Abstract
The budding yeast Saccharomyces cerevisiae is a major model organism for important biological processes such as mitotic growth and meiotic development, it can be a human pathogen, and it is widely used in the food-, and biotechnology industries. Consequently, the genomes of numerous strains have been sequenced and a very large amount of RNA profiling data is available. Moreover, it has recently become possible to quantitatively analyze the entire yeast proteome; however, efficient and cost-effective high-throughput protein profiling remains a challenge. We report here a new approach to direct and label-free large-scale yeast protein identification using a tandem buffer system for protein extraction, two-step protein prefractionation and enzymatic digestion, and detection of peptides by iterative mass spectrometry. Our profiling study of diploid cells undergoing rapid mitotic growth identified 86% of the known proteins and its output was found to be widely concordant with genome-wide mRNA concentrations and DNA variations between yeast strains. This paves the way for comprehensive and straightforward yeast proteome profiling across a wide variety of experimental conditions.
The budding yeast Saccharomyces cerevisiae is important for the food industry (1), biotechnology (2), clinical research (3, 4), and basic sciences (5). Over the past 15 years this versatile organism has been an excellent model organism for the development of methods to study genome evolution (3, 6), the transcriptome (7, 8), the proteome (9–12), as well as protein-protein networks (13) and protein-DNA interactions (14).
More recently, major technological advances have yielded quantitative information on the yeast proteome in both haploid and diploid cycling cells (15). However, these methods are cumbersome and technically challenging and they rely on cellular uptake of amino acid analogs for protein labeling (stable-isotope labeling by amino acids in cell culture, SILAC1 (16)), thus hampering efforts to study the dynamic proteome under environmental conditions that alter the ability of cells to absorb and to process nutrients such as stress response and gametogenesis (17, 18). This is a critical issue for efforts to complement the rapidly growing body of data on DNA and RNA with reliable information on most, if not all proteins under many conditions and in different strain backgrounds. A promising solution for this experimental challenge is selected reaction monitoring (SRM), a highly sensitive method with a large dynamic range, that has been used to detect 100 proteins in a single run (19).
We report the development of Direct Iterative Protein Profiling (DIPP), an innovative, robust and highly sensitive method for protein profiling. Critically, DIPP does not require the uptake of amino acid analogs making it suitable for the analysis of a wide range of experimental conditions and mutant strains. The procedure includes a tandem buffer system for protein extraction and a simple acrylamide-gel based step for protein prefractionation and cleavage, followed by three consecutive rounds of peptide detection and protein identification using mass spectrometry and algorithms implemented in Mascot and SEQUEST. We have employed DIPP to study duplicate samples from diploid SK1 MATa/α cells undergoing rapid mitotic growth and division in rich medium. The vast majority of the proteins predicted in the yeast genome were identified at least once (86%) (20). For many proteins not detected we observed very little or no mRNA expression (21) or we identified strain-specific DNA variations likely deleterious for the proteins (22). Our simplified and versatile method covers the yeast proteome to a level that is comparable to the most sophisticated approach available today (15). DIPP paves the way for future efforts to study the dynamic budding yeast proteome under many experimental conditions in distinct strains.
EXPERIMENTAL PROCEDURES
Yeast Strain and Media
We analyzed a diploid SK1 MATa/MATα ho::LYS2/ho::LYS2 ura3/ura3 lys2/lys2 leu2::hisG/leu2::hisG arg4-Nsp/arg4-Bgl his4x::LEU2-URA3/his4B::LEU2 strain previously used in expression profiling studies (21, 23). Two independent fresh yeast colonies were inoculated into 5 ml YPD (yeast extract, peptone, and dextrose) and cultured at 30 °C at 180 rpm over night (Innova 44/44R rotary shaker, New Brunswick). Cells were resuspended in 100 ml YPD prewarmed to 30 °C at a cell density of 2 × 106 cells/ml and cultured until they reached 3 × 107 cells/ml, before they were harvested in two 50 ml aliquots each, washed with sterile water, snap-frozen in liquid nitrogen, and kept at −80 °C.
Preparation of Protein Extracts
Duplicate proteome samples were prepared from two cell pellets each containing 1.5 × 109 cells. The pellets were washed twice in cold 1 × phosphate-buffered saline (PBS) and suspended either in 750 μl 50 mm Tris pH 8, 10 mm MgCl2, 1 mm EDTA (Sigma), 1 u/ml of Complete protease inhibitor mix (Roche), or in 8 m Urea, 4% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (Chaps), 30 mm Tris-base, 1 mm EDTA, 0.5 mm dithiothreitol (Sigma), 1 u/ml of Complete protease inhibitor mix. Protein extracts were prepared using the MM301 grinder (Retsch) and steel beads with a 20 mm diameter. Cells were broken up in liquid nitrogen during five consecutive cycles lasting 2 mins each at a frequency of 10 Hz. After thawing, the extracts were centrifuged first at 14,000 × g for 20 min in a 4417R centrifuge (Eppendorf, New York, NY) and then at 105000 × g for 30 min in an ultracentrifuge (Sorval M120SE). The protein concentration of the supernatant was determined using a Pierce 660 protein assay kit (Thermo Scientific); aliquots were stored at −80 °C.
Protein Prefractionation and Digestion
Two hundred micrograms of soluble protein extracts obtained with the Tris and Urea/Chaps buffers were pooled, boiled for 5 min in a 5 × Laemmli buffer (10% SDS, 40% glycerol, 0.025 m dithiotreitol, 0.02% bromphenol blue, 0.25 m Tris, pH 6.8) and separated on a custom-made 10% SDS-PAGE gel (gel size of 16 × 18 cm, 1.5 mm thick, Hoeffer 600 electrophoresis unit) overnight at 45 V. Gels were stained with EZBlue (Sigma) for 30 min and destained with water over night. Each gel lane was manually cut into 30 slices of approximately the same size; care was taken to cover the front defined by Coomassie stain with the last three slices to include smaller proteins of <20 kDa. The slices were first treated with 50 mm NH4HCO3 in acetonitrile/water 1:1 (v/v), dehydrated with 100% acetonitrile and rehydrated in 100 mm NH4HCO3. Next they were washed again with 50 mm NH4HCO3 in acetonitrile/water, 1:1 (v/v) and dehydrated with 100% acetonitrile. The slices were then treated with 65 mm DTT for 15 min at 37 °C, and with 135 mm iodoacetamide in the dark at room temperature. Finally, the samples were washed with 100 mm NH4HCO3 in acetonitrile/water, 1:1 (v/v), and dehydrated with 100% acetonitrile before being rehydrated in 100 mm NH4HCO3, washed with 100 mm NH4HCO3 in acetonitrile/water, 1:1 (v/v) and then dehydrated again with 100% acetonitrile. Proteins were digested overnight at 37 °C with 4 ng/μl of modified trypsin (Promega, Madison, WI) in 50 mm NH4HCO3. Peptides were extracted by incubating the slices first in 80 μl of acetonitrile/water/trifluoroacetic acid (70/30/0.1; v/v/v) for 20 min, and then in 40 μl of 100% acetonitrile for 5 min and finally in 40 μl of acetonitrile/water/trifluoroacetic acid (70/30/0.1; v/v/v) for 15 min. Supernatants were transferred into fresh tubes and concentrated in a SpeedVac (Thermo Scientific) for 15 min to a final volume of 60 μl.
MS Analysis
The MS measurements were performed with a nanoflow high-performance liquid chromatography (HPLC) system (Dionex, LC Packings Ultimate 3000) connected to a hybrid LTQ-OrbiTrap XL (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (New Objective). The HPLC system consists of a solvent degasser nanoflow pump, a thermostated column oven kept at 30 °C, and a thermostated autosampler kept at 8 °C to reduce sample evaporation. Mobile A (99.9% MilliQ water and 0.1% formic acid (v:v)) and B (99.9% acetonitrile and 0.1% formic acid (v:v)) phases for HPLC were delivered by the Ultimate 3000 nanoflow LC system (Dionex, LC Packings). Ten microliters of prepared peptide mixture was loaded on a trapping precolumn (5 mm × 300 μm i.d., 300 Å pore size, Pepmap C18, 5 μm) for 3 min in 2% buffer B at a flow rate of 25 μl/minute. This step was followed by reverse-phase separations at a flow rate of 0.250 μl/minute using an analytical column (15 cm × 300 μm i.d., 300 Å pore size, Pepmap C18, 5 μm, Dionex, LC Packings). We ran a gradient ranging from 2 to 35% buffer B for the first 60 min, 35 to 60% buffer B from minutes 60–85, and 60 to 90% buffer B from minutes 85–105. Finally, the column was washed with 90% buffer B for 16 min, and with 2% buffer B for 19 min prior to loading of the next sample. The peptides were detected by directly eluting them from the HPLC column into the electrospray ion source of the mass spectrometer. An electrospray ionization voltage of 1.5 kV was applied to the HPLC buffer using the liquid junction provided by the nanoelectrospray ion source and the ion transfer tube temperature was set to 200 °C.
The MS instrument was operated in its data-dependent mode by automatically switching between full survey scan MS and consecutive MS/MS acquisition. Survey full scan MS spectra (mass range 400–2000) were acquired in the OrbiTrap section of the instrument with a resolution of r = 60,000 at m/z 400; ion injection times are calculated for each spectrum to allow for accumulation of 106 ions in the OrbiTrap. The seven most intense peptide ions in each survey scan with an intensity above 2000 counts (to avoid triggering fragmentation too early during the peptide elution profile) and a charge state ≥2 were sequentially isolated at a target value of 10,000 and fragmented in the linear ion trap by collision induced dissociation. Normalized collision energy was set to 35% with an activation time of 30 milliseconds. Peaks selected for fragmentation were automatically put on a dynamic exclusion list for 120 s with a mass tolerance of ±10 ppm to avoid selecting the same ion for fragmentation more than once. The following parameters were used: the repeat count was set to 1, the exclusion list size limit was 500, singly charged precursors were rejected, and a maximum injection time wet was set at 500 ms and 300 ms for full MS and MS/MS scan events, respectively. For an optimal duty cycle the fragment ion spectra were recorded in the LTQ mass spectrometer in parallel with the OrbiTrap full scan detection. For OrbiTrap measurements, an external calibration was used before each injection series ensuring an overall error mass accuracy below 5 ppm for the detected peptides. MS data were saved in RAW file format (Thermo Fisher Scientific) using XCalibur 2.0.7 with tune 2.4.
Data Processing, Generation of Exclusion Lists and Identification of Peptides and Proteins
The data analysis was performed with the Proteome Discoverer 1.2 software supported by Mascot (Matrixscience) and SEQUEST database search engines for peptide and protein identification. MS/MS spectra were first compared with all predicted budding yeast proteins (data provided by Saccharomyces Genome Database release 06/01/2010; number of residues: 3020761, number of sequences: 6717) (20). Mass tolerance for MS and MS/MS was set at 10 ppm and 0.5 Dalton, respectively. The enzyme selectivity was set to full trypsin with one miss cleavage allowed. Protein modifications were fixed carbamidomethylation of cysteines, variable oxidation of methionine, variable acetylation of lysine, and variable phosphorylation of serine, threonine and tyrosine. Identified peptides were filtered based on Xcorr values and the Mascot score to obtain a false discovery rate of 1% and a false positive rate of 5%. We employed Proteome Discoverer to generate lists of peptides identified in the first and second run that are excluded in subsequent LC-MS/MS analyses. Prior to the third analysis, peptide exclusion files from the first two runs are combined. Lists of peptides not filtered out are exported as a text file containing uncharged and accurate mass values (at five decimals) and a retention time window of approximately 1 min. The instrument is configured to work with uncharged masses and to automatically calculate the mass of a peptide based on its exact mass and charge state. A mass tolerance of ±10 ppm is used to reject previously identified peptides within the specified retention time window. Using lower values can lead to reselection of masses because in the parallel mode of operation on an LTQ OrbiTrap XL, the parent ion selection for an ion trap MS/MS is based on an OrbiTrap preview scan that is acquired at a lower resolution (RP 15,000) than the final OrbiTrap full scan, and therefore the masses are less accurate. The list of identified proteins is provided in Supplemental Files 4 (YPD1) and 5 (YPD2).
Tiling Array Expression Data
DNA-strand specific whole-genome expression data obtained with tiling arrays and duplicate samples from diploid SK1 cells cultured in rich medium (YPD) were integrated and compared with the mass spectrometry measurements; data processing methods and expression threshold level parameters were as published (21). For each gene listed in the reference genome, we selected the segments derived from Sc_tiling experiments overlapping by at least 50 bp. When a gene was overlapping with several segments, it was considered as expressed if at least one of the segments was expressed above threshold.
Protein Abundance Data
The relative abundance data of proteins expressed in log-phase growth were extracted from the quantitative Western blot analysis of tandem affinity purification-tagged strains available via Saccharomyces Genome Database (SGD) (http://yeastgfp.yeastgenome.org/) (24).
DNA Variations Between Yeast Strains S288c and SK1
The variations between the haploid reference S. cerevisiae strain S288c and haploid SK1 were obtained from the Yeast Population Genomics project (21). For each gene, we extracted the reference sequence and the corresponding sequence in SK1. Both sequences were translated (synonymous mutations were thus ignored) and sequences were aligned using a classical Needleman and Wunsch algorithm. We then identified deletions, single nucleotide polymorphisms that create stop codons, and nonsynonymous variations in the SK1 genome. To distinguish between nonsynonymous variations occurring in conserved or nonconserved positions we used the fungal alignment provided by SGD (19, 22). Proteins lacking homologs across yeast species and proteins for which the reference sequence has changed in SGD since the study by Liti et al. (22) was published were excluded. A bilateral statistical test was used to determine if undetectable proteins are more often mutated in conserved positions than observed proteins.
MIAPE Compliance
The raw MS spectra were uploaded to the EBI's PRIDE repository and are available at http://www.ebi.ac.uk/pride/.
RESULTS
Experimental Design and Workflow
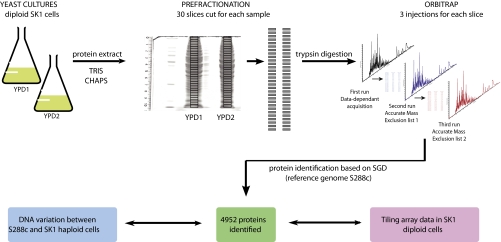
It is our ultimate goal to study the proteome of the budding yeast life cycle. To establish a suitable method we first sought to determine the complete proteome of diploid budding yeast cells undergoing rapid mitotic growth and division in the presence of glucose (fermentation). To this end, we inoculated two cultures with independent colonies of SK1 MATa/α cells and grew them to mid-log phase in rich medium (YPD1 and YPD2). We chose SK1 because it displays normal mitotic growth properties and, as opposed to the reference strain S288c, it undergoes meiosis and gametogenesis efficiently. Moreover, SK1's genome sequence is available (albeit poorly annotated) (22) and we have a large mitotic and meiotic tiling array expression data set for this strain background (21). To maximize protein solubility and peptide detection we prepared extracts using two different buffer systems and then separated the combined protein samples based on their molecular weight via SDS-PAGE. Next, we digested protein fractions present in 30 slices from each of the two lanes with trypsin, and analyzed the peptides with a mass spectrometer during three consecutive rounds of injection; accurate mass exclusion lists of identified peptides were established at each round. Samples were analyzed in duplicate to estimate DIPP's level of reproducibility. Finally, proteome data were interpreted in the context of information on the degree of DNA sequence conservation (25), and DNA mutations such as insertions and deletions (indels) and single nucleotide polymorphisms (22) as well as genome-wide RNA concentrations available for the SK1 strain (Fig. 1, see Materials and Methods) (21).
Fig. 1.
Experimental design and workflow. A schematic diagram of the procedure used to detect 4952 proteins in duplicate diploid SK1 cells cultured in rich medium (YPD 1 and YPD2) is given.
The Core Budding Yeast Proteome of Mitotic Growth
According to the SGD (release 18/10/2010), the 16 chromosomes of the budding yeast genome contain 6685 protein coding genes comprising 4864 verified open reading frames (ORFs, including four silenced genes), 910 uncharacterized ORFs, 801 dubious ORFs, and 110 unclassified ORFs (20). We have identified at least once 4952 out of 6685 theoretically predicted proteins (74%) as being present in mitotically growing cells. Importantly, when taking only the verified genes into account, the output of our experiment covers 86% of the predicted yeast proteome (4175 proteins as compared to 4864 ORFs). This suggests that we have achieved essentially complete coverage of the protein profile in fermenting cells because several hundred proteins are involved in processes not included in our analysis, such as haploid-specific pheromone signal transduction and mating, filamentous growth, stress-response, respiration (mitotic growth in the presence of a nonfermentable carbon source), and sporulation (supplemental File S1).
As expected, the vast majority of the proteins identified fall into the class of verified ORFs (4403 and 4069 in YPD1 and YPD2, respectively) but we also detected proteins corresponding to uncharacterized genes (YPD1: 442; YPD2: 394), dubious ORFs (YPD1: 116; YPD2: 138), and unclassified loci (YPD1: 22; YPD2: 18) (Table I). The surprisingly large number of proteins associated with poorly characterized genes or dubious loci that are not conserved or that overlap with larger validated genes emphasizes that the budding yeast S288c reference genome—15 years after its initial publication (26)—is not yet exhaustively annotated.
Table I. Numbers of proteins detected in duplicate samples. The table summarizes the numbers of ORFs in the yeast genome falling into four different categories as provided by SGD. The output of two independent protein profiling studies in rich medium (YPD1, YPD2) is given.
| ORF category | SGD | YPD1 | YPD2 |
|---|---|---|---|
| All | 6685 | 4403 | 4069 |
| Verified | 4864 | 3823 | 3519 |
| Uncharacterized | 910 | 442 | 394 |
| Dubious | 801 | 116 | 138 |
| Unclassified | 110 | 22 | 18 |
A confounding aspect of the yeast genome annotation project is the finding that backgrounds such as SK1 may not only lack genes present in the reference strain (23) but they may also contain protein-coding genes that are missing in S288c (22). To test this idea we investigated 17 hypothetical ORFs absent in the reference strain but present and conserved in the genomes of several S. cerevisiae strains including SK1 (21). Mapping the peptides identified in YPD1 and YPD2 samples confirmed the presence of gene products in all cases to variable degrees of confidence depending on how many peptides were found for each putative protein and how reproducible protein detection was (supplemental File S2). We conclude that comparative genomics is indeed facilitating the discovery of bona fide protein-coding genes in S. cerevisiae and that efforts to identify the full complement of genes present in the budding yeast genome will require information from many different strain backgrounds.
SDS-Gel Based Protein Prefractionation is Robust and Reproducible
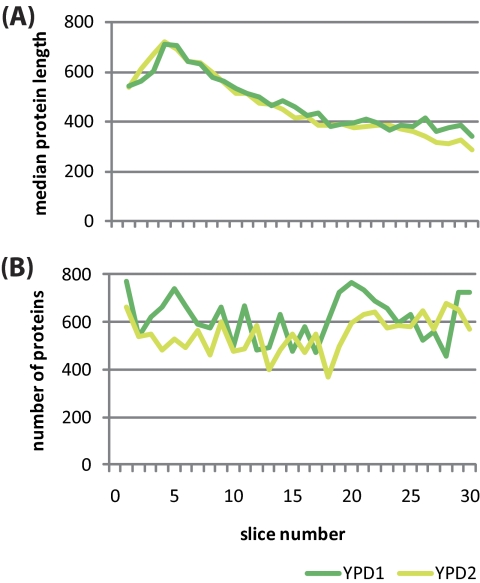
We next explored how efficient and reproducible our simple SDS-PAGE based approach to fractionation of the yeast proteome was. To this end, we plotted the size of the proteins (as the median number of amino acids) over the 30 slices from the top (slice 1) to the bottom (slice 30) of the gel. Somewhat unexpectedly, we observed a negative (albeit reproducible) correlation between molecular weight and migration speed within the top four slices; as opposed to that, a clear and highly reproducible correlation between the molecular weight and the migration position was apparent in slices 5–30 (Fig. 2A). We also found by and large similar numbers of proteins—varying between 400 and 800—within the two sets of 30 slices (Fig. 2B). These results indicate that although we reached the limit of protein separation via size at the very high molecular weight range, manually cut slices yielded consistent results throughout the entire range of the running gel.
Fig. 2.
Prefractionation results and distribution of protein numbers and sizes in gel slices. A, The slice number is plotted (x axis) against the median length of proteins given in amino acids (aa; y axis). B, Slices (x axis) are plotted against the numbers of proteins identified (y axis). Replicate samples from cells grown in rich medium (YPD) are color coded in green (YPD1) and yellow (YPD2).
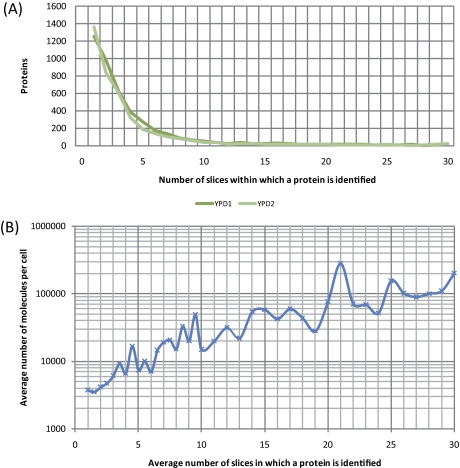
We then asked how efficient proteins were separated based on their molecular weight given that a large amount of protein extract was loaded onto the SDS gel to increase the concentration of peptides injected into the mass spectrometer (see Materials and Methods). Although ∼1300 proteins were found in one slice each (1252 in YPD1 and 1365 in YPD2), all other proteins were present in more than one slice and around 400 proteins were found on average in 10–30 slices, with 97 proteins being present in 20–30 slices (Fig. 3A). The distribution of proteins within slices was found to be highly reproducible (correlation coefficient >0.89 between YPD1 and YPD2). One likely explanation for this phenomenon was that highly abundant proteins would saturate the gel system. To test this idea we plotted the average number of slices in which a protein was detected against its concentration in molecules per cell (24) and found that cellular protein abundance was strikingly correlated with the tendency of a protein to be detected in more than one band (Fig. 3B). Concordantly, the group of 97 proteins found in 20–30 slices for which Gene Ontology (27) annotation data were available was significantly enriched statistically for, among others, Translation (p value 1.72 × 10−20), Glucose metabolic process (1.94 × 10−20), and Protein metabolic process (6.19 × 10−12). We conclude that our method is suitable for prefractionation of most yeast proteins and, specifically, that abundant proteins (including those which completely saturate the gel) are prevented from saturating the MS system.
Fig. 3.
Protein abundance and presence in multiple slices. A, The number of slices in which a protein was detected (x axis) is plotted against the number of proteins (y axis). Replicates are color coded. B, The average number of slices in which a protein was detected is plotted against the average number of molecules per cell (log10 scale, y axis) as determined by quantitative Western blot analysis of tandem affinity purification-tagged strains (24).
DIPP Yields a Core Protein Complement Across Duplicate Experiments
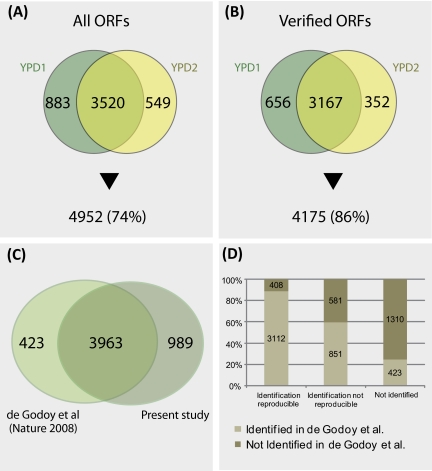
An important precondition for profiling multiple experimental conditions is to ensure that replicates within a given condition are sufficiently reproducible so that meaningful results can be obtained. To test the robustness of DIPP we compared the output of two independent profiling studies (YPD1 and YPD2) first by taking all predicted ORFs into account. Among 4403 proteins detected in YPD1 and 4069 found in YPD2 we identified 3520 twice whereas 883 and 549 were detected only in YPD1 or YPD2, respectively (Fig. 4A). Among verified ORFs we scored 3823 proteins in YPD1 and 3519 in YPD2 as present; 3167 proteins were detected in both samples whereas 656 and 352 were identified only in YPD1 or YPD2, respectively (Fig. 4B).
Fig. 4.
Comparison of predicted proteome and observed protein profile. A, A Venn diagram shows the proteins detected in single samples or both replicates (YPD1 and YPD2) as compared with all predicted open reading frames (ORFs). B, Idem taking only the verified ORFs into consideration. The percentage of detected proteins is indicated. C, A Venn diagram comparing the proteins identified in SK1 and in S288c (15) is shown. D, A graph summarizes the numbers of proteins identified twice in SK1 and in S288c (first column), proteins identified once in SK1 and in S288c (second column) and proteins not identified in this study but in S288c (third column).
Recent work using SILAC has yielded quantitative information on the mitotic proteome in fermenting haploid and diploid cells from the S288c reference strain background (15). In this study, 4386 proteins were identified based on a combination of three different prefractionation strategies. We compared this experiment with our simplified method and found that 3963 proteins were detected in both studies, 423 were reported only by de Godoy et al., and 989 proteins were identified only by DIPP (Figs. 4C and 4D). Furthermore, we compared our results with the output of SRM, a highly sensitive protein profiling method that was applied to groups of proteins present over a very wide range of concentrations: we identified all of ten proteins present at <128 copies/cell (six were detected twice and four only once), all of five proteins reported to be present at <50 copies/cell, and all 15 proteins found by SRM but not by quantitative Western blotting (19, 24). Moreover, among 15 proteins not identified by SRM but known to be present we detect six twice and one only once (19). Finally, we scored as present seven out of 10 proteins (four twice, three once) found by SRM although no peptide is associated with them in PeptideAtlas (supplemental File S1) (28). Taken together, these results highlight the robustness and sensitivity of DIPP.
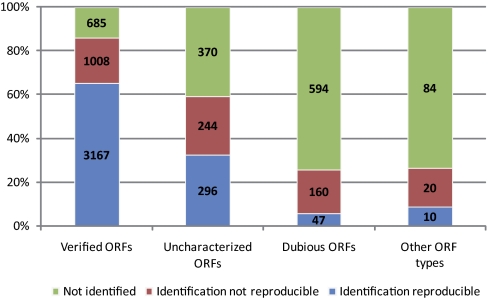
There is a clear correlation between our ability to detect proteins and the level of confidence associated with their biological relevance: among 4860 bona fide genes we detected proteins for 3167 loci twice (65%), for 1008 loci once (21%), and for 685 cases (14%) in neither of the samples. For the group of uncharacterized ORFs we found proteins for 296 loci twice (32%), for 244 loci once (26%), and for 370 cases (40%) we failed to detect a protein. This tendency is even more apparent in the group of dubious ORFs: among 801 cases we find proteins twice for 47 ORFs (6%), once for 160 ORFs (20%), and never for 594 ORFs (74%). Likewise, among 114 cases of unclassified or silenced loci we detected proteins twice for 10 ORFs (9%), once for 20 ORFs (18%), and in none of the samples for 84 ORFs (73%) (Table II; Fig. 5).
Table II. Detection patterns in duplicate samples. The table summarizes the numbers of ORFs in the yeast genome encoding proteins detected in YPD1 and YPD2 or in none of the samples. The percentage of the detected proteins over the total number of annotated genes is given.
| Detection | Detection pattern | All ORFs | % | Verified ORFs | % |
|---|---|---|---|---|---|
| Reproducible | [+ +] | 3520 | 53 | 3167 | 65 |
| Not reproducible | [+ −] | 1432 | 21 | 1008 | 21 |
| Not detected | [− −] | 1733 | 25 | 685 | 14 |
Fig. 5.
Protein identification versus ORF classification. A color coded bar diagram shows the percentage of loci (y axis) falling into four classes as indicated (x axis) that encode proteins identified in both replicates (blue), in only one sample (red), or in none of the samples (green).
The corollary is that DIPP of fermenting diploid cells reproducibly detects approximately two thirds of the proteins encoded by validated genes and one third of the proteins encoded by uncharacterized loci. Furthermore, the vast majority of dubious ORFs do not seem to encode proteins expressed to a level allowing for their detection in asynchronously growing cells.
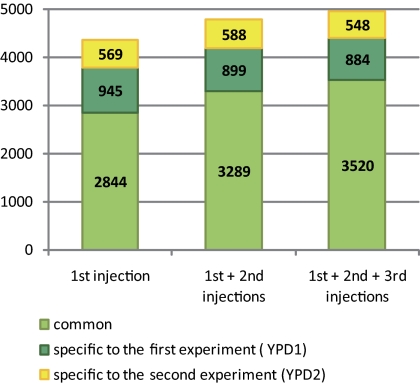
Iterative Injections Increase the Protein Yield
Efficient protein profiling of highly complex samples is hampered by saturation of the MS system. A key element of our method apart from protein prefractionation is to consecutively filter the most abundant peptides that obscure MS signals during iterative rounds of injections. 4358 out of 4952 proteins (88%) are detected after the first injection. However, we found 418 additional proteins during the second round and 176 proteins during the third round (Fig. 6). The data indicate that our approach facilitates the production of interpretable spectra and thereby increases the number of proteins detected. Moreover, we also found that iterative injections increases the reproducibility of our data: with a single injection 65% of the proteins identified are found in both replicates whereas this was the case for 71% after three rounds of iterative injections. Interestingly, a pilot experiment to the present study has shown that a triplicate injection of the same yeast extract without the use of an exclusion list strategy, only lead to an ∼4% increase in the number of proteins identified (data not shown).
Fig. 6.
The effect of iterative sample injection. A color coded bar diagram shows the number of proteins (y axis) identified during three rounds of injections (x axis) in both samples (light green) or in either YPD1 (dark green) or YPD2 (yellow) samples.
Information About DNA Mutations and RNA Expression May Help Predict Protein Stability
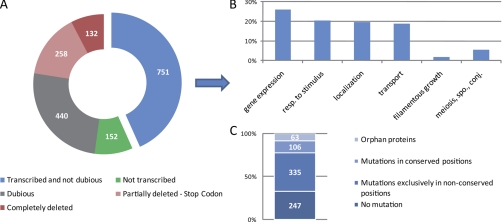
An intriguing outcome of our experiment was that 1733 predicted proteins, including 685 encoded by bona fide genes, were not detected in the replicate DIPP analyses. We therefore set out to explain their absence by integrating proteome data with information on DNA variations between S288c and SK1 strains and RNA expression profiles as determined by microarrays in the SK1 strain background (Fig. 7A) (21, 22).
Fig. 7.
DNA and RNA analysis of predicted proteins not found. A, A color coded doughnut diagram classifies 1733 predicted proteins not found in the replicate samples into five categories as shown in the legend. The percentage of genes falling into each class is shown. B, The major Gene Ontology Biological Process annotations of the 751 genes not deleted and not transcriptionally repressed. C, Analysis of the mutations identified in the 751 genes not deleted and not transcriptionally repressed. Four categories were identified: no mutations, mutations only in nonconserved residues, mutations in at least one conserved residue, and orphan proteins. The conservation was determined using the fungal alignment available on SGD (20, 48), and only identical positions were considered as conserved.
132 genes (8%) were found to be entirely deleted and an additional 258 (15%) lack at least one fifth of their primary sequence because they were partially deleted or because they contained variations that created stop codons leading to the translation of C-terminally truncated proteins in the SK1 background. As expected, C-terminal deletions are mostly small in stable proteins (those identified by DIPP) whereas they are frequently large in the case of proteins not identified (supplemental Fig. S3). Among the genes for which no protein was detected we found 440 dubious loci (25%); this is consistent with the profiling data because they are typically (albeit not exclusively (29)) annotation artifacts that are not expected to encode proteins. Furthermore, for 152 genes (9%) we did not observe expression signals above the threshold level of detection in a tiling array experiment (21) indicating that they are transcriptionally repressed in diploid cells undergoing mitotic growth in rich medium. In the remaining 751 cases (43%) we measured mRNA concentrations above the threshold level, which is consistent with the notion that these genes are post-translationally regulated. Coherently, many of the genes in this group are involved in inducible biological processes such as Response to stimulus (20%), Transport (19%), Meiosis/Sporulation/Conjugation (5%), or Filamentous growth (2%) (Fig. 7B).
We next investigated if the group for which mRNAs but no proteins were found in the cells showed a tendency to contain point mutations in conserved amino acids that might destabilize them. Among 751 cases, we found 247 proteins whose sequences were identical in SK1 and the reference strain S288c and for 63 “orphan” proteins where we were unable to determine sequence conservation because their genes appeared to have no fungal homolog (details in Materials and Methods). Among the remaining 441 cases we found that 335 proteins displayed amino acid substitutions in SK1 exclusively at variable positions, and 106 (24%) proteins contained mutations in at least one highly conserved position (Fig. 7C). Critically, when we determined the frequency of these types of mutations in a randomly selected control group of 441 stable proteins that were detected, we found only 77 proteins (17%) with mutations of highly conserved amino acids. A bilateral test revealed this difference to be statistically significant (confidence interval 0.05, see Materials and Methods). Overall these results suggest that the DNA variations between the reference yeast strain S288c and SK1 may in part explain protein instability observed in SK1. We have initiated experiments to test this idea.
DISCUSSION
We have developed DIPP, a novel robust and straightforward method to profile the proteome in simple eukaryotes and employed it to study mitotic growth in the presence of glucose (fermentation) in the budding yeast S. cerevisiae. DIPP requires only basic equipment for culturing cells, and for processing protein extracts. Peptides were detected using the LTQ-OrbiTrap mass spectrometer—currently the most popular platform in proteomics—and an innovative approach based on iterative rounds of injection followed by masking detected peptides. Critically, our method does not need cell labeling, which means it is suitable for profiling the proteome under all conceivable experimental conditions, including those that entail metabolic changes that interfere with efficient SILAC labeling. We detected essentially all of the known proteins and the vast majority of them were found in duplicate samples, indicating that DIPP is likely efficient enough to carry out meaningful large-scale protein profiling experiments across distinct culture conditions.
A key question is how to increase the protein yield without saturating the system? In a pilot study analyzing a total protein extract in a single round of injections into a hybrid LTQ-OrbiTrap XL mass spectrometer we identified only ∼5% of the predicted proteome, and using only one extraction buffer also yielded suboptimal results (data not shown). Protein fractionation was thus, unsurprisingly, found to be a critical step in large-scale profiling using current MS technology; however, it is tedious and costly in terms of equipment, reagents, and man hours. We therefore sought a simplified solution and found that using mechanical disruption of frozen samples and two buffers with distinct chaotropic properties helped recover proteins over a broad range of solubility (including many genes annotated as encoding membrane proteins). Rather than being analyzed separately, the protein solutions were then mixed prior to high-resolution SDS-gel prefractionation followed by in-gel digestion and iterative injection into the MS system.
Over the past years, shotgun proteomics emerged as a key method in the field (30–32) and various strategies were employed to tackle complex samples including using different MS instruments (33–35), new fragmentation techniques (36), inclusion lists (37), or repeated sample injections (38). However, none of these methods was as efficient as the approach based on peptide mass exclusion lists (39). A critical aspect of iterative mass spectrometry analysis is indeed that already detected peptides are masked during the consecutive step thereby rendering protein detection more effective. To improve our ability to detect proteins we optimized the standard shotgun liquid chromatography-tandem MS (LC-MS/MS) approach using accurate masses and retention time of identified peptides to establish such exclusion lists (39). Our results show that after establishing the first list of identified peptides, the second injection yields a substantial number of proteins not found in the first round whereas the third injection produces a smaller yield probably indicating a plateau effect. It is unclear how far the method could be extended but preliminary results seem to suggest that a fourth round of injection does not lead to the detection of a sufficiently large number of proteins to justify the cost and effort (R. Lavigne and C. Pineau, unpublished). A key question is whether the improvement in protein identification rate is because of the exclusion list strategy rather than chance sampling. It is acknowledged that repeated injections of the same sample improve the protein identification coverage by about 10%. However, this finding is genuinely relevant only for proteome samples of low and medium complexity. In the case of a yeast total cell lysate, it is difficult if not impossible to increase the number of proteins identified without peptide mass exclusion lists. Indeed, a previous study of the yeast proteome using cell lysates shows that at least 10 replicate injections of the same sample are necessary to cover the proteome roughly as extensively as DIPP (32). In this context it should be noted that Piening et al. (38) needed as many as 31 consecutive LC-MS/MS analyses of a yeast cell lysate to reach the number of unique peptides that was identified in a similar sample using only six injections and a mass exclusion-based DDA strategy (i.e. 4550 versus 4490, respectively) (39).
An intriguing outcome of our analysis is that DIPP appears to be very robust and extremely sensitive: we identified proteins such as Erg20, Gcy1, Num1, Pdi1, and Uga2, which were thought to be detectable only by organelle-specific proteomics or extremely elaborate protein fractionation techniques and MS-based peptide detection methods with a threshold as low as 50 molecules per cell (19, 24).
Why do we fail to detect 1733 predicted proteins in the proteome of diploid fermenting cells? One reason is that the SK1 genome contains deletions that remove entire genes and their products and DNA variations that lead to the synthesis of truncated proteins that are likely unstable and subject to rapid degradation for example via the unfolded protein response (22, 40). Although complete or partial deletions obviously provide an excellent explanation for the absence of a protein—provided that our diploid SK1 strain is homozygous for these mutations originally defined in a haploid SK1 background (22)—nonsynonymous mutations represent a weaker, yet still plausible explanation. It is noteworthy in this context that proteins not detected by DIPP are more frequently associated with mutations affecting highly conserved amino acids than a randomly selected group of proteins that were detected at least once. In this context it should be noted that numerous ORFs among the 1733 cases might be annotation artifacts (especially the dubious ones) not encoding functional proteins (25, 41).
The absence of a protein in extracts from cells cultured in YPD may also be because of the fact that many genes involved in processes such as stress-response, filamentous growth, and gametogenesis are transcriptionally repressed or post-translationally regulated during mitotic growth (17, 42–44). Furthermore, it is possible that loci that are transcribed to a level detectable by microarrays may encode proteins that are particularly unstable in vegetatively growing cells (45). Other proteins may escape our detection system because they are not soluble under the conditions we used or they are too small to be captured on the SDS gel system we employed. We note that the latter issue is likely not critical because we detected 669 proteins of less than 150 amino acids at least once. Other potential issues are that peptides may ionize at a low frequency or not at all, that hydrophobic peptides can suppress the ion signal of hydrophilic peptides, and that highly concentrated peptides can mask the ion signals of less abundant ones, making it impossible to detect them (39, 46, 47). It is conceivable that increasing the number of slices beyond 30 may lead to the detection of additional proteins. However, given the fact that the current DIPP approach detects the vast majority of the proteins expected to be present in mitotic cells growing under optimal conditions it is unclear if a potentially marginal improvement justifies the additional cost and labor.
The output of our profiling study is mostly coherent with the level of confidence attributed to different classes of annotated genes because most proteins we find in both samples fall into the group of verified ORFs whereas predicted proteins consistently absent are often encoded by dubious genes (see supplemental File S1). We did, however, detect a surprisingly large number of proteins that appear to be associated with loci that do not fulfill the classical criteria for a bona fide yeast gene (such as a minimal number of codons, lack of overlap with another gene on the opposite stand, and sequence conservation). It is safe to assume that efforts to comprehensively annotate the budding yeast genome will require the output of comparative genomics as well as RNA-, and protein profiling work.
A critical issue of DIPP is its limited ability to quantify protein concentrations. Current MS data yield indirect information about abundance via the number of peptides associated with a given protein but this measure is imprecise. As a consequence DIPP is not a quantitative method but this is compensated by the fact that it is applicable to a large number of experimental conditions, which may be difficult or impossible to study by approaches based on SILAC. Moreover, its workload is comparatively moderate putting proteomics on a large scale within the reach of many laboratories with access to standard MS equipment. Finally, we are currently implementing a new approach known as peptide-count or absolute quantification that will help quantify protein concentrations based on how often a given peptide was detected; this will further enhance the analytical power of our method and open up the avenue for the yeast field to rapid, cost-effective and robust analysis of a very wide range of experimental conditions akin to those that have been studied with microarrays for the past 16 years.
Footnotes
* This work was supported by an FRM fourth-year PhD fellowship awarded to Y. Liu and Inserm Avenir grant N0 R07216NS and Région Bretagne CREATE grant N0 R11016NN to M. Primig. Work performed at the Proteomics Core Facility Biogenouest was supported by grants from Région Bretagne and IBiSA to C. Pineau.
 This article contains supplemental Files S1 to S3.
This article contains supplemental Files S1 to S3.
1 The abbreviations used are:
- SILAC
- stable-isotope labeling by amino acids in cell culture
- DIPP
- direct iterative protein profiling
- ORF
- open reading frame
- SGD
- Saccharomyces Genome Database
- SRM
- selected reaction monitoring
- YPD
- yeast extract, peptone, dextrose.
REFERENCES
- 1. Donalies U. E., Nguyen H. T., Stahl U., Nevoigt E. (2008) Improvement of Saccharomyces yeast strains used in brewing, wine making and baking. Adv. Biochem. Eng. Biotechnol. 111, 67–98 [DOI] [PubMed] [Google Scholar]
- 2. Hirasawa T., Furusawa C., Shimizu H. (2010) Saccharomyces cerevisiae and DNA microarray analyses: what did we learn from it for a better understanding and exploitation of yeast biotechnology? Appl. Microbiol. Biotechnol. 87, 391–400 [DOI] [PubMed] [Google Scholar]
- 3. Wei W., McCusker J. H., Hyman R. W., Jones T., Ning Y., Cao Z., Gu Z., Bruno D., Miranda M., Nguyen M., Wilhelmy J., Komp C., Tamse R., Wang X., Jia P., Luedi P., Oefner P. J., David L., Dietrich F. S., Li Y., Davis R. W., Steinmetz L. M. (2007) Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. U. S. A. 104, 12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siggers K. A., Lesser C. F. (2008) The Yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe 4, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goranov A. I., Amon A. (2010) Growth and division–not a one-way road. Curr. Opin. Cell Biol. 22, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dietrich F. S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., Wing R. A., Flavier A., Gaffney T. D., Philippsen P. (2004) The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304, 304–307 [DOI] [PubMed] [Google Scholar]
- 7. Schena M., Shalon D., Davis R. W., Brown P. O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 [DOI] [PubMed] [Google Scholar]
- 8. Wang Z., Gerstein M., Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Premsler T., Zahedi R. P., Lewandrowski U., Sickmann A. (2009) Recent advances in yeast organelle and membrane proteomics. Proteomics 9, 4731–4743 [DOI] [PubMed] [Google Scholar]
- 10. Chen R., Snyder M. Yeast proteomics and protein microarrays. J. Proteomics 73, 2147–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Massoni A., Moes S., Perrot M., Jenoe P., Boucherie H. (2009) Exploring the dynamics of the yeast proteome by means of 2-DE. Proteomics 9, 4674–4685 [DOI] [PubMed] [Google Scholar]
- 12. Fenn J. B., Mann M., Meng C. K., Wong S. F., Whitehouse C. M. (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 [DOI] [PubMed] [Google Scholar]
- 13. Williamson M. P., Sutcliffe M. J. (2010) Protein-protein interactions. Biochem. Soc. Trans. 38, 875–878 [DOI] [PubMed] [Google Scholar]
- 14. Xie Z., Hu S., Qian J., Blackshaw S., Zhu H. (2011) Systematic characterization of protein-DNA interactions. Cell. Mol. Life Sci. 68, 1657–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 16. de Godoy L. M., Olsen J. V., de Souza G. A., Li G., Mortensen P., Mann M. (2006) Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 7, R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zaman S., Lippman S. I., Zhao X., Broach J. R. (2008) How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42, 27–81 [DOI] [PubMed] [Google Scholar]
- 18. Mills D. (1972) Effect of pH on adenine and amino acid uptake during sporulation in Saccharomyces cerevisiae. J. Bacteriol. 112, 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Engel S. R., Balakrishnan R., Binkley G., Christie K. R., Costanzo M. C., Dwight S. S., Fisk D. G., Hirschman J. E., Hitz B. C., Hong E. L., Krieger C. J., Livstone M. S., Miyasato S. R., Nash R., Oughtred R., Park J., Skrzypek M. S., Weng S., Wong E. D., Dolinski K., Botstein D., Cherry J. M. (2010) Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 38, D433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lardenois A., Liu Y., Walther T., Chalmel F., Evrard B., Granovskaia M., Chu A., Davis R. W., Steinmetz L. M., Primig M. (2011) Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc. Natl. Acad. Sci. U. S. A. 108, 1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., James S. A., Davey R. P., Roberts I. N., Burt A., Koufopanou V., Tsai I. J., Bergman C. M., Bensasson D., O'Kelly M. J., van Oudenaarden A., Barton D. B., Bailes E., Nguyen A. N., Jones M., Quail M. A., Goodhead I., Sims S., Smith F., Blomberg A., Durbin R., Louis E. J. (2009) Population genomics of domestic and wild yeasts. Nature 458, 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., Hwang S. Y., Davis R. W., Esposito R. E. (2000) The core meiotic transcriptome in budding yeasts. Nat. Genet. 26, 415–423 [DOI] [PubMed] [Google Scholar]
- 24. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 25. Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254 [DOI] [PubMed] [Google Scholar]
- 26. Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston M., Louis E. J., Mewes H. W., Murakami Y., Philippsen P., Tettelin H., Oliver S. G. (1996) Life with 6000 genes. Science 274, 546, 563–567 [DOI] [PubMed] [Google Scholar]
- 27. Consortium (2010) The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 38, D331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., Wise K. J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Gerstein M., Dumont M. E., Phizicky E. M., Snyder M., Grayhack E. J. (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Durr E., Yu J., Krasinska K. M., Carver L. A., Yates J. R., Testa J. E., Oh P., Schnitzer J. E. (2004) Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat. Biotechnol. 22, 985–992 [DOI] [PubMed] [Google Scholar]
- 31. Florens L., Washburn M. P., Raine J. D., Anthony R. M., Grainger M., Haynes J. D., Moch J. K., Muster N., Sacci J. B., Tabb D. L., Witney A. A., Wolters D., Wu Y., Gardner M. J., Holder A. A., Sinden R. E., Yates J. R., Carucci D. J. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526 [DOI] [PubMed] [Google Scholar]
- 32. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 33. Scherl A., Francois P., Converset V., Bento M., Burgess J. A., Sanchez J. C., Hochstrasser D. F., Schrenzel J., Corthals G. L. (2004) Nonredundant mass spectrometry: a strategy to integrate mass spectrometry acquisition and analysis. Proteomics 4, 917–927 [DOI] [PubMed] [Google Scholar]
- 34. Seo J., Jeong J., Kim Y. M., Hwang N., Paek E., Lee K. J. (2008) Strategy for comprehensive identification of post-translational modifications in cellular proteins, including low abundant modifications: application to glyceraldehyde-3-phosphate dehydrogenase. J Proteome Res. 7, 587–602 [DOI] [PubMed] [Google Scholar]
- 35. Wang N., Li L. (2008) Exploring the precursor ion exclusion feature of liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry for improving protein identification in shotgun proteome analysis. Anal. Chem. 80, 4696–4710 [DOI] [PubMed] [Google Scholar]
- 36. Davis M. T., Spahr C. S., McGinley M. D., Robinson J. H., Bures E. J., Beierle J., Mort J., Yu W., Luethy R., Patterson S. D. (2001) Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. II. Limitations of complex mixture analyses. Proteomics 1, 93–107 [DOI] [PubMed] [Google Scholar]
- 37. Schmidt A., Gehlenborg N., Bodenmiller B., Mueller L. N., Campbell D., Mueller M., Aebersold R., Domon B. (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol. Cell. Proteomics 7, 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Piening B. D., Wang P., Bangur C. S., Whiteaker J., Zhang H., Feng L. C., Keane J. F., Eng J. K., Tang H., Prakash A., McIntosh M. W., Paulovich A. (2006) Quality control metrics for LC-MS feature detection tools demonstrated on Saccharomyces cerevisiae proteomic profiles. J. Proteome Res. 5, 1527–1534 [DOI] [PubMed] [Google Scholar]
- 39. Rudomin E. L., Carr S. A., Jaffe J. D. (2009) Directed sample interrogation utilizing an accurate mass exclusion-based data-dependent acquisition strategy (AMEx). J. Proteome Res. 8, 3154–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mori K. (2009) Signalling pathways in the unfolded protein response: development from yeast to mammals. J. Biochem. 146, 743–750 [DOI] [PubMed] [Google Scholar]
- 41. Brachat S., Dietrich F. S., Voegeli S., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P. (2003) Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 4, R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Waltermann C., Klipp E. (2010) Signal integration in budding yeast. Biochem. Soc. Trans. 38, 1257–1264 [DOI] [PubMed] [Google Scholar]
- 43. Prinz S., Avila-Campillo I., Aldridge C., Srinivasan A., Dimitrov K., Siegel A. F., Galitski T. (2004) Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res. 14, 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kassir Y., Adir N., Boger-Nadjar E., Raviv N. G., Rubin-Bejerano I., Sagee S., Shenhar G. (2003) Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224, 111–171 [DOI] [PubMed] [Google Scholar]
- 45. Mallory M. J., Cooper K. F., Strich R. (2007) Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol. Cell. 27, 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wilm M. (2011) Principles of electrospray ionization. Mol. Cell. Proteomics 1 July;10(7):M111.009407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilm M., Mann M. (1996) Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68, 1–8 [DOI] [PubMed] [Google Scholar]
- 48. Kellis M., Birren B. W., Lander E. S. (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624 [DOI] [PubMed] [Google Scholar]