Abstract
Expression of caveolin-1 is up-regulated in prostate cancer metastasis and is associated with aggressive recurrence of the disease. Intriguingly, caveolin-1 is also secreted from prostate cancer cell lines and has been identified in secreted prostasomes. Caveolin-1 is the major structural component of the plasma membrane invaginations called caveolae. Co-expression of the coat protein Polymerase I and transcript release factor (PTRF) is required for caveolae formation. We recently found that expression of caveolin-1 in the aggressive prostate cancer cell line PC-3 is not accompanied by PTRF, leading to noncaveolar caveolin-1 lipid rafts. Moreover, ectopic expression of PTRF in PC-3 cells sequesters caveolin-1 into caveolae. Here we quantitatively analyzed the effect of PTRF expression on the PC-3 proteome using stable isotope labeling by amino acids in culture and subcellular proteomics. We show that PTRF reduced the secretion of a subset of proteins including secreted proteases, cytokines, and growth regulatory proteins, partly via a reduction in prostasome secretion. To determine the cellular mechanism accounting for the observed reduction in secreted proteins we analyzed total membrane and the detergent-resistant membrane fractions. Our data show that PTRF expression selectively impaired the recruitment of actin cytoskeletal proteins to the detergent-resistant membrane, which correlated with altered cholesterol distribution in PC-3 cells expressing PTRF. Consistent with this, modulating cellular cholesterol altered the actin cytoskeleton and protein secretion in PC-3 cells. Intriguingly, several proteins that function in ER to Golgi trafficking were reduced by PTRF expression. Taken together, these results suggest that the noncaveolar caveolin-1 found in prostate cancer cells generates a lipid raft microenvironment that accentuates secretion pathways, possibly at the step of ER sorting/exit. Importantly, these effects could be modulated by PTRF expression.
Prostate cancer is the most commonly diagnosed cancer in men and the second leading cause of cancer related deaths in developed countries. Although localized prostate cancer is treatable, metastatic recurrence, together with development of androgen-independence, leads to advanced prostate cancer, which currently has a low survival rate. Caveolin-1, a cholesterol-binding integral membrane protein, has been shown to be up-regulated in prostate cancer metastasis and is associated with androgen-independence and aggressive recurrence of the disease (1). A prostate cancer mouse model showed that genetic ablation of caveolin-1 delays the onset of advanced prostate cancer (2), underscoring its importance in prostate cancer progression. Hence understanding caveolin-1 action in prostate cancer progression is crucial for the design of novel intervention strategies to manage this devastating disease.
Caveolin-1 is a major structural component of caveolae, specialized lipid raft microdomains of the plasma membrane characterized by their flask shaped invaginations (3). Lipid rafts and caveolae are thought to participate in a variety of cellular processes including lipid regulation, endocytosis, cell adhesion, and signal transduction (4). However, a clear delineation between lipid rafts and caveolae function remains to be made. Recently, it was revealed that the protein Polymerase I and transcript release factor (PTRF)1, also known as cavin-1, is an essential cofactor required for stabilization of caveolae at the plasma membrane (5, 6). The loss of PTRF results in the loss of caveolae, as well as a decrease in caveolin-1 stability (5). These results demonstrate a crucial role for PTRF in caveolae structure and function. Given these new insights, the role of PTRF in relation to its function in caveolae is only beginning to be examined in prostate cancer (7).
Intriguingly, although caveolin-1 is predominately a membrane protein, aggressive prostate cancer cell lines have been shown to secrete biologically active caveolin-1 (8). Furthermore, secreted caveolin-1 stimulates cell growth and angiogenesis in tumor models (8, 9). Serum caveolin-1 has been detected in recurrent prostate cancer after radical prostatectomy and has been proposed to be a marker for disease recurrence (8, 10). In addition to classical secretion, prostate cells also secrete proteins via membranous storage vesicles referred to as prostasomes. It is thought that prostasomes are formed in a similar manner to exosomes, that is, via multivesicular bodies that have encapsulated cytoplasmic proteins (11). Prostate epithelial cells secrete prostasomes into the prostate fluid where they have important functions related to fertility. More recently it has become apparent that prostate cancer cell lines also secrete prostasomes and it is thought they may act to modulate the local tumor environment (12–14). Prostasome secretion is modulated by cholesterol levels and caveolin-1 has been identified as a component of prostasomes in the human prostate carcinoma PC-3 cell line (13). These data have prompted us to examine the role of caveolin-1 and PTRF in secretion of prostate cancer cells.
We have previously shown that PC-3 cells express abundant caveolin-1 but lack PTRF, and that ectopic expression of PTRF in PC-3 cells results in caveolae formation (5). Furthermore, PTRF expression in PC-3 cells resulted in reduced directional migration, partly because of attenuation of secreted MMP9 (15). Given that caveolin-1 and caveolae participate in a range of cellular processes, we undertook quantitative subcellular proteomics analysis using our established PC-3 cell lines, which express GFP-PTRF or GFP (5), in order to acquire a global analysis of changes induced by PTRF expression. Systems analysis of the subcellular proteomes points to a role for PTRF in modulating cholesterol dynamics and regulation of the actin cytoskeleton, impacting the trafficking of multiple secretion pathways. Importantly, PTRF reduced the secretion of a specific subset of proteins including secreted proteases, cytokines, and growth factors. Our study reveals the potential for PTRF as a therapeutic option that could target a spectrum of secreted factors rather than a single molecule.
EXPERIMENTAL PROCEDURES
Materials
The following materials were obtained from the indicated commercial sources: Roswell Park Memorial Institute (RPMI)-1640 medium, l-glutamine, Geneticin (G418), and cell culture trypsin were from Invitrogen (Carlsbad, CA); Fetal bovine serum was from Bovogen (Melbourne, Australia); l-lysine, l-arginine, and l-leucine deficient RPMI 1640, l-leucine, Triton X-100, sodium deoxycholate, 2-(N-morpholino)ethanesulfonic acid, dithiotheitol, iodoacetamide, filipin III, water-soluble cholesterol, protease inhibitors, and antibodies against IL-6 and β-actin were from Sigma (St. Louis, MO); 13C615N2-lysine and 13C615N4-arginine were from Silantes (Munchen, Germany); Sequencing grade modified porcine trypsin was from Promega (Madison, WI); Antibody against kallikrein 6 was purchased from Santa Cruz (Santa Cruz, CA); Antibodies against caveolin-1 and flotillin were from BD Biosciences; Antibody against filamin A was from Millipore (Billerica, MA); PTRF antibodies were produced by immunizing rabbits with a synthetic peptide corresponding to the C-terminal 12 amino acids of mouse PTRF, and affinity purified from serum (16). GFP-PTRF has been described previously (5).
Cell Culture and Stable Isotope Labeling by Amino Acids in Culture (SILAC)
PC-3, PC-3 GFP, and PC-3 GFP-PTRF stable cells were grown and maintained in RPMI 1640 medium containing 10% fetal bovine serum. Stable cell lines were maintained in 0.1 mg/ml G418. For SILAC experiments, GFP and GFP-PTRF PC-3 cells were maintained in media lacking Lysine and Arginine with dialyzed FBS and supplemented with the following amino acids: “0/0 ” for the normal isotopic Lys and Arg and “8/10 ” for 13C615N2-Lys and 13C615N4-Arg. Cell populations were amplified 200-fold in the labeling media to achieve >99% incorporation as confirmed by liquid chromatography-tandem MS (LC-MS/MS). Where required, cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For each analysis, two to three 15-cm plates of PC-3 cells were used per condition for total membrane, detergent-resistant membrane extractions, secreted proteome and prostasome extraction.
Isolation of Secretome and Prostasomes
Briefly, PC-3 cells grown to 60 to 70% confluency were washed five times with phosphate-buffered saline (PBS) and then incubated for 24 h in serum free media. Any cell debris was removed by centrifugation at 600 × g for 10 min at 4 °C. Cleared cell culture supernatant was concentrated through an Amicon 10 kDa cutoff spin column and protein concentration was measured using BCA assay. Equal protein levels of secretome prepared from the two cell lines were mixed for isolation of prostasomes or in-solution digest. Isolation of prostasomes from cleared conditioned media involved sequential centrifugation; 10,000 × g 30 min 4 °C spin to remove any cell fragments not pelleted at 600 × g, then centrifugation of the supernatant at 100,000 × g, 2 h, 4 °C. Prostasome pellets were washed with PBS and again pelleted at 100,000 × g, 2 h, 4 °C. Pellets were resuspended in 1% sodium deoxycholate.
P100 and Detergent-Resistant Membrane Preparation
The P100 fraction was isolated from cells as previously described (17). Detergent resistant membranes (DRMs) were prepared by solubilizing cells in lysis buffer (1% Triton X-100, 25 mm 2-(N-morpholino) ethanesulfonate pH 6.5, 150 mm NaCl, (2-(N-morpholino) ethanesulfonate -buffered saline or MBS) plus protease inhibitors) and incubated on ice for 30 min. Relative protein concentrations were determined using Bradford assay and equal protein levels from each SILAC condition were mixed together. Lysates were adjusted to 40% sucrose by addition of an equal volume of 80% sucrose (in MBS), followed by successive layers of 30 and 5% sucrose. The gradients were centrifuged for 18 h at 160,000 × g, 4 °C and the white, light-scattering band appearing between 30 and 5% sucrose collected. The extracted layer was diluted out ∼threefold with MBS and pelleted by centrifugation at 160,000 × g for 2 h, 4 °C. All steps above were carried out at 4 °C.
In-Solution Digest and Strong Cation Exchange Chromatography Fractionation
Samples were resuspended or diluted with 1% sodium deoxycholate in 50 mm NH4HCO3 and heat solubilized for 10 min at 95 °C. Fifty micrograms of sample was reduced with 1 μg dithiotreitol, alkylated with 5 μg iodoacetamide and trypsin digested. An equal volume of 3% acetonitrile, 1% trifluoroacetic acid, 0.5% acetic acid was added to acidify the sample. In some experiments, digested peptides were also further fractionated by strong cation exchange chromatography into five fractions using 0, 20, 50, 100, and 500 mm NH4CH3COO.
LC-MS/MS, Database Searching and SILAC Quantitation
Samples (4 μg) were analyzed using a 1200 Series nano HPLC and Chip-Cube Q-TOF 6510 (Agilent Technologies). Peptides were resolved by 160 nl (75 mm * 150 μm) high capacity C18 reverse phase chip with 55 min gradient from 0 to 45% acetonitrile with the Vcap 1850 V, fragmentor 175 V. Precursor ions were selected in the range of 100–3200 m/z and fragment ions at 59–3200 m/z; reference ion mix was applied.
SILAC samples were analyzed in auto MS/MS mode, with 8 MS and 4 MS/MS per second. Raw MS data are available at PRIDE, accession numbers 19891–19906. Mass spectra extraction, database searching, and quantitative ratios were performed using Spectrum Mill software (Agilent, A03.03) against Swissprot Human database (release-2010_03 containing 23,000 entries). Cysteine carbamidomethylation and SILAC amino acids N-Lys, 13C615N2-Lys, N-Arg, and 13C615N4-Arg were used as a fixed/mix modifications as appropriate and oxidized methionine was selected as variable modification. Other parameters include up to 2 miss cleavages for trypsin; ±20 ppm and ±50 ppm accuracy for MS and MS/MS measurements respectively. Positive identification required a protein score >11, peptide score >10, and >60% scored peak intensity. Single peptide identifications were excluded from further analysis. Mean SILAC ratio and standard deviation was calculated using all the peptide ratios matched to a protein, and p values were calculated using the peptide SILAC ratios. False Discovery Rates were calculated by dividing the number of false positives (identified by searching against a reverse decoy database (18)), by the total number of proteins identified. For each replicate the false discovery rate was <1.25%.
Functional Enrichment and Network Analysis
Proteins with a SILAC p value < 0.05 were submitted to GeneGo for identification of Gene Ontology (GO) terms over represented in each list. Correction for multiple hypothesis testing was performed by controlling for the false discovery rate at p = 0.05. GeneGo networks were built using the shortest path and two connections algorithm. Low trust interactions and unconnected nodes were removed.
Western Blotting
Samples were resolved on SDS-PAGE gels and transferred to polyvinylidene difluoride using wet transfer. The membranes were blocked in 5% milk powder and then probed with primary antibody for 1 h. After washing, the membrane was incubated with the appropriate horseradish peroxidase-coupled secondary antibody for 1 h and then developed using SuperSignal West Pico chemiluminescence and captured on film (Kodak).
Immunofluorescence
Cells were grown on coverslips, transiently transfected and fixed in 4% paraformaldehyde after 24 h. For cholesterol staining, the coverslip was incubated with 50 μg/ml filipin for 2 h in the dark. Coverslips were washed three times with PBS, mounted, and visualized. For all other staining the coverslips were fixed in 4% paraformaldehyde, then permeabilized and blocked in 0.1% Triton-X, 1% BSA in PBS for 30 min. Anti-caveolin-1, anti-filamin A, or Texas Red conjugated phalloidin was incubated on the coverslips for 1 h. Where necessary, after washing in PBS, the appropriate secondary antibody was incubated for another hour. Coverslips were washed in PBS, mounted using Vectashield and visualized using a Zeiss Meta 510 confocal microscope.
Cholesterol Assay
Cholesterol assay was performed using the Amplex Red cholesterol assay kit (Invitrogen) according to the manufacturer's instructions.
RESULTS
PTRF Expression Alters the PC-3 Proteome
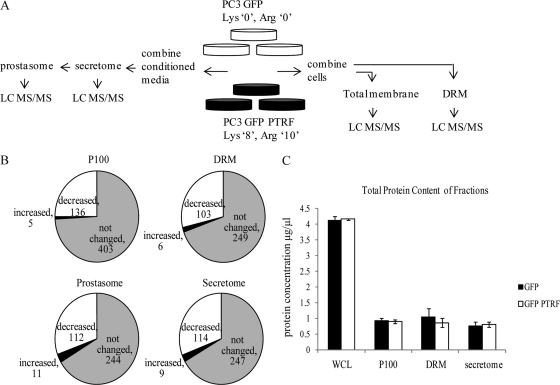
To gain a comprehensive view of cellular changes induced upon PTRF expression, we used SILAC with subcellular fractionation to quantitatively analyze the effect of PTRF expression on the PC-3 subproteomes. Based on the potential caveolar role of PTRF, we focused on four fractions: secreted proteins, secreted vesicles (prostasomes), total membrane fraction prepared as a pellet after 100,000 × g spin (P100), or DRM, a fraction enriched in lipid raft microdomains (Fig. 1A). Equal amount of total cell lysate from SILAC-labeled PC-3 cells stably expressing GFP (no PTRF expression) or GFP-PTRF (PTRF mainly localized to caveolae) (data not shown) were combined. The P100 or the DRM fractions were isolated and then analyzed by mass spectrometry (Fig. 1A). The secretome was collected by growing cells in serum free media for 24 h and using equal protein amount of conditioned media for each cell type. No significant difference was observed in the viability of cells grown in serum free media (93.12 ± 7.62%) compared with cells grown in 10% serum containing media (100%). The vesicular component of the secretome (also termed prostasomes), was further fractionated from the secretome and then both fractions were analyzed by mass spectrometry (Fig. 1A). An overall mean was calculated for each protein using SILAC ratios for peptides from the four independent measurements (supplemental Tables S1 and S3) and then analyzed for proteins significantly changed (p value ≤ 0.05) upon PTRF expression.
Fig. 1.
PTRF expression induced changes in the PC-3 proteome. A, SILAC labeling was performed by incorporating Lys“0”, Arg“0 ” into PC-3 GFP cells and Lys“8”, Arg“10 ” into PC-3 GFP-PTRF cells. Cells were fractionated into secretome, prostasome, P100, and DRM compartments, trypsin digested, desalted using SCX/C18, and analyzed by LC MS/MS. B, Pie charts show the number of proteins with significantly different (p ≤ 0.05) abundance in the P100, DRM, secretome, and prostasome fractions upon PTRF expression, and proteins not changed. C, Protein content was determined using BCA assay for whole cell lysates (WCL), P100, DRM, and secretome fraction of PC-3 GFP and GFP-PTRF cells. The graph shows the mean protein concentrations ± S.E. (n = 3).
We found that PTRF expression significantly decreased 136 (25%) from the 544 proteins identified in the P100 fraction (Fig. 1B). In the DRM fraction, 103 proteins (29%) were decreased with PTRF expression, from a total of 358 proteins quantified (Fig. 1B). Very few proteins were increased in abundance in the P100 or DRM fractions, five and six respectively (Fig. 1B). PTRF expression also caused a significant reduction in proteins from the secreted and prostasome fractions: 114 proteins (31%) were secreted less with PTRF expression from a total of 370 proteins identified, and 112 proteins (30%) decreased from a total of 367 proteins in the prostasome (Fig. 1B). In contrast, very few proteins were increased in the secretome and prostasome, nine and 11 respectively (Fig. 1B). Importantly PTRF expression did not alter the total protein content of any of the fractions (Fig. 1C). These results reveal a specific change in the membrane targeting and/or secretion of a subset of proteins upon PTRF expression in PC-3 cells.
PTRF Modulates Secretion Pathways to Reduce the Secretion of a Subset of Proteins
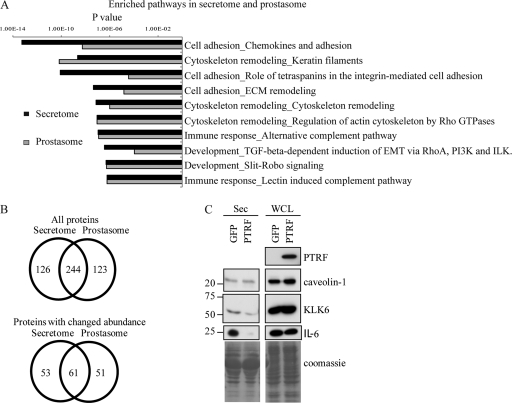
Previously we reported PTRF expression reduced PC-3 cell migration though collagen-coated Boyden chamber partly via reduction of MMP9 secretion (15). Because cell migration and invasion requires co-ordination between cellular events and secreted proteases, we examined the quantitative proteomics data for proteins with a role in cancer and cell migration. Pathways enrichment analysis using GeneGo software revealed that PTRF expression most significantly reduces the secretion of proteins involved in cell adhesion and cytoskeleton remodeling (Fig. 2A). Interestingly, PTRF modulated similar pathways in the prostasome fraction. Because the prostasome is a subfraction of the secretome, one possible explanation for the overlap is that PTRF expression specifically attenuated the secretion of vesicles. To examine this possibility, we analyzed the proteins common to the secretome and prostasome fraction (Fig. 2B). The result shows that 244 proteins (66%) were identified in both fractions, indicating that prostasomes contribute a significant portion of total secreted proteins. In the subset of proteins significantly changed by PTRF expression, 61 (∼50%) were common to the secretome and prostasome, meaning that the reduction in secreted proteins upon PTRF expression could only be partially explained by a reduction in prostasomal proteins.
Fig. 2.
PTRF expression affects multiple secretory pathways to reduce a subset of secreted proteins that function in cell adhesion and cytoskeleton remodeling. A, Bar chart of GeneGo analysis displaying the most highly enriched molecular pathways for the secretome and prostasome fraction. B, Venn diagrams displaying proteins common to the secretome and prostasome fraction (top) and those common in the list of proteins significantly changed (p ≤ 0.05) upon PTRF expression (bottom). C, PC-3 cells were transiently transfected with GFP or GFP-PTRF. The secretome was collected from serum free media incubated on the cells for 24 h. Whole cells lysates were isolated in parallel. Western blotting was performed to analyze changes in protein expression by probing with antibodies against PTRF, caveolin-1, KLK-6, and IL-6. Equal loading was shown by Coomassie stain. Whole cells lysates showed no difference in protein levels between GFP and GFP-PTRF cells however, secreted levels of KLK-6 and IL-6 were reduced in GFP-PTRF cells.
Closer inspection of the proteins reduced in both the secretome and prostasome fractions by PTRF expression identified a number of proteins with established links to cancer and in particular metastasis, including extracellular matrix proteins, secreted proteases, and cytokines (Table I). We selected two proteins that have been previously implicated in prostate cancer (19–21) for verification by Western blotting, namely kallikrein 6 (KLK6) and interleukin-6 (IL-6). To cross-validate the stable cell line quantitative proteomics results, we chose transient transfection of wild-type PC-3 cells for these experiments. Conditioned media collected from PC-3 cells transiently transfected with control GFP or GFP-PTRF plasmids were immunoblotted with antibodies against KLK-6 and IL-6. This result confirmed that expression of PTRF reduced secreted KLK-6 and IL-6 (Fig. 2C, left panel). Whole cell lysates of GFP and GFP-PTRF cells showed no change in expression levels (Fig. 2C, right panel) indicating that PTRF expression does not affect total cellular levels of KLK-6 and IL-6 but rather specifically modulates their secretion.
Table I. List of selected proteins that were reduced in the secretome and prostasome fraction of SILAC PC-3 cells expressing PTRF. Protein levels were quantified by mean SILAC (heavy/light) ratio with PTRF expression over four independent experiments. No. Ob = Number of SILAC ratios used for quantification. No. Pep = Number of unique peptides observed. Proteins were classified by function.
| Accession | Protein name | Secretome |
Prostasome |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. Ob | No. Pep | Ratio ± S.D. | p value | No. Ob | No. Pep | Ratio ± S.D. | p value | ||
| Extracellular Matrix Protein | |||||||||
| P12109 | Collagen alpha-1(VI) chain | 103 | 16 | 0.42 ± 0.12 | <0.001 | 62 | 13 | 0.42 ± 0.09 | 0.015 |
| Secreted Proteases | |||||||||
| Q9Y337 | Kallikrein-5 | 3 | 2 | 0.28 ± 0.02 | 0.007 | 2 | 1 | 0.27 ± 0.01 | 0.010 |
| Q92876 | Kallikrein-6 | 27 | 2 | 0.54 ± 0.07 | 0.012 | 19 | 2 | 0.55 ± 0.08 | 0.020 |
| P00749 | Urokinase-type plasminogen activator | 179 | 12 | 0.40 ± 0.08 | <0.001 | 139 | 11 | 0.45 ± 0.02 | 0.030 |
| P80188 | Neutrophil gelatinase-associated lipocalin | 161 | 10 | 0.42 ± 0.12 | <0.001 | 209 | 11 | 0.42 ± 0.08 | 0.050 |
| Cytokines/Growth Factors | |||||||||
| P05231 | Interleukin-6 | 41 | 7 | 0.10 ± 0.06 | <0.001 | 28 | 6 | 0.10 ± 0.07 | 0.006 |
| P08476 | Inhibin βA chain | 111 | 13 | 0.29 ± 0.12 | <0.001 | 30 | 11 | 0.24 ± 0.13 | 0.006 |
| P61812 | Transforming growth factor β-2 | 62 | 7 | 0.60 ± 0.11 | 0.006 | 55 | 4 | 0.59 ± 0.07 | 0.020 |
| O00622 | Protein CYR61 | 30 | 6 | 0.40 ± 0.21 | 0.004 | 14 | 4 | 0.31 ± 0.11 | 0.006 |
PTRF Expression Alters the DRM-Associated Pool of Actin Cytoskeletal Proteins
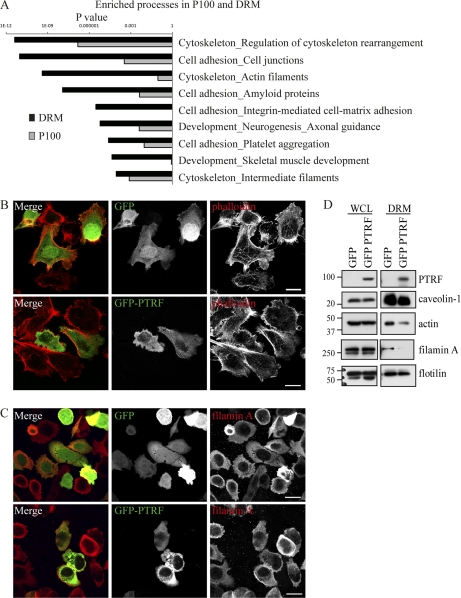
Quantitative changes in the P100 and DRM fractions were further analyzed to determine potential cellular mechanisms linking PTRF expression to the observed reduction in secreted proteins. Proteins that were significantly changed (p value ≤ 0.05) by PTRF in the P100 and DRM fractions were analyzed using GeneGo software. This analysis identified regulation of cytoskeleton rearrangement as the most significant pathway (Fig. 3A), with a p value of 2.09 × 10−11 for the DRM and 1.51 × 10−7 for the P100 fraction. In agreement with previous reports (22, 23), our DRM fraction was highly enriched in cytoskeletal proteins and furthermore, many were reduced by PTRF expression (Table II). This includes structural and regulatory proteins such as actin, 14–3-3, epiplakin, filamin A, myosin 9 and 11, plastin, spectrin, and stomatin like protein 2. In addition, many of the structural cytoskeletal proteins such as actin, myosin, and spectrin were changed in the DRM fraction and not in the P100 fraction. This result raises the possibility that PTRF expression altered the cytoskeleton link to lipid raft membrane microdomains. Consistent with this, one of the proteins identified in our proteomic study to be reduced in the DRM by PTRF expression was filamin A, a filamentous actin binding protein that is thought to link caveolae to the actin cytoskeleton (24).
Fig. 3.
PTRF expression alters the DRM-associated pool of actin cytoskeletal proteins. A, Bar chart of GeneGo analysis displaying the most highly enriched biological processes for the P100 and DRM fraction. Cells were stained with phalloidin (B) or antibody against filamin A (C). Localization was analyzed by fluorescent confocal microscopy. No obvious change in distribution was observed between GFP and GFP-PTRF expressing cells. Scale bar = 20 μm. D, Whole cell lysates or DRMs were isolated and analyzed by Western blotting. Blots were probed with antibody against PTRF to show transfection, caveolin-1, β-actin, filamin A, and flotillin. The whole cells lysates showed no difference in protein levels between GFP and GFP-PTRF cells however, the DRM fraction showed a reduction in β-actin and filamin A in GFP-PTRF cells.
Table II. List of cytoskeletal proteins reduced in abundance in the DRM and the corresponding ratio from the P100 fraction upon PTRF expression. Protein levels were quantified by mean SILAC (heavy/light) ratio with PTRF expression over four independent experiments. No. Ob = Number of SILAC ratios used for quantification. No. Pep = Number of unique peptides observed.
| Accession | Protein name | DRM |
P100 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. Ob | No. Pep | Ratio ± S.D. | p value | No. Ob | No. Pep | Ratio ± S.D. | p value | ||
| P63104 | 14-3-3 protein ζ/Δ | 25 | 5 | 0.49 ± 0.22 | 0.028 | 43 | 6 | 0.55 ± 0.15 | 0.010 |
| P62736 | Actin | 116 | 19 | 0.46 ± 0.17 | 0.039 | 100 | 18 | 0.81 ± 0.46 | 0.941 |
| P60709 | Actin, cytoplasmic 1 | 120 | 19 | 0.48 ± 0.17 | 0.045 | 98 | 18 | 0.67 ± 0.15 | 0.591 |
| P50995 | Annexin A11 | 85 | 13 | 0.55 ± 0.15 | 0.040 | Not found | |||
| P80723 | Brain acid soluble protein 1 | 48 | 9 | 0.54 ± 0.14 | 0.040 | 35 | 8 | 0.30 ± 0.11 | 0.003 |
| P12830 | Cadherin-1 | 4 | 3 | 0.07 ± 0.03 | 0.002 | Not found | |||
| P58107 | Epiplakin | 55 | 151 | 0.51 ± 0.25 | 0.029 | 20 | 76 | 0.52 ± 0.11 | 0.020 |
| P21333 | Filamin-A | 15 | 5 | 0.46 ± 0.23 | 0.030 | 14 | 4 | 0.46 ± 0.14 | 0.008 |
| O75369 | Filamin-B | 14 | 5 | 0.35 ± 0.18 | 0.011 | Not found | |||
| P05556 | Integrin β-1 | 3 | 2 | 0.27 ± 0.08 | 0.019 | 82 | 10 | 0.41 ± 0.12 | 0.007 |
| Q9UHB6 | LIM domain and actin-binding protein 1 | 6 | 4 | 0.42 ± 0.27 | 0.054 | Not found | |||
| P35749 | Myosin-11 | 2 | 4 | 0.15 ± 0.09 | 0.007 | Not found | |||
| P35579 | Myosin-9 | 6 | 4 | 0.33 ± 0.02 | 0.013 | 3 | 3 | 0.55 ± 0.32 | 0.247 |
| P13796 | Plastin-2 | 2 | 1 | 0.13 ± 0.00 | 0.005 | 2 | 1 | 0.32 ± 0.03 | 0.023 |
| P61225 | Rap-2b | 21 | 3 | 0.51 ± 0.14 | 0.048 | 3 | 2 | 0.69 ± 0.23 | 0.647 |
| Q13813 | Spectrin α chain | 79 | 14 | 0.49 ± 0.22 | 0.032 | 7 | 2 | 0.65 ± 0.09 | 0.528 |
| Q01082 | Spectrin β chain, brain 1 | 11 | 5 | 0.40 ± 0.21 | 0.015 | Not found | |||
| O15020 | Spectrin β chain, brain 2 | 2 | 1 | 0.18 ± 0.01 | 0.012 | Not found | |||
| Q9UJZ1 | Stomatin-like protein 2 | 79 | 9 | 0.52 ± 0.28 | 0.032 | 15 | 2 | 0.40 ± 0.22 | 0.007 |
To verify the quantitative proteomics data, we first utilized fluorescence microscopy to examine the morphology of the actin cytoskeleton by staining with phalloidin, which stains filamentous actin. No obvious difference was observed in the distribution between GFP and GFP-PTRF expressing cells (Fig. 3B). Similarly, immunofluorescence microscopy using an antibody to filamin A showed no obvious difference under these conditions (Fig. 3C). In agreement, Western blots of whole cell lysate isolated from transient GFP or GFP-PTRF expressing cells showed no difference in total protein levels of β-actin or filamin A (Fig. 3D, left panel). In stark contrast, Western blots of the DRM fractions demonstrated a substantial reduction in β-actin and filamin A protein levels in GFP-PTRF expressing cells, but no change in flotillin, a marker of lipid rafts (Fig. 3D, right panel). These results confirmed our proteomic findings and suggest that PTRF expression attenuated the recruitment of cytoskeletal proteins β-actin and filamin A to DRMs in PC-3 cells.
Recent literature supports a role for the actin cytoskeleton in the early stages of secretion pathways. Secretory pathways rely on Golgi derived carrier vesicles to deliver their cargo to the required destination. Furthermore, during the sorting process, cargo proteins are segregated into distinct membrane microdomains. To gain insight into secretory pathways affected by PTRF expression, proteins identified by proteomics to be significantly changed in the P100 and DRM fractions were categorized according to function and those with a reported role in membrane trafficking were further analyzed for pathways associated with secretion (Table III). This analysis revealed an enrichment of proteins associated with trafficking from the endoplasmic reticulum (ER), Golgi, and lysosome. In particular, aminopeptidase N, protein LYRIC, SEC24C, reticulon-4, TMED 10, TMED 2, TMED 4, vesicle-fusing ATPase, and SEC22b have been associated with the ER-Golgi intermediate compartment (ERGIC) or trafficking between the ER and Golgi, and were all reduced in the P100 fraction upon PTRF expression. Fewer trafficking proteins were regulated by PTRF expression in the DRM fraction, with aminopeptidase N, kinectin and reticulon-4 the only ER associated proteins reduced in this fraction. Together our data suggests that PTRF alters the lipid raft associated actin cytoskeleton which may in turn affect trafficking between the ER and the Golgi.
Table III. List of proteins associated with trafficking that were significantly changed in the P100 and DRM fractions of SILAC PC-3 cells expressing PTRF. Protein levels were quantified by mean SILAC (heavy/light) ratio with PTRF expression over four independent experiments. No. Ob = Number of SILAC ratios used for quantification. No. Pep = Number of unique peptides observed.
| Accession number | Protein name | Function | No. Ob | No. Pep | Ratio | St Dev | p value |
|---|---|---|---|---|---|---|---|
| P100 fraction | |||||||
| P08195 | 4F2 cell-surface antigen heavy chain | Targeting of LAT1 LAT2 to PM (CD98) | 39 | 6 | 0.42 | 0.12 | 0.003 |
| Q96BM9 | ADP-ribosylation factor-like protein 8A | Lysosome motility, LE | 9 | 3 | 0.35 | 0.12 | 0.005 |
| P15144 | Aminopeptidase N | ERGIC (CD13) | 36 | 8 | 0.29 | 0.11 | 0.003 |
| P80723 | Brain acid soluble protein 1 | Cytoskeleton-PM link (NAP22) | 35 | 8 | 0.30 | 0.11 | 0.003 |
| P08962 | CD63 antigen | Lysosome, LE (LAMP3) | 4 | 2 | 0.41 | 0.10 | 0.022 |
| Q14108 | Lysosome membrane protein 2 | Lysosome (CD36) | 3 | 1 | 0.26 | 0.03 | 0.004 |
| P13473 | Lysosome-associated membrane glycoprotein 2 | Lysosome (CD107b), Adhesion | 19 | 3 | 0.51 | 0.16 | 0.015 |
| P29966 | Myristoylated alanine-rich C-kinase substrate | Actin-PM link | 10 | 4 | 0.15 | 0.07 | 0.001 |
| P13796 | Plastin-2 | Actin-PM link, membrane ruffles (LCP-1) | 2 | 1 | 0.32 | 0.03 | 0.023 |
| Q15149 | Plectin-1 | Intermediate filament and microtubule link, dynamics regulation, hemidesmosomes | 264 | 76 | 0.52 | 0.32 | 0.000 |
| Q86UE4 | Protein LYRIC | ER, tight junction | 19 | 6 | 0.47 | 0.15 | 0.009 |
| P53992 | Protein transport protein Sec24C | ER, COPII coat | 29 | 3 | 0.56 | 0.08 | 0.028 |
| P59190 | Ras-related protein Rab-15 | synaptic membrane, interacts with Rab3A | 3 | 5 | 0.22 | 0.28 | 0.003 |
| Q9NQC3 | Reticulon-4 | ER tubules, PM, regulates migration via ICAM and fibulin-5 (Nogo) | 16 | 3 | 0.53 | 0.19 | 0.030 |
| P49755 | Transmembrane emp24 domain-containing protein 10 | ERGIC (TMED10) | 23 | 3 | 0.54 | 0.13 | 0.023 |
| Q15363 | Transmembrane emp24 domain-containing protein 2 | ERGIC, COPI vesicles, interacts with ARFGAP, (complex with SURF4, TMED2, TMED10) | 18 | 3 | 0.54 | 0.15 | 0.040 |
| Q7Z7H5 | Transmembrane emp24 domain-containing protein 4 | ER (TMED4) | 4 | 2 | 0.17 | 0.09 | 0.002 |
| P46459 | Vesicle-fusing ATPase | ER-Golgi and intra-Golgi vesicle fusion (NSF) | 2 | 1 | 0.25 | 0.21 | 0.009 |
| O75396 | Vesicle-trafficking protein SEC22b | ER-Golgi SNARE | 2 | 1 | 3.06 | 2.56 | 0.026 |
| Q10567 | AP-1 complex subunit beta-1 | Golgi vesicle budding | 26 | 2 | 0.42 | 0.08 | 0.006 |
| DRM fraction | |||||||
| P08195 | 4F2 cell-surface antigen heavy chain | Targeting of LAT1 LAT2 to PM (CD98) | 5 | 2 | 0.18 | 0.10 | 0.004 |
| P15144 | Aminopeptidase N | ERGIC (CD13) | 140 | 18 | 0.48 | 0.14 | 0.043 |
| Q03135 | Caveolin-1 | Caveolae formation | 23 | 2 | 1.99 | 0.41 | 0.038 |
| Q86UP2 | Kinectin | ER vesicle, kinesin-driven vesicle transport, accumulates at integrin adhesions | 20 | 5 | 0.45 | 0.24 | 0.020 |
| Q9NX40 | OCIA domain-containing protein 1 | Endosome | 8 | 1 | 0.38 | 0.27 | 0.017 |
| Q56VL3 | OCIA domain-containing protein 2 | Endosome | 6 | 2 | 0.41 | 0.13 | 0.046 |
| P13796 | Plastin-2 | Actin-PM link, membrane ruffles (LCP-1) | 2 | 1 | 0.13 | 0.00 | 0.005 |
| Q969L2 | Protein MAL2 | Endosome-Apical PM transport | 7 | 1 | 0.34 | 0.17 | 0.011 |
| Q9NQC3 | Reticulon-4 | ER tubules, PM, regulates migration via ICAM and fibulin-5 (Nogo) | 13 | 3 | 0.41 | 0.17 | 0.015 |
PTRF Expression Modifies Cholesterol Dynamics in DRMs
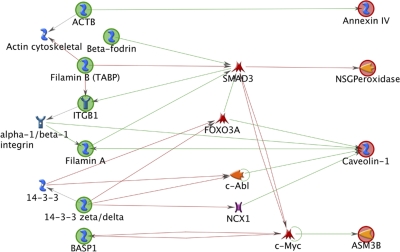
One factor that regulates the association between lipid rafts and the actin cytoskeleton is cholesterol (25–27). Caveolin-1 binds cholesterol and is a major mediator of cholesterol transport to the plasma membrane (28, 29). Therefore we investigated whether PTRF expression modifies the cholesterol dynamics in lipid rafts, possibly accounting for the changes in the actin cytoskeleton. Our proteomics study identified a small subset of proteins from the DRM fraction that play a role in cholesterol transport (Table IV) and none in the P100 fraction. GeneGo was used to uncover potential interactions between the subset of cholesterol trafficking proteins and the subset of cytoskeletal proteins identified in the DRM fraction (Table II and IV). The network diagram illustrated a direct relationship between key cytoskeletal proteins such as β-actin (ACTB) and filamin A with cholesterol regulating proteins caveolin-1 and Annexin IV (Fig. 4). In addition, many of the cytoskeletal proteins regulate transcription factors SMAD3, FOXO3A, c-Abl, NCX1, and c-Myc that in turn regulate the expression of cholesterol regulating proteins.
Table IV. Proteins associated with cholesterol transport in the DRM fraction that was significantly changed upon PTRF expression. Protein levels were quantified by mean SILAC (heavy/light) ratio with PTRF expression over four independent experiments. No. Ob = Number of SILAC ratios used for quantification. No. Pep = Number of unique peptides observed.
| Accession number | Protein name | No. Ob | No. Pep | Ratio | St Dev | p value |
|---|---|---|---|---|---|---|
| Q92485 | Acid sphingomyelinase-like phosphodiesterase 3b | 5 | 4 | 0.68 | 0.61 | 0.644 |
| O14734 | Acyl-coenzyme A thioesterase 8 | 3 | 1 | 0.21 | 0.08 | 0.008 |
| P09525 | Annexin A4 | 33 | 7 | 0.49 | 0.15 | 0.025 |
| P02647 | Apolipoprotein A-I | 2 | 1 | 0.23 | 0.06 | 0.027 |
| Q03135 | Caveolin-1 | 23 | 2 | 1.99 | 0.41 | 0.038 |
| O75131 | Copine-3 | 42 | 6 | 0.53 | 0.15 | 0.040 |
| P30041 | Peroxiredoxin-6 | 2 | 2 | 0.10 | 0.07 | 0.003 |
Fig. 4.
Network analysis to uncover relationships between actin cytoskeletal proteins and cholesterol regulating proteins. The network diagram illustrates a direct relationship between key cytoskeletal proteins such as actin (ACTB) and filamin A with cholesterol regulating proteins caveolin-1 and annexin IV (Fig. 7). In addition, many of the cytoskeletal proteins regulate transcription factors SMAD3, FOXO3A, c-abl, NCX1, and c-myc that in turn regulate the expression of cholesterol regulating proteins. Legend: Green circles, objects used to build pathway from. Red circles, objects used to build pathway to. Green arrows, positive effect. Red arrow, negative effect. Blue symbol, generic binding protein. Blue receptor symbol, generic receptor. Orange reverse arrow symbol, protein kinase. Yellow symbol, generic enzyme. Red symbol, transcription factor. Purple symbol, generic channel.
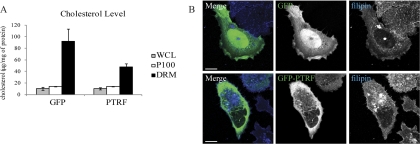
We went on to measure cholesterol levels in whole cell lysate, P100, and the DRM fraction of PC-3 cells transiently expressing GFP or GFP-PTRF. PTRF expression did not affect cholesterol levels in the whole cell lysate or the P100 fraction, but induced a decrease in cholesterol levels in the DRM fraction (Fig. 5A). Curiously, this was not accompanied by a significant change in the protein content of the DRM fractions (Fig. 1C). To examine the subcellular location of cholesterol-enriched membranes, PC-3 GFP and PC-3 GFP-PTRF cells were stained with filipin, a UV-light fluorescent stain that binds sterols. Control cells transfected with GFP exhibited distinct plasma membrane staining as well as an intracellular vesicular pool of cholesterol (Fig. 5B). PTRF expressing cells also displayed strong plasma membrane staining but in contrast to control cells, the intracellular vesicular pool was more pronounced (Fig. 5B). Taken together, these data indicate that PTRF expression alters cholesterol distribution.
Fig. 5.
PTRF expression changes cholesterol dynamics in lipid rafts. PC-3 cells were transiently transfected with GFP or GFP-PTRF. A, Whole cell lysates, P100, or DRM fractions were isolated from the cells and a cholesterol assay performed. The graph shows the mean cholesterol level ± S.E. (n = 3). B, GFP and GFP-PTRF cells were stained with filipin and analyzed by fluorescent confocal microscopy to detect cholesterol localization. GFP-PTRF expressing cells displayed a more pronounced intracellular pool of cholesterol. Scale bar = 20 μm.
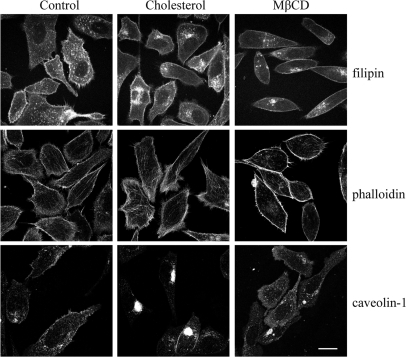
To corroborate decreased cholesterol levels in the DRM with changes in the actin cytoskeleton, we explored the effects of additional cholesterol or depleting cholesterol from the plasma membrane using methyl-β-cyclodextrin (MβCD). We examined the distribution of cholesterol, caveolin-1, and filamentous actin by using filipin, anti-caveolin-1 antibody, and phalloidin, respectively. PC-3 cells treated with 8 μm cholesterol showed substantially elevated intracellular cholesterol levels, and concurrently a greater pool of intracellular caveolin-1 was observed compared with control cells (Fig. 6). Strikingly phalloidin staining illustrated less distinct cortical actin and more dispersed filamentous actin in cholesterol treated cells compared with control cells (Fig. 6). Treatment with MβCD resulted in a clear decrease in filipin staining, and plasma membrane caveolin-1 but a greater intracellular pool for both (Fig. 6). The corresponding phalloidin stain displayed enhanced cortical actin staining (Fig. 6). These results demonstrate that manipulation of cholesterol levels in PC-3 cells does in fact impact on regulation of the actin cytoskeleton.
Fig. 6.
Changes in cholesterol affects the actin cytoskeleton and caveolin-1 localization. PC-3 cells were treated with vehicle (control), 8 μm cholesterol, or 500 μm MβCD for 24 h. Cells were analyzed for the localization of cholesterol with filipin, caveolin-1, and filamentous actin with phalloidin using fluorescent confocal microscopy. Scale bar = 20 μm.
Disruption to Cholesterol Alters Secretion
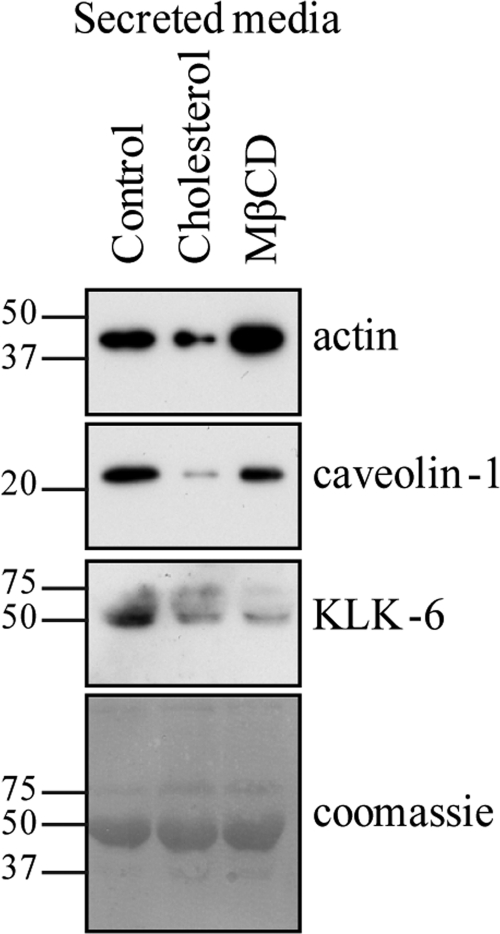
Recent literature highlights the important role cholesterol levels and the actin cytoskeleton play in multiple secretion pathways (30–32). Therefore we speculated that the observed changes in subcellular cholesterol and actin cytoskeletal distribution induced by PTRF expression would also account for the perturbed secretion. To test this hypothesis, we next incubated PC-3 cells with vehicle, 8 μm cholesterol or 500 μm MβCD for 24 h in serum free media, collected the secreted fraction, and immunoblotted for actin, caveolin-1, and KLK-6. The addition of cholesterol reduced the secreted levels of all three proteins (Fig. 7). In striking contrast, MβCD had differential effects depending on the protein. Secreted levels of actin were clearly increased with MβCD treatment (Fig. 7). Secreted caveolin-1 was slightly reduced with MβCD but not nearly as much as seen with cholesterol addition. Finally, KLK-6 levels were substantially decreased in the secretome with MβCD (Fig. 7). The conflicting results likely reflect the different secretory pathways of each protein. It is probable that adding cholesterol affects multiple secretory pathways whereas MβCD may target a more specific subset.
Fig. 7.
Cholesterol dynamics modulates secretion. The secretome was isolated from PC-3 cells incubated with serum free media for 24 h. During the final 6 h of incubation, cells were supplemented with vehicle (control), 8 μm cholesterol, or 500 μm MβCD. Changes in secretion were analyzed by Western blot analysis. Antibodies against β-actin, caveolin-1, and KLK-6 were used to detect secreted protein levels. Equal loading was shown by coomassie stain.
DISCUSSION
In this study we found that expression of PTRF in the caveolin-1 positive cell line PC-3 reduced the secretion of a specific subset of proteins including extracellular matrix proteins, secreted proteases, cytokines, and growth factors. Many of these secreted proteins are known to be involved in prostate cancer progression. As an example, elevated levels of IL-6 have been reported in the sera of prostate cancer patients (19, 20). Despite evidence linking IL-6 levels to the progression of prostate cancer, preliminary studies using IL-6 monoclonal antibodies to specifically target IL-6 in advanced prostate cancer have given poor outcomes in the clinic (33). Therefore, targeting a single molecule may not represent the ideal solution to attack prostate cancer. Here we show that by expressing PTRF in PC-3 cells, secretion of multiple factors associated with prostate cancer progression is reduced. These results reveal an exciting, novel therapeutic option that could potentially target a broad spectrum of cancer promoting factors secreted by tumor cells, namely, modulation of lipid raft composition and function by expression of PTRF. Thus, PTRF therapy has the potential to target the tumor microenvironment as well as the cancer cells because many of the secreted factors reduced by PTRF expression have pleiotropic activities on the surrounding stromal cells as well as the cancer cells.
PTRF specifically reduced the secretion of IL-6 and KLK6 with no affect on total cellular levels indicating that PTRF influences secretory pathways. This is most likely explained by the PTRF induced sequestration of noncaveolar caveolin-1 to form caveolae. However, we cannot exclude the possibility that PTRF may also function outside of caveolae. Further evidence of caveolin-1 mediated effects on secretion comes from studies showing that modulation of caveolin-1 expression levels affects IL-6 secretion. Ectopic expression of caveolin-1 in mouse lung alveolar type-1 (AT-1) cells increased LPS-modulated IL-6 production (34). However, in contrast caveolin-1 expression in the macrophage cell line RAW264.7 decreased LPS modulated IL-6 secretion levels (35). These inconsistencies may reflect differential caveolin-1 and PTRF expression in the two cell types.
To unveil specific secretory pathways altered by PTRF, our analysis of the P100 and DRM fractions demonstrated that PTRF induced a change in the expression and/or membrane recruitment of proteins that are associated with secretion at several cell compartments, namely the ER, Golgi, and lysosomes. However, we observed particular enrichment of proteins that function in ER to Golgi transport. Furthermore, PTRF regulated the recruitment of cytoskeletal proteins actin and spectrin to DRMs. Actin is important for Golgi-derived carrier vesicle biogenesis as well as transport from the Golgi to the ER. Spectrin βII functions in ER to Golgi transport and Golgi architecture (36). Therefore it is likely that PTRF expression impacts the actin cytoskeleton associated with the ER and Golgi. An important role for actin dynamics at the Golgi and in early stages of secretory pathways derived from the Golgi is well documented, reviewed in (37), however, actin based mechanisms between the ER and Golgi are less well defined. Different classes of Golgi derived vesicles bind to distinct sets of actin binding proteins (38). Given that PTRF modifies the abundance of a third of proteins recruited to membranes, we speculate that this would disrupt a number of vesicles types, and thereby explain why we observed a reduction in protein secreted by more than one pathway.
The actin cytoskeleton is linked with many aspects of membrane trafficking and its function is particularly well documented at the plasma membrane (39). Intriguingly, our results suggest that PTRF induced changes to the actin cytoskeleton are concentrated at ER and Golgi trafficking, in turn affecting secretion pathways. A number of factors may contribute to this trend: First, the Golgi is central to many membrane trafficking pathways, being the meeting point for endocytic and exocytic vesicles. Moreover, Golgi derived vesicles are directed to many cellular compartments including endosomes, ER, and plasma membrane. Therefore disruption of membrane trafficking may have the greatest influence on the Golgi. Second, these effects are likely to be compounded by the multiple roles for actin at the Golgi including maintenance of architecture, homeostasis, signaling, and transport (36). Finally, PTRF induced changes to lipid rafts and membrane trafficking might upset the delicate balance of membrane internalized at the plasma membrane and synthesized at the ER. The Golgi would be extremely sensitive to these changes because of the large amount of cargo continuously passing through.
An association between the actin cytoskeleton and lipid rafts is well documented (40, 41). Caveolae lipid rafts are thought to tether to the cortical actin cytoskeleton through binding of filamin A and filamin B (24). Studies indicate that in a phosphorylation dependent manner, filamin A regulates a number of caveolin-1 functions: the distribution of caveolin-1 along actin stress fibers, anchoring of caveolin-1 vesicles at the plasma membrane, and internalization of caveolae (42, 43). We have shown that PTRF expression reduced the abundance of filamin A and filamin B in the DRM fraction, possibly via caveolae formation. Therefore, it is possible that noncaveolar caveolin-1 domains attach to the cytoskeleton differently from caveolae. Furthermore, stable over expression of caveolin-1 in a breast cancer cell line increased expression levels of filamin A (43). Our data suggests that in addition to caveolin-1 levels, caveolin-1 microdomain structure also modulates the abundance of filamin A and B in lipid rafts.
One of the major constituents of lipid rafts is cholesterol. Cholesterol levels are elevated in PC-3 cells compared with normal prostate epithelial cell lines and the caveolin-1 deficient cell LNCaP (44). Caveolin-1 binds directly to cholesterol and regulates its trafficking (25–29). Expression of PTRF significantly impacts lipid raft structure in PC-3 cells by inducing the formation of caveolae (5). We found that PTRF expression specifically reduced cholesterol in lipid rafts and appeared to redistribute it partly to intracellular vesicles. Moreover, by modulating cellular cholesterol levels we demonstrated altered secretion of actin, IL-6, and KLK6 indicating that cholesterol dynamics in turn regulate secretion pathways. Consistent with our observations for a role of cholesterol in secretion, others have shown that cholesterol depletion blocked the formation of regulatory and secretory vesicles from AtT-20 cells (31), reduced zymogen granules and increased amylase secretion in pituitary cells (45), and the release of prostasomes from PC-3 cells (14). Although our study and others clearly link changes in cholesterol and cytoskeletal dynamics to alterations in a range of secretory pathways, the mechanisms remain to be elucidated. Further insight has come from a study that shows that PIP2 induced actin polymerization around lipid raft enriched membrane vesicles, forming comet tails that directed them to the cytoplasm (46). In addition, the authors show that depletion of membrane cholesterol using MβCD reduced actin comets, indicating an essential role for cholesterol and lipid rafts in vesicular trafficking. The correct balance of cholesterol on vesicles is critical because cholesterol accumulation on late endocytic compartments impairs their mobility by interfering with Rab7 activity (47). Together these studies and ours indicate that noncaveolar caveolin-1 in PC-3 cells have elevated lipid raft cholesterol, which impacts on regulation of actin cytoskeleton and secretory pathways.
Taken together, our data are consistent with a model in which the cholesterol-rich noncaveolar caveolin-1 microdomain recruits elevated levels of cyoskeletal proteins, in particular filamin A and B, which are likely to be vital in tethering caveolin-1 to actin filaments. PTRF induced sequestration of noncaveolar caveolin-1 to form caveolae, altering cholesterol dynamics and lipid raft composition in PC-3 cells. As a result, cytoskeletal proteins recruited to lipid rafts are modified, impacting multiple secretory pathways. Therefore, the unique noncaveolae caveolin-1 microdomains of PC-3 cells, created by expression of caveolin-1 but not PTRF, has allowed these cells to develop a mechanism to increase secretion of proteases, cytokines, and growth factors, potentially increasing their metastatic ability. Most importantly, we show these effects were neutralized by PTRF expression. Cell motility is intimately associated with cytoskeletal remodeling and membrane trafficking, because in order for cells to migrate, actin polymerization is required for membrane extensions and membrane turnover. In agreement with our model, PTRF expression was shown to reduce cell migration in PC-3 cells (15). Our study brings to light the different cellular mechanisms resulting from noncaveolae and caveolae caveolin-1 and the potential for developing novel therapies designed to target caveolin-1 and its microdomain structure on the plasma membrane as a means to improve patient outcome. Although we investigated prostate cancer cells in this study, this type of strategy has the potential to be applied to other tumor types involving caveolin-1.
Acknowledgments
We thank Sarah Williams (QFAB) for technical assistance and Prof. Pamela Russell (Queensland University of Technology) for critical reading of the manuscript.
Footnotes
* This study was supported by grants from the Prostate Cancer Foundation Australia, Prostate Cancer Canada, and Association for International Cancer Research. MMH is supported by a National Health and Medical Research Council of Australia (NHMRC) Career Development Fellowship (App ID 569512). RGP is supported by an NHMRC Australia Fellowship and project grant ID 631371. Access to GeneGo was provided by the Queensland Facility for Advanced Bioinformatics (QFAB) though LIEF Grant LE098933.
 This article contains supplemental Tables S1 to S3.
This article contains supplemental Tables S1 to S3.
1 The abbreviations used are:
- PTRF
- polymerase I and transcript release factor
- DRMs
- detergent-resistant membranes
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- SILAC
- stable isotope labeling by amino acids in cell culture
- MβCD
- methyl-β-cyclodextrin
- IL-6
- Interleukin-6
- KLK-6
- Kallikrein 6
- PBS
- phosphate-buffered saline.
REFERENCES
- 1. Bennett N., Hooper J. D., Lee C. S., Gobe G. C. (2009) Androgen receptor and caveolin-1 in prostate cancer. IUBMB Life 61, 961–970 [DOI] [PubMed] [Google Scholar]
- 2. Williams T. M., Hassan G. S., Li J., Cohen A. W., Medina F., Frank P. G., Pestell R. G., Di Vizio D., Loda M., Lisanti M. P. (2005) Caveolin-1 promotes tumor progression in an autochthonous mouse model of prostate cancer: genetic ablation of Cav-1 delays advanced prostate tumor development in tramp mice. J. Biol. Chem. 280, 25134–25145 [DOI] [PubMed] [Google Scholar]
- 3. Parton R. G., Simons K. (2007) The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 8, 185–194 [DOI] [PubMed] [Google Scholar]
- 4. Parton R. G., Richards A. A. (2003) Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms. Traffic 4, 724–738 [DOI] [PubMed] [Google Scholar]
- 5. Hill M. M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S. J., Walser P., Abankwa D., Oorschot V. M., Martin S., Hancock J. F., Parton R. G. (2008) PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu L., Brown D., McKee M., Lebrasseur N. K., Yang D., Albrecht K. H., Ravid K., Pilch P. F. (2008) Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab. 8, 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gould M. L., Williams G., Nicholson H. D. (2010) Changes in caveolae, caveolin, and polymerase 1 and transcript release factor (PTRF) expression in prostate cancer progression. Prostate 70, 1609–1621 [DOI] [PubMed] [Google Scholar]
- 8. Tahir S. A., Yang G., Ebara S., Timme T. L., Satoh T., Li L., Goltsov A., Ittmann M., Morrisett J. D., Thompson T. C. (2001) Secreted caveolin-1 stimulates cell survival/clonal growth and contributes to metastasis in androgen-insensitive prostate cancer. Cancer Res. 61, 3882–3885 [PubMed] [Google Scholar]
- 9. Tahir S. A., Yang G., Goltsov A. A., Watanabe M., Tabata K., Addai J., Fattah el M. A., Kadmon D., Thompson T. C. (2008) Tumor cell-secreted caveolin-1 has proangiogenic activities in prostate cancer. Cancer Res. 68, 731–739 [DOI] [PubMed] [Google Scholar]
- 10. Tahir S. A., Ren C., Timme T. L., Gdor Y., Hoogeveen R., Morrisett J. D., Frolov A., Ayala G., Wheeler T. M., Thompson T. C. (2003) Development of an immunoassay for serum caveolin-1: a novel biomarker for prostate cancer. Clin. Cancer Res. 9, 3653–3659 [PubMed] [Google Scholar]
- 11. Théry C., Zitvogel L., Amigorena S. (2002) Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 12. Jansen F. H., Krijgsveld J., van Rijswijk A., van den Bemd G. J., van den Berg M. S., van Weerden W. M., Willemsen R., Dekker L. J., Luider T. M., Jenster G. (2009) Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol. Cell. Proteomics 8, 1192–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Llorente A., de Marco M. C., Alonso M. A. (2004) Caveolin-1 and MAL are located on prostasomes secreted by the prostate cancer PC-3 cell line. J. Cell Sci. 117, 5343–5351 [DOI] [PubMed] [Google Scholar]
- 14. Llorente A., van Deurs B., Sandvig K. (2007) Cholesterol regulates prostasome release from secretory lysosomes in PC-3 human prostate cancer cells. Eur. J. Cell. Biol. 86, 405–415 [DOI] [PubMed] [Google Scholar]
- 15. Aung C. S., Hill M. M., Bastiani M., Parton R. G., Parat M. O. (2011) PTRF-cavin-1 expression decreases the migration of PC3 prostate cancer cells: role of matrix metalloprotease 9. Eur J. Cell Biol. 90, 136–142 [DOI] [PubMed] [Google Scholar]
- 16. Bastiani M., Liu L., Hill M. M., Jedrychowski M. P., Nixon S. J., Lo H. P., Abankwa D., Luetterforst R., Fernandez-Rojo M., Breen M. R., Gygi S. P., Vinten J., Walser P. J., North K. N., Hancock J. F., Pilch P. F., Parton R. G. (2009) MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J. Cell Biol. 185, 1259–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inder K. L., Lau C., Loo D., Chaudhary N., Goodall A., Martin S., Jones A., van der Hoeven D., Parton R. G., Hill M. M., Hancock J. F. (2009) Nucleophosmin and nucleolin regulate K-Ras plasma membrane interactions and MAPK signal transduction. J. Biol. Chem. 284, 28410–28419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reidegeld K. A., Eisenacher M., Kohl M., Chamrad D., Körting G., Blüggel M., Meyer H. E., Stephan C. (2008) An easy-to-use Decoy Database Builder software tool, implementing different decoy strategies for false discovery rate calculation in automated MS/MS protein identifications. Proteomics 8, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 19. Nakashima J., Tachibana M., Horiguchi Y., Oya M., Ohigashi T., Asakura H., Murai M. (2000) Serum interleukin 6 as a prognostic factor in patients with prostate cancer. Clin. Cancer Res. 6, 2702–2706 [PubMed] [Google Scholar]
- 20. Twillie D. A., Eisenberger M. A., Carducci M. A., Hseih W. S., Kim W. Y., Simons J. W. (1995) Interleukin-6: a candidate mediator of human prostate cancer morbidity. Urology 45, 542–549 [DOI] [PubMed] [Google Scholar]
- 21. Sardana G., Marshall J., Diamandis E. P. (2007) Discovery of candidate tumor markers for prostate cancer via proteomic analysis of cell culture-conditioned medium. Clin. Chem. 53, 429–437 [DOI] [PubMed] [Google Scholar]
- 22. Nebl T., Pestonjamasp K. N., Leszyk J. D., Crowley J. L., Oh S. W., Luna E. J. (2002) Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J. Biol. Chem. 277, 43399–43409 [DOI] [PubMed] [Google Scholar]
- 23. Ponce J., Brea D., Carrascal M., Guirao V., Degregorio-Rocasolano N., Sobrino T., Castillo J., Dávalos A., Gasull T. (2010) The effect of simvastatin on the proteome of detergent-resistant membrane domains: decreases of specific proteins previously related to cytoskeleton regulation, calcium homeostasis and cell fate. Proteomics 10, 1954–1965 [DOI] [PubMed] [Google Scholar]
- 24. Stahlhut M., van Deurs B. (2000) Identification of filamin as a novel ligand for caveolin-1: evidence for the organization of caveolin-1-associated membrane domains by the actin cytoskeleton. Mol. Biol. Cell 11, 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farina H. G., Bublik D. R., Alonso D. F., Gomez D. E. (2002) Lovastatin alters cytoskeleton organization and inhibits experimental metastasis of mammary carcinoma cells. Clin. Exp. Metastasis 19, 551–559 [DOI] [PubMed] [Google Scholar]
- 26. Romanenko V. G., Roser K. S., Melvin J. E., Begenisich T. (2009) The role of cell cholesterol and the cytoskeleton in the interaction between IK1 and maxi-K channels. Am. J. Physiol. Cell Physiol 296, C878–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun M., Northup N., Marga F., Huber T., Byfield F. J., Levitan I., Forgacs G. (2007) The effect of cellular cholesterol on membrane-cytoskeleton adhesion. J. Cell Sci. 120, 2223–2231 [DOI] [PubMed] [Google Scholar]
- 28. Murata M., Peränen J., Schreiner R., Wieland F., Kurzchalia T. V., Simons K. (1995) VIP21/caveolin is a cholesterol-binding protein. Proc. Natl. Acad. Sci. U.S.A. 92, 10339–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smart E. J., Ying Y., Donzell W. C., Anderson R. G. (1996) A role for caveolin in transport of cholesterol from endoplasmic reticulum to plasma membrane. J. Biol. Chem. 271, 29427–29435 [DOI] [PubMed] [Google Scholar]
- 30. Dhanvantari S., Loh Y. P. (2000) Lipid raft association of carboxypeptidase E is necessary for its function as a regulated secretory pathway sorting receptor. J. Biol. Chem. 275, 29887–29893 [DOI] [PubMed] [Google Scholar]
- 31. Wang Y., Thiele C., Huttner W. B. (2000) Cholesterol is required for the formation of regulated and constitutive secretory vesicles from the trans-Golgi network. Traffic 1, 952–962 [DOI] [PubMed] [Google Scholar]
- 32. Lelkes P. I., Friedman J. E., Rosenheck K., Oplatka A. (1986) Destabilization of actin filaments as a requirement for the secretion of catecholamines from permeabilized chromaffin cells. FEBS Lett. 208, 357–363 [DOI] [PubMed] [Google Scholar]
- 33. Dorff T. B., Goldman B., Pinski J. K., Mack P. C., Lara P. N., Jr., Van Veldhuizen P. J., Jr., Quinn D. I., Vogelzang N. J., Thompson I. M., Jr., Hussain M. H. (2010) Clinical and correlative results of SWOG S0354: a phase II trial of CNTO328 (siltuximab), a monoclonal antibody against interleukin-6, in chemotherapy-pretreated patients with castration-resistant prostate cancer. Clin. Cancer Res. 16, 3028–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lv X. J., Li Y. Y., Zhang Y. J., Mao M., Qian G. S. (2010) Over-expression of caveolin-1 aggravate LPS-induced inflammatory response in AT-1 cells via up-regulation of cPLA2/p38 MAPK. Inflamm. Res. 59, 531–541 [DOI] [PubMed] [Google Scholar]
- 35. Wang X. M., Kim H. P., Song R., Choi A. M. (2006) Caveolin-1 confers antiinflammatory effects in murine macrophages via the MKK3/p38 MAPK pathway. Am. J. Respir. Cell Mol. Biol. 34, 434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Egea G., Lázaro-Dieguez F., Vilella M. (2006) Actin dynamics at the Golgi complex in mammalian cells. Curr. Opin. Cell Biol. 18, 168–178 [DOI] [PubMed] [Google Scholar]
- 37. Stamnes M. (2002) Regulating the actin cytoskeleton during vesicular transport. Curr. Opin. Cell Biol. 14, 428–433 [DOI] [PubMed] [Google Scholar]
- 38. Heimann K., Percival J. M., Weinberger R., Gunning P., Stow J. L. (1999) Specific isoforms of actin-binding proteins on distinct populations of Golgi-derived vesicles. J. Biol. Chem. 274, 10743–10750 [DOI] [PubMed] [Google Scholar]
- 39. Lanzetti L. (2007) Actin in membrane trafficking. Curr. Opin. Cell Biol. 19, 453–458 [DOI] [PubMed] [Google Scholar]
- 40. Chichili G. R., Rodgers W. (2009) Cytoskeleton-membrane interactions in membrane raft structure. Cell Mol. Life Sci. 66, 2319–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ikonen E. (2001) Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13, 470–477 [DOI] [PubMed] [Google Scholar]
- 42. Muriel O., Echarri A., Hellriegel C., Pavón D. M., Beccari L., Del Pozo M. A. (2011) Phosphorylated filamin A regulates actin-linked caveolae dynamics. J. Cell Sci. 124, 2763–2776 [DOI] [PubMed] [Google Scholar]
- 43. Ravid D., Chuderland D., Landsman L., Lavie Y., Reich R., Liscovitch M. (2008) Filamin A is a novel caveolin-1-dependent target in IGF-I-stimulated cancer cell migration. Exp. Cell Res. 314, 2762–2773 [DOI] [PubMed] [Google Scholar]
- 44. Li Y. C., Park M. J., Ye S. K., Kim C. W., Kim Y. N. Elevated levels of cholesterol-rich lipid rafts in cancer cells are correlated with apoptosis sensitivity induced by cholesterol-depleting agents. Am. J. Pathol. 168:1107–1118, 2006; quiz 1404–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schmidt K., Schrader M., Kern H. F., Kleene R. (2001) Regulated apical secretion of zymogens in rat pancreas. Involvement of the glycosylphosphatidylinositol-anchored glycoprotein GP-2, the lectin ZG16p, and cholesterol-glycosphingolipid-enriched microdomains. J. Biol. Chem. 276, 14315–14323 [DOI] [PubMed] [Google Scholar]
- 46. Rozelle A. L., Machesky L. M., Yamamoto M., Driessens M. H., Insall R. H., Roth M. G., Luby-Phelps K., Marriott G., Hall A., Yin H. L. (2000) Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10, 311–320 [DOI] [PubMed] [Google Scholar]
- 47. Lebrand C., Corti M., Goodson H., Cosson P., Cavalli V., Mayran N., Fauré J., Gruenberg J. (2002) Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J. 21, 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]