Mundo-Paredes E. (2005) Cytochrome P450 ubiquitination: Branding for the proteolytic slaughter? Annu. Rev. Pharmacol. Toxicol. 45, 439–464 [DOI] [PubMed] [Google Scholar]
- 4. Correia M. A., Liao M. (2007) Cellular proteolytic systems in P450 degradation: Evolutionary conservation from Saccharomyces cerevisiae to mammalian liver. Expert Opin. Drug Metab. Toxicol. 3, 33–49 [DOI] [PubMed] [Google Scholar]
- 5. Correia M. A., Davoll S. H., Wrighton S. A., Thomas P. E. (1992) Degradation of rat liver cytochromes P450 3A after their inactivation by 3,5-dicarbethoxy-2,6-dimethyl-4-ethyl-1,4-dihydropyridine: Characterization of the proteolytic system. Arch. Biochem. Biophys. 297, 228–238 [DOI] [PubMed] [Google Scholar]
- 6. Wang H. F., Figueiredo Pereira M. E., Correia M. A. (1999) Cytochrome P450 3A degradation in isolated rat hepatocytes: 26S proteasome inhibitors as probes. Arch. Biochem. Biophys. 365, 45–53 [DOI] [PubMed] [Google Scholar]
- 7. Faouzi S., Medzihradszky K. F., Hefner C., Maher J. J., Correia M. A. (2007) Characterization of the physiological turnover of native and inactivated cytochromes P450 3A in cultured rat hepatocytes: A role for the cytosolic AAA ATPase p97? Biochemistry 46, 7793–7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Acharya P., Liao M., Engel J. C., Correia M. A. (2011) Liver cytochrome P450 3A endoplasmic reticulum-associated degradation: A major role for the p97 AAA ATPase in cytochrome P450 3A extraction into the cytosol. J. Biol. Chem. 286, 3815–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Korsmeyer K. K., Davoll S., Figueiredo-Pereira M. E., Correia M. A. (1999) Proteolytic degradation of heme-modified hepatic cytochromes P450: A role for phosphorylation, ubiquitination, and the 26S proteasome? Arch. Biochem. Biophys. 365, 31–44 [DOI] [PubMed] [Google Scholar]
- 10. Wang X., Medzihradszky K. F., Maltby D., Correia M. A. (2001) Phosphorylation of native and heme-modified CYP3A4 by protein kinase C: A mass spectrometric characterization of the phosphorylated peptides. Biochemistry 40, 11318–11326 [DOI] [PubMed] [Google Scholar]
- 11. Wang Y., Liao M., Hoe N., Acharya P., Deng C., Krutchinsky A. N., Correia M. A. (2009) A role for protein phosphorylation in cytochrome P450 3A4 ubiquitin-dependent proteasomal degradation. J. Biol. Chem. 284, 5671–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y., Guan S., Acharya P., Koop D. R., Liu Y., Liao M., Burlingame A. L., Correia M. A. (2011) The ubiquitin-dependent proteasomal degradation (UPD) of human liver cytochrome P450 2E1: Identification of sites targeted for phosphorylation and ubiquitination. J. Biol. Chem. 286, 9443–9456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pabarcus M. K., Hoe N., Sadeghi S., Patterson C., Wiertz E., Correia M. A. (2009) CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases. Arch. Biochem. Biophys. 483, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim S. M., Acharya P., Engel J. C., Correia M. A. (2010) Liver cytochrome P450 3A ubiquitination in vivo by GP78/autocrine motility factor receptor (AMFR) and C terminus of HSP70-interacting protein (chip) E3 ubiquitin ligases: Physiological and pharmacological relevance. J. Biol. Chem. 285, 35866–35877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aguiar M., Masse R., Gibbs B. F. (2005) Regulation of cytochrome P450 by posttranslational modification. Drug Metab. Rev. 37, 379–404 [DOI] [PubMed] [Google Scholar]
- 16. Redlich G., Zanger U. M., Riedmaier S., Bache N., Giessing A. B., Eisenacher M., Stephan C., Meyer H. E., Jensen O. N., Marcus K. (2008) Distinction between human cytochrome P450 (CYP) isoforms and identification of new phosphorylation sites by mass spectrometry. J. Proteome Res. 7, 4678–4688 [DOI] [PubMed] [Google Scholar]
- 17. Weiner M., Buterbaugh G. G., Blake D. A. (1972) Inhibition of hepatic drug metabolism by cyclic 3′, 5′-adenosine monophosphate. Res. Commun. Chem. Pathol. Pharmacol. 3, 249–263 [PubMed] [Google Scholar]
- 18. Ross W. E., Simrell C., Oppelt W. W. (1973) Sex-dependent effects of cyclic AMP on the hepatic mixed function oxidase system. Res. Commun. Chem. Pathol. Pharmacol. 5, 319–332 [PubMed] [Google Scholar]
- 19. Sanghvi A., Grassi E., Warty V., Diven W., Wight C., Lester R. (1981) Reversible activation-inactivation of cholesterol 7α-hydroxylase possibly due to phosphorylation-dephosphorylation. Biochem. Biophys. Res. Commun. 103, 886–892 [DOI] [PubMed] [Google Scholar]
- 20. Pyerin W., Wolf C. R., Kinzel V., Kübler D., Oesch F. (1983) Phosphorylation of cytochrome-P450-dependent monooxygenase components. Carcinogenesis 4, 573–576 [DOI] [PubMed] [Google Scholar]
- 21. Müller R., Schmidt W. E., Stier A. (1985) The site of cyclic AMP-dependent protein kinase catalyzed phosphorylation of cytochrome P450 LM2. FEBS Lett. 187, 21–24 [DOI] [PubMed] [Google Scholar]
- 22. Koch J. A., Waxman D. J. (1989) Posttranslational modification of hepatic cytochrome P450: Phosphorylation of phenobarbital-inducible P450 forms PB-4 (IIB1) and PB-5 (IIB2) in isolated rat hepatocytes and in vivo. Biochemistry 28, 3145–3152 [DOI] [PubMed] [Google Scholar]
- 23. Menez J. F., Machu T. K., Song B. J., Browning M. D., Deitrich R. A. (1993) Phosphorylation of cytochrome P4502E1 (CYP2E1) by calmodulin dependent protein kinase, protein kinase C and cAMP dependent protein kinase. Alcohol Alcohol. 28, 445–451 [PubMed] [Google Scholar]
- 24. Löhr J. B., Kühn-Velten W. N. (1997) Protein phosphorylation changes ligand-binding efficiency of cytochrome P450c17 (CYP17) and accelerates its proteolytic degradation: putative relevance for hormonal regulation of CYP17 activity. Biochem. Biophys. Res. Commun. 231, 403–408 [DOI] [PubMed] [Google Scholar]
- 25. Oesch-Bartlomowicz B., Oesch F. (2005) Phosphorylation of cytochromes P450: First discovery of a posttranslational modification of a drug-metabolizing enzyme. Biochem. Biophys. Res. Commun. 338, 446–449 [DOI] [PubMed] [Google Scholar]
- 26. Anandatheerthavarada H. K., Biswas G., Mullick J., Sepuri N. B., Otvos L., Pain D., Avadhani N. G. (1999) Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP-dependent phosphorylation at Ser128. EMBO J. 18, 5494–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robin M. A., Anandatheerthavarada H. K., Biswas G., Sepuri N. B., Gordon D. M., Pain D., Avadhani N. G. (2002) Bimodal targeting of microsomal CYP2E1 to mitochondria through activation of an N-terminal chimeric signal by cAMP-mediated phosphorylation. J. Biol. Chem. 277, 40583–40593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sepuri N. B., Yadav S., Anandatheerthavarada H. K., Avadhani N. G. (2007) Mitochondrial targeting of intact CYP2B1 and CYP2E1 and N-terminal truncated CYP1A1 proteins in Saccharomyces cerevisiae: Role of protein kinase A in the mitochondrial targeting of CYP2E1. FEBS J. 274, 4615–4630 [DOI] [PubMed] [Google Scholar]
- 29. Eliasson E., Johansson I., Ingelman-Sundberg M. (1990) Substrate-, hormone-, and cAMP-regulated cytochrome P450 degradation. Proc. Natl. Acad. Sci. U.S.A. 87, 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eliasson E., Mkrtchian S., Halpert J. R., Ingelman-Sundberg M. (1994) Substrate-regulated, cAMP-dependent phosphorylation, denaturation, and degradation of glucocorticoid-inducible rat liver cytochrome P450 3A1. J. Biol. Chem. 269, 18378–18383 [PubMed] [Google Scholar]
- 31. Zhukov A., Werlinder V., Ingelman-Sundberg M. (1993) Purification and characterization of two membrane bound serine proteinases from rat liver microsomes active in degradation of cytochrome P450. Biochem. Biophys. Res. Commun. 197, 221–228 [DOI] [PubMed] [Google Scholar]
- 32. Pearce R. E., Lu W., Wang Y., Uetrecht J. P., Correia M. A., Leeder J. S. (2008) Pathways of carbamazepine bioactivation in vitro: III. The role of human cytochrome P450 enzymes in the formation of 2,3-dihydroxycarbamazepine. Drug Metab. Dispos. 36, 1637–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brimer C., Dalton J. T., Zhu Z., Schuetz J., Yasuda K., Vanin E., Relling M. V., Lu Y., Schuetz E. G. (2000) Creation of polarized cells coexpressing CYP3A4, NADPH cytochrome P450 reductase and MDR1/P-glycoprotein. Pharm. Res. 17, 803–810 [DOI] [PubMed] [Google Scholar]
- 34. Brimer-Cline C., Schuetz E. G. (2002) Polarized cell cultures for integrated studies of drug metabolism and transport. Methods Enzymol. 357, 321–329 [DOI] [PubMed] [Google Scholar]
- 35. Cheng Q., Sohl C. D., Guengerich F. P. (2009) High-throughput fluorescence assay of cytochrome P450 3A4. Nat. Protoc. 4, 1258–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chalkley R. J., Baker P. R., Medzihradszky K. F., Lynn A. J., Burlingame A. L. (2008) In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell Proteomics 7, 2386–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clauser K. R., Baker P., Burlingame A. L. (1999) Role of accurate mass measurement (+/- 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71, 2871–2882 [DOI] [PubMed] [Google Scholar]
- 38. Balgley B. M., Laudeman T., Yang L., Song T., Lee C. S. (2007) Comparative evaluation of tandem MS search algorithms using a target-decoy search strategy. Mol. Cell Proteomics 6, 1599–1608 [DOI] [PubMed] [Google Scholar]
- 39. Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 [DOI] [PubMed] [Google Scholar]
- 40. Steen H., Jebanathirajah J. A., Springer M., Kirschner M. W. (2005) Stable isotope-free relative and absolute quantitation of protein phosphorylation stoichiometry by MS. Proc. Natl. Acad. Sci. U.S.A. 102, 3948–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steen J. A., Steen H., Georgi A., Parker K., Springer M., Kirchner M., Hamprecht F., Kirschner M. W. (2008) Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: A quantitative proteomic analysis. Proc. Natl. Acad. Sci. U.S.A. 105, 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nielsen M. L., Vermeulen M., Bonaldi T., Cox J., Moroder L., Mann M. (2008) Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods 5, 459–460 [DOI] [PubMed] [Google Scholar]
- 43. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 44. Wang H., Dick R., Yin H., Licad-Coles E., Kroetz D. L., Szklarz G., Harlow G., Halpert J. R., Correia M. A. (1998) Structure-function relationships of human liver cytochromes P450 3A: Aflatoxin B1 metabolism as a probe. Biochemistry 37, 12536–12545 [DOI] [PubMed] [Google Scholar]
- 45. Freeman J. E., Wolf C. R. (1994) Evidence against a role for serine 129 in determining murine cytochrome P450 Cyp2E1 protein levels. Biochemistry 33, 13963–13966 [DOI] [PubMed] [Google Scholar]
- 46. Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007) Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 [DOI] [PubMed] [Google Scholar]
- 47. Reed S. I. (2006) The ubiquitin-proteasome pathway in cell cycle control. Results Probl. Cell Differ. 42, 147–181 [DOI] [PubMed] [Google Scholar]
- 48. Klotz K., Cepeda D., Tan Y., Sun D., Sangfelt O., Spruck C. (2009) SCF(Fbxw7/hCdc4) targets cyclin E2 for ubiquitin-dependent proteolysis. Exp. Cell Res. 315, 1832–1839 [DOI] [PubMed] [Google Scholar]
- 49. Cripps D., Thomas S. N., Jeng Y., Yang F., Davies P., Yang A. J. (2006) Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J. Biol. Chem. 281, 10825–10838 [DOI] [PubMed] [Google Scholar]
- 50. Dickey C. A., Kamal A., Lundgren K., Klosak N., Bailey R. M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C. B., Patterson C., Dickson D. W., Nahman N. S., Jr., Hutton M., Burrows F., Petrucelli L. (2007) The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 117, 648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rees I., Lee S., Kim H., Tsai F. T. (2006) The E3 ubiquitin ligase CHIP binds the androgen receptor in a phosphorylation-dependent manner. Biochim. Biophys. Acta 1764, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 52. Tsai Y. C., Weissman A. M. The unfolded protein response, degradation from endoplasmic reticulum and cancer. Genes Cancer 1, 764–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. (2006) The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. U.S.A. 103, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. (2004) AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J. Biol. Chem. 279, 45676–45684 [DOI] [PubMed] [Google Scholar]
- 56. Song B. L., Sever N., DeBose-Boyd R. A. (2005) Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 57. Shen Y., Ballar P., Fang S. (2006) Ubiquitin ligase gp78 increases solubility and facilitates degradation of the Z variant of α-1-antitrypsin. Biochem. Biophys. Res. Commun. 349, 1285–1293 [DOI] [PubMed] [Google Scholar]
- 58. Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., Nagata K. (2008) Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol. Biol. Cell 19, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yano J. K., Wester M. R., Schoch G. A., Griffin K. J., Stout C. D., Johnson E. F. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J. Biol. Chem. 279, 38091–38094 [DOI] [PubMed] [Google Scholar]
- 60. Ekroos M., Sjögren T. (2006) Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Catic A., Collins C., Church G. M., Ploegh H. L. (2004) Preferred in vivo ubiquitination sites. Bioinformatics 20, 3302–3307 [DOI] [PubMed] [Google Scholar]
- 62. Miao H., Jiang W., Ge L., Li B., Song B. (2010) Tetra-glutamic acid residues adjacent to Lys248 in HMG-CoA reductase are critical for the ubiquitination mediated by gp78 and UBE2G2. Acta Biochim. Biophys. Sin. 42, 303–310 [DOI] [PubMed] [Google Scholar]
- 63. Kuehl P., Zhang J., Lin Y., Lamba J., Assem M., Schuetz J., Watkins P. B., Daly A., Wrighton S. A., Hall S. D., Maurel P., Relling M., Brimer C., Yasuda K., Venkataramanan R., Strom S., Thummel K., Boguski M. S., Schuetz E. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 27, 383–391 [DOI] [PubMed] [Google Scholar]
- 64. Hustert E., Haberl M., Burk O., Wolbold R., He Y. Q., Klein K., Nuessler A. C., Neuhaus P., Klattig J., Eiselt R., Koch I., Zibat A., Brockmöller J., Halpert J. R., Zanger U. M., Wojnowski L. (2001) The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11, 773–779 [DOI] [PubMed] [Google Scholar]