Abstract
Bacteria in the genus Streptomyces are soil-dwelling oligotrophs and important producers of secondary metabolites. Previously, we showed that global messenger RNA expression was subject to a series of metabolic and regulatory switches during the lifetime of a fermentor batch culture of Streptomyces coelicolor M145. Here we analyze the proteome from eight time points from the same fermentor culture and, because phosphate availability is an important regulator of secondary metabolite production, compare this to the proteome of a similar time course from an S. coelicolor mutant, INB201 (ΔphoP), defective in the control of phosphate utilization. The proteomes provide a detailed view of enzymes involved in central carbon and nitrogen metabolism. Trends in protein expression over the time courses were deduced from a protein abundance index, which also revealed the importance of stress pathway proteins in both cultures. As expected, the ΔphoP mutant was deficient in expression of PhoP-dependent genes, and several putatively compensatory metabolic and regulatory pathways for phosphate scavenging were detected. Notably there is a succession of switches that coordinately induce the production of enzymes for five different secondary metabolite biosynthesis pathways over the course of the batch cultures.
Streptomycetes are prolific producers of complex secondary metabolites with pharmaceutically important activities including antibiotics and anti-cancer compounds. In a batch culture growth system, secondary metabolism is often observed when the culture has entered the stationary phase and well after rapid vegetative growth has ceased. Well known examples are the production of actinorhodin (Act)1 and undecylprodigiosin (Red) in Streptomyces coelicolor (1). This timing implies that secondary metabolism is not simply a consequence of nutrient limitation, but there is an ordered pathway of metabolic and regulatory switches. Many of these have been characterized using a classical genetic approach of generating mutants that no longer produce the antibiotics and studying the genes affected (1–3). Thus it is understood that antibiotic production is controlled generally on at least two levels: global regulation by transcription factors (AfsK, AfsS, AfsR, AbsA1/AbsA2, PhoP, etc.) and signaling molecules (ppGpp, γ-butyrolactones), and pathway-specific regulation by activators (e.g. actII-ORF4, RedD, and members of the SARP family) (1–3). Although these studies have provided considerable understanding of the later stages of the regulation of antibiotic production, the earlier events that prime these regulatory pathways and networks are poorly understood. Moreover, it is evident from Nieselt et al. (4) that secondary metabolites are not uniquely expressed in stationary phase; a cluster of genes in S. coelicolor SCO6269-SCO6288 thought to be required for the synthesis of CPK, a type I polyketide are induced, whereas the culture is still undergoing rapid growth (4, 5). Thus the physiological conditions required for the activation of secondary metabolite pathways vary.
In this study we have built on the work of Nieselt et al. (4) that describes the transcriptional switches that occur over the lifetime of a fermentor culture of S. coelicolor. In the paper by Nieselt et al., S. coelicolor was grown in batch culture in a defined medium with glutamate and glucose as carbon sources and glutamate as the sole source of nitrogen. Phosphate depletion was the trigger for the cessation of rapid growth and metabolic switching. Although biomass was still rapidly increasing, there was repression of many glutamate/glutamine metabolism genes and induction of CPK genes. On phosphate depletion (35 h post-inoculation), there was a coordinated and rapid activation of all phosphate utilization proteins, and this was shortly followed by another coordinated downshift of expression of all ribosomal protein genes. Late in the stationary phase, genes for biosynthesis of Act and Red were switched on. This dynamic series of switches was shown to be reproducible between fermentors and biological replicates.
Here we have used the same biomass from one of these fermentors to study the proteome using gel-enhanced LC-MS/MS at eight time points. We use spectral counting in the form of EmPAI values to provide a semi-quantitative analysis of protein abundance (6). The protein data set was used to predict the most likely flow of nutrients through carbon and nitrogen metabolism and how metabolism changes over the lifetime of the fermentor. Moreover, we compared the proteome from wild type S. coelicolor strain M145 with S. coelicolor INB201 containing a deletion of the phoP gene (7) and discuss the possible compensatory pathways employed by the phoP mutant for phosphate scavenging and metabolic imbalance.
EXPERIMENTAL PROCEDURES
Bacterial Cultures
Two strains of S. coelicolor A3(2) were used in this study; the parent strain M145 and a ΔphoP knock-out mutant of M145, INB201 (7, 8). The cultivation of S. coelicolor M145 is described in detail by Nieselt et al. (4), and the cultivation of INB201 was performed using identical conditions. Briefly S. coelicolor was grown for 60 h in defined medium with glucose and glutamate as major carbon and nitrogen sources in 3-liter fermentors with a starting volume of 1.8 liters. For proteome analysis, 5-ml samples were taken at specific time intervals (see Fig. 1) and centrifuged (3200 × g, 5 min, 4 °C), and the resulting cell pellets were frozen rapidly at −80 °C. The cell pellets were subsequently resuspended in 3 ml of 20 mm Tris buffer (pH 8.0, containing protease inhibitors; Roche Applied Science). The cell suspension was disrupted in a French pressure cell (Aminco); each sample was passed through the press twice at 124 MPa (18000 p.s.i.). The efficiency of lysis was determined by microscopy comparing the number of mycelial pellets in unlysed versus lysed cells. The lysed cells were centrifuged (5,000 × g, 30 min, 4 °C) to obtain the cleared cell extract.
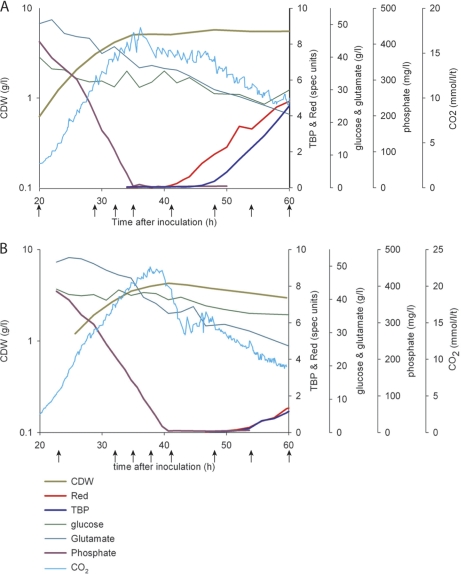
Fig. 1.
Growth parameters for fermentor cultures of S. coelicolor M145 and the ΔphoP mutant INB201. Pregerminated spores of M145 (A) and INB201 (B) were used to inoculate 3 liters of fermentors containing 1.8 liters of defined medium (reference fermentation numbers F199 and F335, respectively) at time 0 as described previously (4). Thereafter, online (CO2 evolution rate) and offline measurements (cell dry weight (CDW), Red, and total blue pigment (TBP; includes Act, glucose, glutamate, and phosphate) were taken to monitor growth. The growth measurements for F199 are redrawn with permission from the data published previously (4). Samples for proteomic analysis were taken at 20, 29, 32, 35, 41, 48, 54, and 60 h post-inoculation from F199 and 23, 32, 35, 38, 41, 48, 54, and 60 h post-inoculation from F335.
Preparation of Tryptic Peptides
Proteins from cell lysates (110 μg of total protein) were precipitated by adding 4× sample volume of ice-cold acetone. Pelleted proteins were resuspended in 20 μl of NuPage sample buffer and 2.5 μl of NuPage reducing agent (Invitrogen) and incubated (72 °C, 10 min). The proteins were then separated on a 4–12% NuPAGE gel, and the gel was stained (3 h) with Colloidal Blue (Invitrogen) and destained in deionized water overnight. Each lane was then cut into 15 approximately equal horizontal sections, and each section further macerated and put into a well in a 96-well digestion plate. In-gel trypsin digestion was performed using an Investigator ProGest robot (Genomics Solutions, Huntingdon, UK). This procedure involved the proteins being reduced with DTT (60 °C, 20 min), S-alkylated with iodoacetamide (25 °C, 10 min), and then digested with trypsin at 37 °C (8 h, sequencing grade modified trypsin; Promega, Southhampton, UK). The samples were then dried by rotary evaporation and resuspended in 12 μl of 0.1% formic acid.
Mass Spectrometry and Data Analysis
Mass spectrometric analysis was performed using an HCT Ultra PTM Discovery System (Bruker Daltonics, Coventry, UK) with an inline UltiMate 3000 LC System (Dionex, Camberley, Surrey, UK). The peptides were separated on a monolithic capillary column (200-μm inner diameter × 5 cm; Dionex). Eluent A was 3% acetonitrile, 0.05% formic acid, and eluent B 80% acetonitrile containing 0.04% formic acid with a gradient of 0–40% B in 40 min at a flow rate of 2.0 μl/min. Peptide fragment mass spectra were acquired in data-dependent AutoMS(n) mode with a scan range of 300–1500 m/z. Three averages and up to three precursor ions were selected from the MS scan (300–3000 m/z). Precursor ions were actively excluded within a 0.5-min window and included 1+, 2+, and 3+ charged ions. Peptide peaks were detected and deconvoluted automatically using DataAnalysis version 3.4 build 192 software (Bruker Daltronics). Mass lists in the form of MASCOT Generic Files were created automatically and used as input for MASCOT MS/MS ion search engine (release date February 2, 2007) of the S. coelicolor A3(2) genome from the NCBI-nr database (in house database update July 31, 2009) RefSeq-Id NC_003888 (8480 protein sequences searched). Searches were performed allowing no missed cleavages but allowing cysteine carbamidomethylation and methionine oxidation as fixed and variable modifications, respectively. The mass tolerance for precursor ions and fragment ions was 1.5 and 0.5 Da, respectively. An expectation value significance threshold of 0.05 was employed to exclude peptides with individual ion scores below the Mascot identity threshold. This reduced the false discovery rate to an acceptable level. An automatically generated decoy database was searched by MASCOT to assay the false discovery rate, which for all searches was <3%. The MASCOT MudPIT protein scoring default settings were used to exclude ion scores that exceeded the significance threshold. In addition, positive protein database hits had to contain at least one peptide with the highest score to a particular spectrum and must not have already been assigned to another protein with a higher score. The number of distinct peptides assigned for each protein varied between 1 and 106 (average = 7.4) and the percentage of coverage of each protein was between 1.3 and 100% (average = 29.7%). Peptides used to identify proteins are listed in supplemental Tables S1–S4.
A pilot experiment was performed to assess the number of replicates required to adequately describe the proteins present in each extract. Two of the extracts (from the M145 fermentation, fermentor reference F199; time points, 29 and 48 h post-inoculation) were analyzed five times (replicates a–e). Approximately 38% of the total proteins detected from all five replicates were present in all five replicates, and 70% of the proteins were present in two or more replicates. The accumulating total of proteins that were recaptured with increasing numbers of replicates was scored (supplemental Fig. S1). With two replicates, 60% of the total proteins detected were recaptured, and this rate of recapture did not significantly increase as the numbers of replicates increased. Because more than two replicates did not greatly enhance the rate of recapture of proteins (supplemental Fig. S1), each extract was analyzed twice.
Web-based tools were employed for the analysis of proteins, in particular StrepDB (http://streptomyces.org.uk), an updated version of ScoCyc (http://www.BioCyc.org/SCO/),2 and KEGG pathways (http://www.genome.jp). The MS data were prepared for submission to the Proteomics Identifications database (PRIDE) data repository at the European Bioinformatics Institute (www.ebi.ac.uk/pride) using PRIDE Converter and can be accessed via accession numbers 18302–18333 (9, 10).
Microarray Analysis of mRNA
The transcriptome data for S. coelicolor M145 were reported previously in Nieselt et al. (4). The transcriptome for the ΔphoP mutant INB201 was acquired and processed using the same procedures as those described previously and deposited in GEO (accession number GSE31068) (4). The mRNA expression data from both M145 and INB201 coding for each of the proteins detected here were plotted and can be accessed in supplemental Fig. S2.
RESULTS AND DISCUSSION
Crude lysates were prepared from samples taken at eight time points from two fermentors: F199 containing the parent strain M145 and F335 containing the ΔphoP mutant INB201 (Fig. 1). Each crude lysate was analyzed twice by gel-enhanced LC-MS/MS. Because only ∼60% of proteins are recaptured in replicate runs from the same extract, the replicates were compiled into two complete replicate time courses (M145:F199A and M145:F199B for the parent strain and INB201:F335A and INB201:F335B for the ΔphoP mutant). The data for replicate time courses were then analyzed separately (in preference to averaging) to reveal reproducible trends of protein abundances where they exist. The entire data set was thus derived from 32 experiments, i.e. eight time points taken from two fermentors and duplicate analysis of each time point. Each experiment yielded on average 487 proteins, and in all the experiments combined, we detected 1535 proteins (supplemental Table S5). Within a complete time course (M145:F199A, M145:F199B, INB201:F335A, and INB201:F335B), we detected 993, 1041, 991, and 1006 proteins, respectively (supplemental Table S6). 650 proteins were present in all four time courses (supplemental Table S7), 167 proteins were present in only three time courses, 212 in two time courses, and 506 proteins identified in only one time course.
S. coelicolor has 7770 protein coding genes, so our total of 1535 observed proteins represents 19% of the total theoretical proteome. The observed proteome had 145 proteins annotated as either membrane proteins or proteins associated with the membrane, and this represents 8% of the theoretical membrane proteome (1779 predicted proteins). The proteins detected had predicted pIs that ranged from 3.7 (SCO3288) to 12.06 (SCO3672) pH units and molecular masses from 4.4 kDa (SCO4726, 50 S ribosomal protein L36) to 799 kDa (SCO3230, CDA PS1), implying a good coverage of physical properties of the proteins (supplemental Table S5). This data set therefore represents the most complete inventory, to our knowledge, of the Streptomyces proteome.
EmPAI or exponentially modified protein abundance index provides a semi-quantitative measure of protein abundance (6). PAI is derived by dividing the number of observed peptides by the number of observable peptides per protein minus one (6). EmPAI values, reported by the MASCOT search (Matrix Science, http://www.matrixscience.com) use, for the observed peptides, peptide matches with scores at or above the homology threshold or identity threshold if there is no homology threshold. MASCOT also only counts unique parent ions and only includes the same peptide when it is present in different charged states. For observable peptides, MASCOT uses a calculated estimate based on the mass of the protein, the average amino acid composition of the database, and the protease specificity.
Because we were interested in how protein abundance changes during the fermentation and the differences in the proteomes from the parent strain M145 and the ΔphoP deletion strain INB201, we wished to compare EmPAI values between time courses. However, EmPAI values vary with the total protein used for each experiment, so the experiments were conducted according to standard operating procedures to enable comparability of EmPAIs from time course to time course. The ratios of total EmPAI values per protein between replicate time courses should be close to one if the time courses are comparable. The individual EmPAI values for each protein from the eight time points were therefore added together to give ΣEmPAIM145:F199A, ΣEmPAIM145:F199B, ΣEmPAIINB201:F335A, and ΣEmPAIINB201:F335B values for each of 650 proteins that were detected in all four time courses (supplemental Table S7). Where a protein was not detected, the EmPAI value was treated as 0. To test whether the summed EmPAI values per protein were comparable between replicate time courses, the per protein ratios ΣEmPAIM145:F199A/B and ΣEmPAIINB201:F335A/B were calculated for M145_F199A compared with M145_F199B and INB201_F335A compared with INB201_F335B, respectively. The average ratios from ΣEmPAIM145:F199A/B and ΣEmPAIINB201:F335A/B were 0.87 ± 0.60 (S.D.) and 0.99 ± 0.6 (S.D.), respectively, indicating that a comparison of EmPAI values between replicates is reasonable, although the large standard deviation indicates a wide variation between replicates (supplemental Table S7). We then summed the total EmPAI values obtained per protein from all time points from both replicate time courses to give the total abundance of each protein in each strain or ΣEmPAIM145 and ΣEmPAIINB201 values. The average ratio between these two values, ΣEmPAIM145/INB201, per protein was 1.09 ± 0.67 (S.D.) (supplemental Table S7). The overall accordance between the total EmPAIs per protein from the replicates and between the two strains was further demonstrated by regression plots; ΣEmPAIM145:F199A versus ΣEmPAIM145:F199B (supplemental Fig. S3A), ΣEmPAIINB201:F335A versus ΣEmPAIINB201:F335B (supplemental Fig. S3B) and ΣEmPAI M145 versus ΣEmPAIINB201 (supplemental Fig. S3C).
The Most Abundant Proteins Include Those Involved in Stress Responses
The top 65 most abundant proteins in M145 consisted of 29 proteins involved in translation, 11 stress proteins, 10 metabolic enzymes or nutrient transport proteins, 3 proteins involved in development or secondary metabolism, 2 histone-like proteins, and 10 unknowns (Table I). In the ΔphoP mutant, INB201, a similar profile of proteins was observed (supplemental Table S7).
Table I. The most abundant proteins in S. coelicolor.
| Gene ID | M145 abundance (ΣEmPAIM145) | INB201 abundance (ΣEmPAIINB201) | Gene name | Product |
|---|---|---|---|---|
| SCO4660 | 98.03 | 86.58 | rspG | 30 S ribosomal protein S7 |
| SCO2368 | 90.81 | 48.17 | Conserved hypothetical protein, TerD-like | |
| SCO4277 | 88.81 | 66.29 | Putative tellurium resistance protein, TerD-like | |
| SCO0641 | 87.87 | 92.56 | terD | Tellurium resistance protein |
| SCO4761 | 85.6 | 77.74 | groES | 10-kDa chaperonin cpn10 |
| SCO4717 | 85.28 | 93.22 | rplF | 50 S ribosomal protein L6 |
| SCO2950 | 74.36 | 79.86 | hup | DNA-binding protein Hu (hs1) |
| SCO3767 | 60.02 | 59.65 | Conserved hypothetical protein, TerB-like | |
| SCO5736 | 58.93 | 58.95 | rpsO | 30 S ribosomal protein S15 |
| SCO2596 | 58.67 | 77.4 | rpmA | 50 S ribosomal protein L27 |
| SCO4719 | 57.36 | 56.8 | rpsE | 30 S ribosomal protein S5 |
| SCO4662 | 56.52 | 56.66 | tuf1 | Elongation factor TU-1 |
| SCO4716 | 54.49 | 57.24 | rpsH | 30 S ribosomal protein S8 |
| SCO4165 | 47.74 | 52.23 | Conserved hypothetical protein, DUF1416 | |
| SCO4296 | 47.22 | 46.03 | groEL2 | Chaperonin 2 |
| SCO4725 | 46.3 | 39.76 | infA | Translational initiation factor IF1 |
| SCO4505 | 45.81 | 47.71 | scoF2 | Cold shock protein |
| SCO4714 | 43.17 | 45.7 | rplE | 50 S ribosomal protein L5 |
| SCO4727 | 41.15 | 41.72 | rpsM | 30 S ribosomal protein S13 |
| SCO5254 | 39.75 | 45.09 | sodN | Superoxide dismutase |
| SCO2887 | 39.4 | 52.89 | Putative membrane protein. | |
| SCO6282 | 39.33 | 38.39 | Putative 3-oxoacyl-[acyl-carrier protein] reductase | |
| SCO4709 | 37.85 | 32.55 | rplP | 50 S ribosomal protein L16 |
| SCO4762 | 37.34 | 33.7 | groEL1 | 60-kDa chaperonin cpn60 |
| SCO4687 | 37.01 | 31.28 | Hypothetical protein SCD31.12c | |
| SCO4710 | 36.27 | 35.26 | rpmC | 50 S ribosomal protein L29 |
| SCO1505 | 35.99 | 37.1 | rpsD | 30 S ribosomal protein S4 |
| SCO5500 | 35.91 | 44.56 | Putative membrane protein | |
| SCO5776 | 35.82 | 49.7 | gluB | Glutamate-binding protein |
| SCO0527 | 35.48 | 34.94 | scoF | Cold shock protein |
| SCO5371 | 35.36 | 51.01 | atpA | ATP synthase alpha chain |
| SCO5477 | 35.29 | 36.16 | Putative oligopeptide-binding lipoprotein | |
| SCO1598 | 33.81 | 34.51 | rplT | 50 S ribosomal protein L20 |
| SCO4703 | 33.52 | 40.05 | rplD | 50 S ribosomal protein L4 |
| SCO4653 | 33.5 | 37.66 | rplL | 50 S ribosomal protein L7/L12 |
| SCO4649 | 33.33 | 44.47 | rlpA | 50 S ribosomal protein L1 |
| SCO4702 | 32.74 | 33.86 | rplC | 50 S ribosomal protein L3 |
| SCO4721 | 32.54 | 39.76 | rplO | 50 S ribosomal protein L15 |
| SCO4718 | 32.23 | 37.2 | rplR | 50 S ribosomal protein L18 |
| SCO5595 | 30.14 | 34.06 | rplS | 50 S ribosomal protein L19 |
| SCO4091 | 29.67 | 21.63 | bldC | Putative DNA-binding protein |
| SCO1480 | 29.29 | 36.74 | Conserved hypothetical protein, MihF-like | |
| SCO5077 | 29.14 | 15.89 | actVA2 | Hypothetical protein |
| SCO1947 | 27.98 | 24.68 | gap1 | Glyceraldehyde-3-phosphate dehydrogenase |
| SCO5624 | 27.88 | 29.81 | rpsB | 30 S ribosomal protein S2 |
| SCO4711 | 27.63 | 21.46 | rpsQ | 30 S ribosomal protein S17 |
| SCO3906 | 27.51 | 25.18 | rpsF | Putative 30 S ribosomal protein S6 |
| SCO5625 | 27.41 | 24.86 | tsf | Elongation factor Ts |
| SCO4809 | 26.55 | 33.44 | sucD | Succinyl CoA synthetase alpha chain |
| SCO3909 | 26.28 | 29.43 | rplI | 50 S ribosomal protein L9 |
| SCO2736 | 26.27 | 34.71 | citA | Citrate synthase. |
| SCO2180 | 26.2 | 33.38 | pdhL | Putative dihydrolipoamide dehydrogenase |
| SCO6279 | 25.29 | 20.39 | Putative diaminobutyrate-pyruvate aminotransferase | |
| SCO4706 | 25.13 | 24 | rpsS | 30 S ribosomal protein S19 |
| SCO3899 | 24.74 | 19.63 | Hypothetical protein/myo-inositol-1-phosphate synthase | |
| SCO3671 | 24.36 | 27.47 | dnaK | Heat shock protein 70 |
| SCO4655 | 24.08 | 27.28 | rpoC | DNA-directed RNA polymerase β chain (fragment) |
| SCO1998 | 23.9 | 27.89 | rpsA | 30 S ribosomal protein S1 |
| SCO4252 | 23.83 | 16.61 | Phate T4 gp19-like/ phage tail protein | |
| SCO4164 | 23.8 | 22.46 | cysA | Putative thiosulfate sulfurtransferase |
| SCO3549 | 23.76 | 27.94 | bldG | Putative anti-sigma factor antagonist |
| SCO2911 | 23.67 | 21.68 | moaD | Conserved hypothetical protein |
| SCO3096 | 23.36 | 34.24 | eno | Enolase |
| SCO0682 | 23.33 | 45.43 | Hypothetical protein SCF15.03c | |
| SCO4253 | 23.15 | 18.01 | Phage tail sheath protein |
Stress response proteins are among the most abundant proteins in the cell (Table I). This observation confirms and extends those made previously that stress response proteins were associated with transition phase during diauxic growth of an S. coelicolor culture (11). The abundant stress response proteins included chaperones, GroEL1, GroEL2, GroES, and DnaK, and two cold shock proteins SCO4505 (scoF2) and SCO0527 (scoF). Cold shock proteins are small proteins that might skew the EmPAI values if they contain a peptide that is particularly easy to detect under our experimental conditions. However, there are seven predicted cold shock proteins in the S. coelicolor genome, five of which were detected in the proteome and whose total EmPAI values placed them in the most abundant 10% of proteins, implying that cold shock proteins are probably present in high abundance. The putative cold shock protein regulators, CspA (SCO4325) and CspB (SCO4325), were also detected but only up to 40 h post-inoculation (supplemental Table S6). GroEL1 and GroEL2 were very abundant in early time points and depleted rapidly as the culture entered transition phase (Fig. 2). This pattern of expression correlated well with the mRNA levels (supplemental Fig. S2), implying that there is some degree of protein instability. Similar protein and mRNA profiles were observed for other heat shock proteins, HtpG, ClpP1, ClpP2, and a Clp homologue, SCO3373 (Fig. 2). Levels of GroES varied greatly, even between replicates. DnaK is involved in negative feedback regulation of a ClpB-like protein (SCO3661) and a lon protease (SCO5285) (12), both of which were detected, but at low abundance, consistent with DnaK remaining at high levels throughout growth. Chaperones are known to have an important role in Streptomyces development and antibiotic production (13), and at least GroEL2 is likely to be essential (14). The 20 S proteasome subunits α and β (SCO1643 and SCO1644, respectively) were also expressed, as was the putative ubiquitin-like protein modifier, Pup (SCO1646) first detected in Mycobacterium tuberculosis, and the proteasome accessory factors PafA and PafA2 (putative Pup ligases encoded by SCO1640 and SCO1647) (15). The proteins expressed by SCO1640 and SCO1648 (the putative proteasome ATPase) showed an increase in abundance as the fermentation progressed, whereas the other proteins of the Pup/20 S proteasome system had no discernable trends of expression (supplemental Table S6).
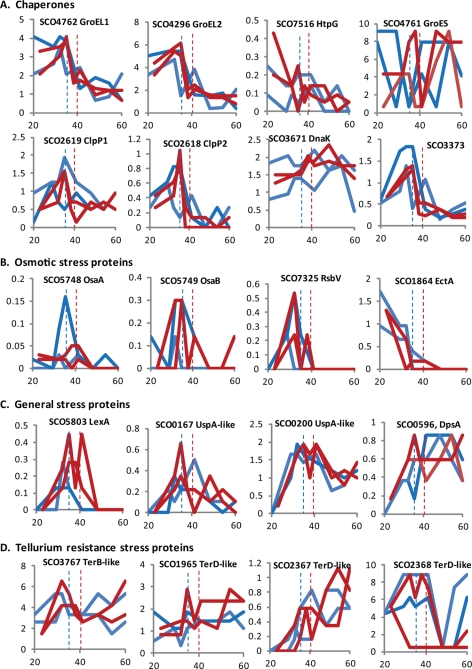
Fig. 2.
Levels of stress response proteins during growth of S. coelicolor cultures. EmPAI values (y axis) were plotted against time from inoculation (hours; x axis) for proteins detected from M145 (time course replicates M145_F199A and M145_F199B; blue) and from the ΔphoP mutant, INB201 (INB201_F335A and INB201_F335B; red). The dashed blue lines and the dashed red lines represent the times of phosphate depletion in M145 and INB201 at 35 and 40 h, respectively. Panels A–D indicate different categories of stress response proteins.
Four of the highly abundant proteins are the TerD-like stress response proteins, SCO2368, SCO4277, SCO0641, and the TerB-like protein SCO3767 (Table I and Fig. 2). The biochemical functions of these proteins are not known, although structures of the TerD-like proteins SCO2368 (Protein Data Bank entry 3IBZ) and SCO6318 (Protein Data Bank entry 2QZ7) both indicate metal binding activities (Midwest Center for Structural Genomics). The Ter proteins are named after their ability to confer resistance to tellurite oxyanions, but S. coelicolor was recently shown to be sensitive to potassium tellurite (16). A knockout of tdd8 (SCO2368) resulted in an increased growth rate compared with parent but was defective in spore development (16). Tdd8 was found to contain a potential recognition sequence for sigmaR, required for control of the oxidative stress regulon (17). Thus the high abundance of TerD-like and TerB-like proteins suggests that control of oxidative stress as a result of general metabolism is an important housekeeping function in rapidly growing cells. Consistent with this hypothesis is the high abundance of superoxide dismutase (SodN, SCO5254) (Table I).
The incidence of stress response proteins among the most highly abundant proteins in our data set prompted us to look for less abundant proteins also involved in stress. Other TerD-like homologues were discovered (e.g. SCO2367, SCO1965) and DpsA (SCO0596), a member of the ferritin family involved in protection of DNA from oxidative stress (18) (Fig. 2 and supplemental Table S6). Surprisingly the mRNA trends for DpsA and the TerD-like protein, SCO2367, gave no indication of the observed accumulation of these proteins as the cells enter transition and stationary phase (supplemental Fig. S2 and Fig. 2). The top 65 most abundant proteins in the ΔphoP mutant INB201 included a putative bacterioferritin (SCO2113) that appeared to accumulate to a greater level in stationary phase in INB201 than in the parent, M145 (supplemental Table S7). In this case the mRNA for SCO2113 was up-regulated in INB201, confirming a previous report (supplemental Fig. S2) (8).
There was a distinct pattern of expression for proteins (and mRNA) belonging to the universal stress protein (Usp) family and LexA, the major regulator of the SOS response, all peaking at the onset of transition phase (Fig. 2). Regulators of osmotic stress, OsaA, OsaB, OsaD, and RsbV (19), were also more likely to be detected at transition phase or early stationary phase time points (Fig. 2 and supplemental Table S6). However, the enzymes (SCO1864-SO1867) for synthesis of ectoine, a solute that can protect against high salt (20), were maximally detected in the earliest time points and steadily depleted as growth progressed (Fig. 2). Similarly a putative glycine betaine transport system permease protein (SCO1620) was detected but only in the first time point (supplemental Table S6). Possibly these proteins are normally expressed during rapid growth to set up protection against osmotic stress that occurs later in the fermentor batch culture with the synthesis of glycogen (21). BldG (SCO3549), expressed throughout growth (supplemental Table S6), is an anti-sigma factor antagonist, required for development and antibiotic production, and activates sigmaH during the osmotic stress response (22, 23). Taken together, it seems that chaperones and ectoine biosynthesis enzymes quickly turn over at transition phase/phosphate depletion; other stress response proteins, notably some of the regulators of osmotic and oxidative stress, are induced at this time; and others, in particular DnaK, DpsA, and the TerD, TerB-like proteins, remain at or accumulate to high levels throughout growth (Fig. 2).
Other abundant proteins include the ribosomal proteins that generally decline in abundance during batch cultivation; the dramatic step down, clearly seen in mRNA levels, was not observed at the protein level (4) (supplemental Table S6 and Fig. S2). Many metabolic enzymes were present at high levels and are discussed further below. Three putative periplasmic amino acid-binding proteins, an oligopeptide transport protein encoded by SCO5477, the branched chain amino acid-binding protein SCO2008 and the glutamate-binding protein GluB SCO5776 were all highly abundant; the first two may be recycling amino acids and peptides from lysed mycelium. There also appears to be constitutive expression of proteins that resemble phage tail proteins (SCO4252 and SCO4253), whose functions are unknown but that could be involved in protein secretion, DNA conjugation, or production of a colicin-like molecule.
Carbon Metabolism
Based on known and predicted activities, detected proteins could be assigned to almost all the enzymatic steps required for functional glycolytic (Fig. 3) and pentose phosphate (supplemental Fig. S4) pathways, and the TCA cycle (Fig. 4). The only reactions for which no candidate enzymes could be detected were two steps in the pentose phosphate pathway, i.e. 6-phosphogluconolactonase (SCO1939) and a transketolase for the conversion of d-erythrose-4-phosphate to d-glyceraldehyde-3-phosphate (SCO6768). A key enzyme in the Entner-Duodoroff pathway, 2-keto-3-deoxy-6-phosphogluconate aldolase, SCO0852, was detected. Pyruvate metabolic enzymes, except pyruvate carboxylase, were detected (Fig. 5). Enzymes for the glyoxylate shunt were not detected. The S. coelicolor genome predicts that many of the catalytic steps are mediated by multiple paralogues (24). Our data clearly show which of these paralogues are most likely to be used for each enzymatic step.
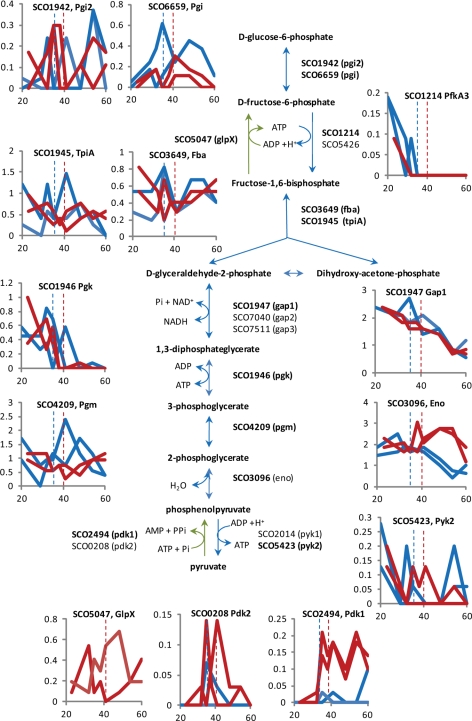
Fig. 3.
Major glycolytic and gluconeogenic enzymes in S. coelicolor. The gene identifications (SCO numbers) for the candidate enzymes for glycolysis and gluconeogenesis are shown next to the steps they are thought to catalyze. Reactions specific to gluconeogenesis are shown with green arrows. Gene identifications in bold are the more abundant paralogues. Graphs to show the general expression trends for some of the enzymes are shown as described in the legend to Fig. 2.
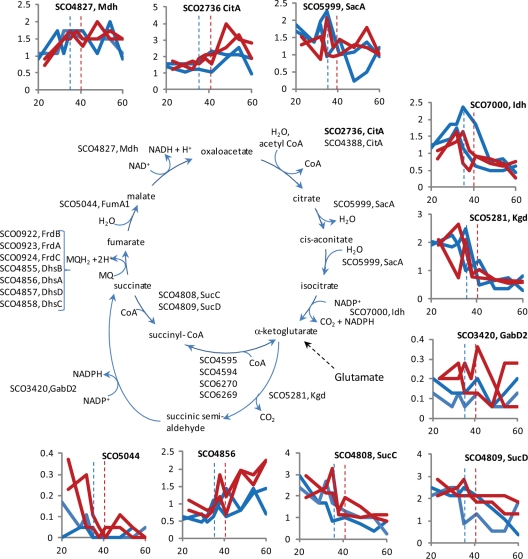
Fig. 4.
Enzymes of the TCA cycle in S. coelicolor. The gene identifications for the candidate enzymes for the TCA cycle are shown next to the steps they are thought to catalyze. Graphs to show the general expression trends for some of the enzymes are shown as described in the legend to Fig. 2.
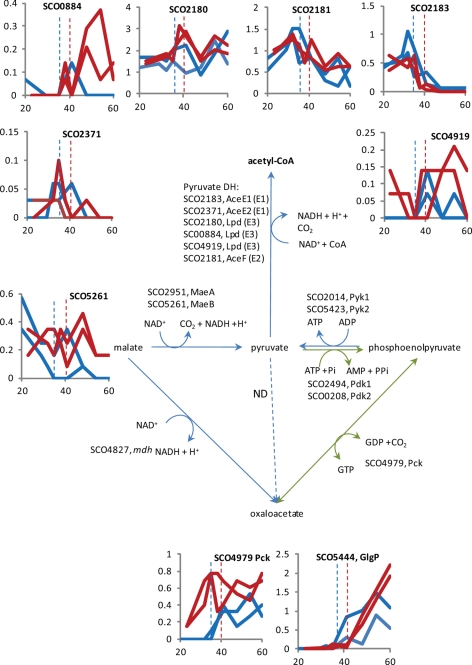
Fig. 5.
Pyruvate metabolic enzymes in S. coelicolor. The gene identifications for the candidate enzymes for pyruvate metabolism are shown next to the steps they are thought to catalyze. Reactions specific to gluconeogenesis are shown with green arrows. Graphs to show the general expression trends for some of the enzymes are shown as described in the legend to Fig. 2. The expression profile of glycogen phosphorylase (GlgP, SCO5444) is also shown.
In terms of protein abundance over the time courses, enzymes for carbon metabolism either deplete or stay approximately level, and only a few become undetectable at later time points (Figs. 3–5 and supplemental Fig. S4). Several proteins increase in abundance over the time course, and these are a glycogen phosphorylase (SCO5444) that retrieves metabolically accessible glucose (glucose-1-phosphate) from glycogen (21) and those involved in gluconeogenesis (Figs. 3 and 6), particularly in the ΔphoP mutant INB201 (discussed further below).
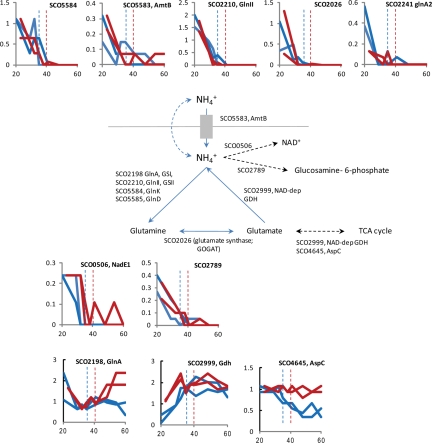
Fig. 6.
Nitrogen metabolic enzymes in S. coelicolor. The gene identifications for the candidate enzymes for the assimilation of glutamate and glutamine are shown next to the steps they are thought to catalyze. Graphs to show the general expression trends for some of the enzymes are shown as described in the legend to Fig. 2.
Although glucose is used as a carbon source, the level of glucose changes little throughout the fermentation (Fig. 1). Glutamate is, however, depleted to ∼50% of the starting concentration (Fig. 1) and is used as the preferred carbon source and the sole nitrogen source. Cultivation experiments confirmed that glucose is not required for growth because M145 can grow on glutamate as the sole carbon and nitrogen source with only a small decrease in growth rate (data not shown). The high levels of glutamate assimilatory enzymes (aspartate aminotransferase, SCO4645, and an NAD-specific glutamate dehydrogenase, SCO2999) are consistent with the flow of carbon directly from glutamate into the TCA cycle (Figs. 4 and 6). The proteins suggest that the TCA cycle in S. coelicolor may then resemble that in M. tuberculosis (25) in that SCO5281 is predicted to encode 2-α-ketoglutarate dehydrogenase that converts α-ketoglutarate to succinyl semi-aldehyde, and SCO3420 converts succinate semi-aldehyde to succinate. SCO3420 is orthologous to GabD2 from M. tuberculosis (25). However unlike M. tuberculosis, S. coelicolor also expresses enzymes that can catalyze the oxidative decarboxylation of α-ketoglutarate to succinyl-CoA (SCO6269 and SCO6270). The synthesis of succinyl-CoA can also occur through succinyl CoA synthetase (SCO4808 and SCO4809).
Electron Transport and Oxidative Phosphorylation
Streptomyces are obligate aerobes and therefore use oxygen as the terminal electron acceptor in the electron transport chain. We observed electron transfer flavoproteins α and β subunits (SCO1081 and SCO1082, respectively), three subunits (SCO4567, SCO4568, and SCO4573 or NuoF, NuoG, and NuoL, respectively) from an NADH dehydrogenase gene cluster (SCO4562-SCO4575) and the membrane-bound succinate dehydrogenase (SCO4855–SCO4858) that are likely to function in the initial entry of electrons into the electron transport chain (supplemental Table S6). We also observed cytochrome c reductase components, (SCO2148, SCO2149, SCO7120, SCO7236, and SCO2150) and components of the cyctochrome oxidase (SCO2155, SCO2156, and SCO2151). The integral membrane components and the cytoplasmic components of the ATPase complex were detected (SCO5366–SCO5374); indeed two of these subunits, AtpA and AtpB, were some of the most abundant proteins in the mycelium. Mostly the components of the electron transport chain remained at fairly constant levels throughout the fermentation, but SCO1081 and ATPase subunits decreased as the culture enters phosphate starvation (supplemental Fig. S5 and Table S6).
A surprising observation was high level expression of several proteins required for respiratory nitrate reduction even though the medium contained neither nitrate nor nitrite. S. coelicolor has three Nar loci each encoding the NarG, NarH, NarI, and NarJ subunits (26). We detected all four subunits from the Nar2 locus (SCO0216–SCO0219), suggested to be the major nitrate reductase complex made in mycelia (26) (supplemental Table S6). The α chain subunit NarG2 (SCO0216) and NarG3 (SCO4947) from the Nar3 locus, also proposed to be a mycelially expressed enzyme, were highly abundant and increased in abundance during the fermentations (supplemental Fig. S5). The α subunit NarG1 SCO6535 from the Nar1 locus, predominately expressed in spores (26),3 was also detected. The reductase capacity of the Nar proteins may be connected to the high levels of the TerD proteins that are predicted to be involved in countering oxidative stress (27). The NirB protein (SCO6102) a nitrite/sulfite reductase was expressed, but its expression pattern was completely opposite to that of the Nar proteins (supplemental Fig. S5). SCO6102 was up-regulated in a ΔglnR mutant, suggesting that it is normally repressed by GlnR (28).
Nitrogen Metabolism
At the time of inoculation, the sole nitrogen source in the growth medium is glutamate, and this is gradually metabolized during the course of the fermentation to just under 50% of the starting concentration (Fig. 1). Despite the high concentration of glutamate, proteins central to nitrogen metabolism were detected in the early time points and rapidly became undetectable as the cultures approached the time of phosphate depletion (Fig. 6 and supplemental Table S6). During rapid growth, therefore, the cultures appeared to be in a physiological state that mimics growth in limited ammonium salts (28, 29). Nieselt et al. (4) showed that there is a dramatic reduction in the transcription of several central nitrogen assimilation genes early in the time course of M145, specifically SCO2198 (glnA), SCO2210 (glnII), SCO2211 (unknown), SCO4159 (glnR), and SCO5584 (glnK). The protein products of three of these genes were detected (GlnA, GlnII, and GlnK), and the protein levels of GlnII and GlnK closely followed the mRNA levels (Fig. 6 and supplemental Fig. S2). The expression patterns of the glutamine synthetase paralogues (SCO2210 and SCO2241), the regulator of GlnA, PII (SCO5584), the ammonium transporter AmtB (SCO5583), and GltB (SCO2026), the large subunit of glutamate synthetase, imply that these enzymes were derepressed during rapid growth (Fig. 6). The genes for these nitrogen assimilation proteins are targets for the major regulator, GlnR (SCO4159) (29).
During rapid growth, we propose that ammonium ions were released via the action of the NAD-specific glutamate dehydrogenase (SCO2999) that feeds the TCA cycle with α-ketoglutarate. Indeed increasing amounts of ammonium were detected in the medium over the fermentation time course, and the production of ammonium during growth on glutamate has also been observed previously (11). An alternative, NADP-specific, glutamate dehygrogenase, GdhA (SCO4683), in S. coelicolor has been described (30, 31), but this was not detected by proteomics presumably because it was repressed by GlnR. Ammonium released could be used to synthesize glutamine via glutamine synthase (SCO2198 and SCO2210). Glutamate is also an important substrate for transaminases; for example, we observed a high level of aspartate aminotransferase, AspC, SCO4645 that transfers the amine group from glutamate to oxaloacetate to generate aspartate. AspC was proposed previously to be activated by GlnR (28, 29).
The rapid fall in the levels of the GlnR-regulated gene products occurred at or just before the cessation of growth, indicating that, without the demand for amino acid, purine, and pyrimidine biosynthesis, the ammonium levels in the medium become increasingly less limiting (Fig. 6). The major glutamine synthetase, GlnA (SCO2198), NAD-specific glutamate dehydrogenase (SCO2999), and AspC (SCO4645) were highly abundant right up to the last time points. Even though GlnA was maintained at a high level, the levels of the PII protein regulator encoded by glnK dropped to below detectable levels, suggesting that modification of GlnA plays a minor role later in growth. We assume that GlnA, SCO2999, and AspC are required for oxidative amino acid assimilation during the breakdown and turnover of proteins during the stationary phase. The perturbation of the regulation of SCO2198 (glnA) by PhoP is clearly demonstrated by the increase in abundance of GlnA in the ΔphoP mutant, INB201 compared with the parent strain M145 (Fig. 6) (32). Examination of the promoter sequence of SCO4645 (aspC) revealed a putative PhoP operator (PHO box) that could explain the sustained high levels of AspC in INB201 compared with M145 (Fig. 6).
Other nitrogen metabolism proteins that changed in abundance over the fermentation time course include a putative amino-butyrate transaminase (SCO5676, gabT) that increased in the stationary phase (supplemental Fig. S6). Although glutamate could be converted to 4-aminobutryate via glutamate decarboxylase, the most likely candidate protein to catalyze this step in S. coelicolor is SCO3416, gad, and was not detected. In the absence of 4-amino butyrate, it seems likely that SCO5676 is degrading β-alanine by reaction with α-ketoglutarate to produce glutamate and malonate semialdehyde. An NAD+ synthetase (SCO0506) that requires ammonium as a substrate decreased in abundance as did SCO2789 (GlmS2; glucosamine fructose-6-phosphate aminotransferase). Both SCO0506 and SCO2789 behave as if they are repressed by GlnR (28, 29) (Fig. 6).
As growth ceases, the levels of enzymes for purine, pyrimidine, and amino acid biosynthesis become reduced (supplemental Table S6). A notable exception given the role of ppGpp in controlling antibiotic biosynthesis (3) is the product of SCO5737, a putative guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase, which increases in abundance over the time course.
Comparison of Proteomes from S. coelicolor M145 and the ΔphoP Mutant, INB201
Proteins detected only in the M145 or the INB201 replicate time courses were identified. Within these sets, proteins that occurred in less than four time points in the replicate time courses were eliminated (supplemental Tables S8 and S9). Proteins that were more abundant in M145 versus INB201 or vice versa were also identified from the total EmPAI values per protein over both replicate time courses as described above but included proteins that were detected in only three of the four time courses as well as proteins detected in all four time courses. Where a protein was not detected, the EmPAI value was treated as 0. As before, the sum of the EmPAI values ΣEmPAIM145, and ΣEmPAIINB201 reflecting the abundance of a protein in the parent strain M145 versus the ΔphoP mutant INB201 were used to calculate ratios. A fold change of greater than 2.8-fold was used as a cut-off (supplemental Tables S5 and S8).
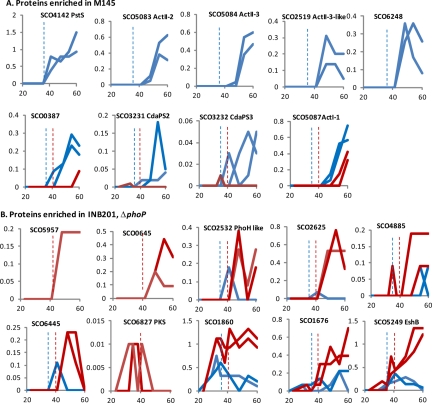
The phosphate starvation response is clearly evident from the M145-specific proteome (supplemental Table S8 and Fig. 7). Proteins that are strongly induced on phosphate depletion in M145 but absent in the ΔphoP mutant INB201 include PhoP (SCO4230) itself, the high affinity phosphate transport system, PstSCAB (SCO4139–4142), polyphosphate kinase (SCO4145), alkaline phosphatase (SCO2286), and enzymes involved in a putative teichuronic acid biosynthesis pathway (includes SCO4883 and SCO4871). Promoters that express these proteins have been characterized previously as PhoP targets (8, 33). Other known or possible PhoP-regulated gene products (encoded by SCO1565, glpQ1; SCO1968, glpQ2; SCO4151; and SCO3790) and two alkaline phosphatase-like proteins (SCO2068 and SCO2286) were observed only in the M145 replicates but at low abundances (supplemental Table S6) (33). A small group of M145-specific proteins were detected only in the last three time points, slightly later than those known to be regulated by PhoP. The ActII-2 and ActII-3 proteins (SCO5083 and SCO5084) mediate actinorhodin transport and are repressed by the product of ActII-1 (34, 35). Biosynthetic intermediates of Act bind to the ActII-1 repressor and derepress the promoter for actII-2 (35). The absence of ActII-2 and ActII-3 in INB201 could be because the levels of Act intermediates never reach the concentrations required to bind the ActII-1 repressor and relieve repression of the actII-2 promoter (35). Indeed M145 contains higher levels of at least one of the Act biosynthesis enzymes (SCO5087; Fig. 7) and correspondingly more Act (and Red) was produced during the fermentation (Fig. 1). SCO2519 is not only a paralogue of ActII-3 (SCO5084) but was also expressed in exactly the same time points, also only in M145. Also present in the last three time points was a putative allantoicase (SCO6248), an enzyme that uses purines as an alternative nitrogen source, ultimately breaking them down to ammonium and carbon dioxide. Although the similar expression profiles of ActII-2, ActII-3, SCO2519, and SCO6248 might suggest they are all regulated by ActII-1, the binding site for ActII-1 was not detected upstream of SCO2519 and SCO6248. The only M145-specific protein that appears to be induced before phosphate depletion is SCO7586, which encodes a putative flavin-dependent, NADPH-dependent oxidoreductase (supplemental Table S8).
Fig. 7.
Proteins enriched in M145 or in the ΔphoP mutant INB201. Trends in the expression profiles of selected proteins from the M145 proteome (A) or the INB201 proteome (B) are shown as graphs prepared as described in the legend to Fig. 2.
Proteins only detected or enriched in INB201 indicate that a number of possible phosphate scavenging proteins are induced but also that there are differences in metabolism compared with M145. SCO2532, a PhoH-like phosphate starvation-inducible protein with predicted ATPase activity was markedly more abundant in INB201 and is perhaps an indicator of the greater level of phosphate depletion in the ΔphoP mutant (Fig. 7). Transcription of SCO2532 was also up-regulated in INB201 compared with the parent M145 (supplemental Fig. S2) (8). The induction of putative transport proteins SCO5957 and SCO5783 (Fig. 7 and supplemental Table S9 and Fig. S2) may enable phosphate uptake in the absence of the PhoP-inducible PstSCAB phosphate transporter. At least three other proteins of unknown function had expression profiles similar to SCO2532 (SCO2625, SCO4884, and SCO6445; Fig. 7 and supplemental Fig. S2). All of these proteins may be responding to phosphate depletion by a PhoP-independent mechanism. In addition, a putative secreted protein, SCO1860, and a nucleotide-binding protein, SCO5249, EshB, that were only detected in M145 over the transition phase, accumulated to much higher levels in the ΔphoP mutant compared with its parent (Fig. 7). An eshB null mutant has no obvious phenotype, but its paralogue, eshA (SCO5249), is required for high levels of actinorhodin production because it is thought to accentuate the physiological role of ppGpp (36). EshA (SCO7699) was only detected in INB201 (supplemental Table S6), and mRNA of both eshA and eshB rose toward the end of the fermentation (supplementalFig. S2). Taken together there appears to be an important role for these proteins in the ΔphoP mutant.
The ΔphoP strain contained high levels of enzymes involved in gluconeogenesis compared with the parent strain M145. Pyruvate phosphate dikinase paralogues (SCO2494 and SCO0208, Pdk1 and Pdk2), phosphoenol pyruvate carboxykinase (SCO4979, Pck), and fructose 1,6 bisphosphatase (SCO5047, GlpX) were all detected repeatedly in the ΔphoP extracts but at fewer time points or not at all in the parent strain (Fig. 3 and supplemental Tables S6 and S9). M145 had higher levels of glycolytic enzymes, phosphofructokinase (SCO1214, PfkA3), and a paralogue of pyruvate kinase (SCO2014, Pyk1) (Fig. 3 and supplemental Tables S6 and S8). As the level of glucose in the fermentors changed little over the time course, the up-regulation of the gluconeogenesis pathway in INB201 is more likely to reflect overall remodeling of metabolism to balance the availability of phosphate rather than sensing glucose depletion (Fig. 1). A plausible hypothesis is that gluconeogenesis is used to correct imbalances in redox levels in particular the ratio of NADH:NAD+. The enzymes involved in gluconeogenesis in the ΔphoP mutant are in abundance from the earliest time points (SCO4979 and SCO5047) or as the growth rate slows (SCO2494 and SCO0208), suggesting that the metabolic stress of growth without the PhoP-regulated gene products extends through all of the phases of growth in the fermentor. When phosphate became depleted during the cultivation of the ΔphoP mutant, the rate of CO2 production fell further and faster compared with that during the cultivation of the parent strain (Fig. 1), probably because of the absence of normal phosphate homeostasis and the effects of phosphate limitation on oxidative phosphorylation. There are several dehydrogenases and reductases more highly expressed in the ΔphoP mutant: SCO0645, SCO7040 (Gap2), GlpA (SCO1661), NuoG (SCO4568), SCO5385, SCO7236 (QcrB3), and SCO7120 (QcrB2) that could be compensating for a redox imbalance caused by phosphate limitation (Fig. 7 and supplemental Table S9).
Synthesis of Secondary Metabolites in M145 and INB201
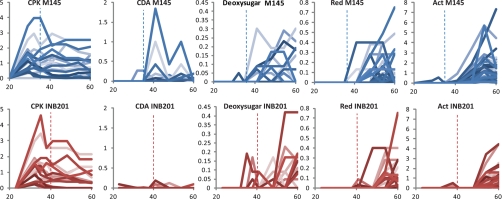
The timing of appearance of enzymes required for all of the secondary metabolic pathways correlated approximately with the time of their mRNA synthesis (4) (Fig. 8 and supplemental Fig. S2). The CPK enzymes were produced during rapid growth; the CDA enzymes and those expressed from the proposed deoxysugar cluster (SCO0381-SCO0401) (37) were produced when the rate of respiration/biomass production slowed; and Red and Act clusters were expressed, in that order, several hours after stationary phase was reached (Fig. 8). Although the CDA enzymes were present at low abundance in both M145 (wild type) and INB201 (ΔphoP mutant), the two nonribosomal peptide synthases, SCO3230 and SCO3232 were more abundant in M145 (Figs. 7 and 8). The mRNA expression for the corresponding CDA proteins were coordinately up-regulated in M145 at ∼35 h post-inoculation, whereas mRNAs for the same proteins in the ΔphoP mutant gradually increased over several hours (supplemental Fig. S2). Clearly the physiological conditions required for expression of the CDA gene cluster were not optimal in the ΔphoP mutant and may reflect perturbations in the expression of regulators. AbrC1/2/3, AbsA1/A2, and CdaR (SCO4598–96, SCO3225/6 and, SCO3217, respectively) are known to affect the expression of cda genes (38–40), but we did not observe differential expression of these regulators at the mRNA level, and none of the gene products were detected in the proteomes.
Fig. 8.
Timing of expression of enzymes for secondary metabolism synthesis in M145 and INB201. The graphs show EmPAI values (y axis) versus time since inoculation (h; x axis) for all the enzymes detected for each secondary metabolite from the M145 replicates (blue) or from the ΔphoP mutant INB201 replicates (red). The dashed blue lines and the dashed red lines represent the times of phosphate depletion in M145 and INB201 at 35 and 40 h, respectively.
There is an obvious temporal overlap between primary and secondary metabolism and secondary metabolic pathway-specific switches are likely to be sensing very different metabolic and regulatory signals. We detected a total of 183 putative regulators in the combined proteomes from M145 and the ΔphoP mutant (supplemental Table S5). Of these, 23 are predicted to be response regulators, and 19 are presumed sensor kinases. The next largest groups are the TetR and MarR families (12 and 11, respectively). Nine sigma factors and five anti-sigma factors were detected. The EmPAI values indicated that the vast majority of these regulatory proteins had no reproducible trends in their expression, and their abundances were very low (supplemental Table S6). Many of the regulators are involved in stress responses (discussed above). Proteins known to be involved in development were observed, and they either had no particular trend of expression (e.g. BldG), or they were, like the regulators of the stress responses, optimally detected over transition and early stationary phase (e.g. ScbR, RarA, BldN, SCO4677 encoding an anti-sigmaF regulator, BldD) (supplemental Table S6). BldC (SCO4091), which is required for high levels of transcription of the pathway-specific activators ActII-Orf4 and RedD (41), was not detected in the last two time points from the INB201 fermentation despite being constitutively transcribed in both M145 and in INB201 (supplemental Fig. S2 and Table S6). This apparent depletion of BldC cannot, however, account for the poor levels of production of total blue pigment (TBP; includes Act) and Red in INB201 because the levels of biosynthetic enzymes produced were similar in M145 and INB201 (supplemental Table S6 and Figs. 1 and 8).
The synthesis of antibiotics is greatly affected by precursor supply. CPK is synthesized during the rapid growth phase when precursor supply for polyketides, notably acetyl and malonyl-CoA, should be plentiful. Act is also synthesized from acetyl-CoA, whereas the precursors for Red are proline and l-serine/l-glycine in addition to acetyl-CoA. However, many of the enzymes required to generate acetyl-CoA such as pyruvate dehydrogenase appeared to be greatly depleted by the time Act and Red are produced (e.g. Figs. 1 and 6). Also important is the synthesis of malonyl-CoA through the activity of acetyl-CoA carboxylase. Previously it was shown that overexpression of these enzymes led to increased expression of Act (42, 43). SCO6271 (AccA1), SCO4921 (AccA2), and SCO5535 (AccB) encode subunits of an acetyl-CoA carboxylase (44). SCO4921 and SCO6271 have identical amino acid sequences, and our study was therefore unable to distinguish between the products of these two genes. SCO5535 and SCO6271/4921 gene products were most abundant early on in growth consistent with their role in FAS (supplemental Fig. S6). No other homologues of these proteins were detected in the proteome.
Acetyl-CoA can be synthesized from other metabolites. A putative pathway, based on increasing levels of SCO5385 and SCO5399 late in the growth cycle, might include an acyl-CoA dehydrogenase (SCO5385) that could convert hydroxylbutanoyl-CoA to acetoacetyl-CoA followed by a putative acetoacetyl-CoA thiolase (SCO5399; supplemental Fig. S6). Although SCO5399 was the only acetyl-CoA acetyltransferase detected in our study there were other putative acyl-CoA dehydrogenases including SCO1591, SCO6026, SCO3834, and SCO6789 detected (supplemental Table S6).
Two other enzymes that could be of relevance to precursor supply based on their increasing abundance late in the fermentation cycle are SCO5676 encoding a putative 4-amino butyrate aminotransferase (GabT) that could generate methylmalonate semi-aldehyde (discussed also above) and SCO2726 or MsdA that converts methyl malonate semi-aldehyde to methylmalonyl CoA (supplemental Fig. S6). Notably the levels SCO2726 were much lower in the ΔphoP mutant, INB201, which correlates with the much lower levels of TBP and Red production in this strain (Fig. 1 and supplemental Fig. S6).
From carbon balance calculations, we determined the yields of both Red and TBP to be in the order of 1% Cmol for the parent strain, M145 (data not shown). The enzymes for the biosynthesis of Act and Red are coordinately expressed (as shown by both the mRNA and the proteome data) at a time that coincides closely with the start of Act and Red production, and they are still increasing in abundance even at the end of the time course. The rise in TBP and Red showed an almost constant rate of production in both parent and ΔphoP mutant strains, implying the existence of a rate-limiting metabolic step, either related to the biosynthesis of Act and Red or in the pathways supplying the respective precursors (45).
Conclusion
We have used a semi-quantitative index of protein abundance, the EmPAI, to find significant changes to the proteome during the fermentation of S. coelicolor M145 in phosphate-limited medium and compared this with a ΔphoP mutant INB201. We have identified enzymes that could mediate all of the reactions required for nitrogen assimilation: the TCA cycle, glycolysis, and pyruvate metabolism, and most enzymes required for the pentose phosphate pathway and oxidative phosphorylation. Streptomyces genomes, more than most other bacterial genomes, contain multiple paralogues, but the proteome indicated that usually only one or two of these candidate proteins is present at detectable levels. Stress response proteins are some of the most abundant proteins in the proteome and have a crucial role in switching on antibiotic synthesis. A comparison of the proteome from M145 with that of the ΔphoP mutant showed the expected absence of induction of PhoP-dependent gene products. Compensation for the lack of normal phosphate homeostasis (uptake, storage, and utilization) appeared to involve an up-regulation of transport proteins, regulators, and, surprisingly, enzymes required for gluconeogenesis. The coordinated appearance of enzymes for secondary metabolite biosynthesis, including the novel deoxysugar derivative proposed by Bentley et al. (37), at various stages of growth emphasizes that very different physiological conditions switch on the different pathways. The proteome and the corresponding transcriptome data are a useful resource for understanding the molecular changes that occur during a batch culture fermentation leading to antibiotic biosynthesis.
Acknowledgments
We are grateful for help, advice, and access to instrumentation from the Aberdeen Proteomics Facility. We acknowledge the excellent technical help of Anders Øverby, Sunniva Hoel, and Elin Hansen in fermentation experiments.
Footnotes
* This work was funded by Biotechnology and Biological Sciences Research Council Grant BB/F003439/1, ERA-NET SysMO Project GEN2006-27745-E/SYS Grant P-UK-01-11-3i, and Research Council of Norway Project 181840/I30. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
2 J. Moore, R. Legaie, V. Armendarez, E. Laing, G. Chandra, M. Bibb, and D. A. Hodgson, manuscript in preparation.
3 G. Sawers personal communication.
1 The abbreviations used are:
- Act
- actinorhodin
- Red
- undecylprodigiosin
- mRNA
- messenger RNA.
REFERENCES
- 1. Bibb M. (1996) 1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142, 1335–1344 [DOI] [PubMed] [Google Scholar]
- 2. Martín J. F., Liras P. (2010) Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr. Opin. Microbiol. 13, 263–273 [DOI] [PubMed] [Google Scholar]
- 3. Bibb M. J. (2005) Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8, 208–215 [DOI] [PubMed] [Google Scholar]
- 4. Nieselt K., Battke F., Herbig A., Bruheim P., Wentzel A., Jakobsen Ø. M., Sletta H., Alam M. T., Merlo M. E., Moore J., Omara W. A., Morrissey E. R., Juarez-Hermosillo M. A., Rodríguez-García A., Nentwich M., Thomas L., Iqbal M., Legaie R., Gaze W. H., Challis G. L., Jansen R. C., Dijkhuizen L., Rand D. A., Wild D. L., Bonin M., Reuther J., Wohlleben W., Smith M. C., Burroughs N. J., Martín J. F., Hodgson D. A., Takano E., Breitling R., Ellingsen T. E., Wellington E. M. (2010) The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics 11, 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pawlik K., Kotowska M., Chater K. F., Kuczek K., Takano E. (2007) A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2). Arch. Microbiol. 187, 87–99 [DOI] [PubMed] [Google Scholar]
- 6. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (EmPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 7. Santos-Beneit F., Rodríguez-García A., Sola-Landa A., Martín J. F. (2009) Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol. Microbiol. 72, 53–68 [DOI] [PubMed] [Google Scholar]
- 8. Rodríguez-García A., Barreiro C., Santos-Beneit F., Sola-Landa A., Martín J. F. (2007) Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics 7, 2410–2429 [DOI] [PubMed] [Google Scholar]
- 9. Barsnes H., Vizcaíno J. A., Eidhammer I., Martens L. (2009) PRIDE Converter: Making proteomics data-sharing easy. Nat. Biotechnol. 27, 598–599 [DOI] [PubMed] [Google Scholar]
- 10. Vizcaíno J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 38, D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Novotna J., Vohradsky J., Berndt P., Gramajo H., Langen H., Li X. M., Minas W., Orsaria L., Roeder D., Thompson C. J. (2003) Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol. Microbiol. 48, 1289–1303 [DOI] [PubMed] [Google Scholar]
- 12. Bucca G., Brassington A. M., Hotchkiss G., Mersinias V., Smith C. P. (2003) Negative feedback regulation of dnaK, clpB and lon expression by the DnaK chaperone machine in Streptomyces coelicolor, identified by transcriptome and in vivo DnaK-depletion analysis. Mol. Microbiol. 50, 153–166 [DOI] [PubMed] [Google Scholar]
- 13. de Crécy-Lagard V., Servant-Moisson P., Viala J., Grandvalet C., Mazodier P. (1999) Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol. Microbiol. 32, 505–517 [DOI] [PubMed] [Google Scholar]
- 14. Servant P., Thompson C., Mazodier P. (1993) Use of new Escherichia coli/Streptomyces conjugative vectors to probe the functions of the two groEL-like genes of Streptomyces albus G by gene disruption. Gene 134, 25–32 [DOI] [PubMed] [Google Scholar]
- 15. Pearce M. J., Mintseris J., Ferreyra J., Gygi S. P., Darwin K. H. (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322, 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanssouci E., Lerat S., Grondin G., Shareck F., Beaulieu C. (2011) tdd8: a TerD domain-encoding gene involved in Streptomyces coelicolor differentiation. Antonie van Leeuwenhoek 100, 385–398 [DOI] [PubMed] [Google Scholar]
- 17. Paget M. S., Kang J. G., Roe J. H., Buttner M. J. (1998) SigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 17, 5776–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Facey P. D., Hitchings M. D., Saavedra-Garcia P., Fernandez-Martinez L., Dyson P. J., Del Sol R. (2009) Streptomyces coelicolor Dps-like proteins: Differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol. Microbiol. 73, 1186–1202 [DOI] [PubMed] [Google Scholar]
- 19. Fernández Martínez L., Bishop A., Parkes L., Del Sol R., Salerno P., Sevcikova B., Mazurakova V., Kormanec J., Dyson P. (2009) Osmoregulation in Streptomyces coelicolor: Modulation of SigB activity by OsaC. Mol. Microbiol. 71, 1250–1262 [DOI] [PubMed] [Google Scholar]
- 20. Bursy J., Kuhlmann A. U., Pittelkow M., Hartmann H., Jebbar M., Pierik A. J., Bremer E. (2008) Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 74, 7286–7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schneider D., Bruton C. J., Chater K. F. (2000) Duplicated gene clusters suggest an interplay of glycogen and trehalose metabolism during sequential stages of aerial mycelium development in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263, 543–553 [DOI] [PubMed] [Google Scholar]
- 22. Sevcikova B., Rezuchova B., Homerova D., Kormanec J. (2010) The anti-anti-sigma factor BldG is involved in activation of the stress response sigma factor sigma(H) in Streptomyces coelicolor A3(2). J. Bacteriol. 192, 5674–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bignell D. R., Warawa J. L., Strap J. L., Chater K. F., Leskiw B. K. (2000) Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology 146, 2161–2173 [DOI] [PubMed] [Google Scholar]
- 24. van Keulen G., Siebring J., Dijkhuizen L. (2011) In Streptomyces: Molecular Biology and Biotechnology (Dyson P. J., ed) pp. 105–123, Caister Academic Press, Norfolk, UK [Google Scholar]
- 25. Tian J., Bryk R., Itoh M., Suematsu M., Nathan C. (2005) Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: Identification of α-ketoglutarate decarboxylase. Proc. Natl. Acad. Sci. U.S.A. 102, 10670–10675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fischer M., Alderson J., van Keulen G., White J., Sawers R. G. (2010) The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 156, 3166–3179 [DOI] [PubMed] [Google Scholar]
- 27. Chasteen T. G., Fuentes D. E., Tantaleán J. C., Vásquez C. C. (2009) Tellurite: History, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33, 820–832 [DOI] [PubMed] [Google Scholar]
- 28. Tiffert Y., Franz-Wachtel M., Fladerer C., Nordheim A., Reuther J., Wohlleben W., Mast Y. (2011) Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl. Microbiol. Biotechnol. 89, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 29. Tiffert Y., Supra P., Wurm R., Wohlleben W., Wagner R., Reuther J. (2008) The Streptomyces coelicolor GlnR regulon: Identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67, 861–880 [DOI] [PubMed] [Google Scholar]
- 30. Fisher S. H. (1989) Glutamate synthesis in Streptomyces coelicolor. J. Bacteriol. 171, 2372–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wohlleben W., Mast Y., Reuther J. (2011) In Streptomyces: Molecular Biology and Biotechnology (Dyson P., ed) pp. 125–136, Caister Academic Press, Norfolk, UK [Google Scholar]
- 32. Rodríguez-García A., Sola-Landa A., Apel K., Santos-Beneit F., Martín J. F. (2009) Phosphate control over nitrogen metabolism in Streptomyces coelicolor: Direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res. 37, 3230–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sola-Landa A., Rodríguez-García A., Apel A. K., Martín J. F. (2008) Target genes and structure of the direct repeats in the DNA-binding sequences of the response regulator PhoP in Streptomyces coelicolor. Nucleic Acids Res. 36, 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fernández-Moreno M. A., Caballero J. L., Hopwood D. A., Malpartida F. (1991) The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66, 769–780 [DOI] [PubMed] [Google Scholar]
- 35. Tahlan K., Ahn S. K., Sing A., Bodnaruk T. D., Willems A. R., Davidson A. R., Nodwell J. R. (2007) Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol. 63, 951–961 [DOI] [PubMed] [Google Scholar]
- 36. Saito N., Xu J., Hosaka T., Okamoto S., Aoki H., Bibb M. J., Ochi K. (2006) EshA accentuates ppGpp accumulation and is conditionally required for antibiotic production in Streptomyces coelicolor A3(2). J. Bacteriol. 188, 4952–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N. R., James K. D., Harris D. E., Quail M. A., Kieser H., Harper D., Bateman A., Brown S., Chandra G., Chen C. W., Collins M., Cronin A., Fraser A., Goble A., Hidalgo J., Hornsby T., Howarth S., Huang C. H., Kieser T., Larke L., Murphy L., Oliver K., O'Neil S., Rabbinowitsch E., Rajandream M. A., Rutherford K., Rutter S., Seeger K., Saunders D., Sharp S., Squares R., Squares S., Taylor K., Warren T., Wietzorrek A., Woodward J., Barrell B. G., Parkhill J., Hopwood D. A. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 [DOI] [PubMed] [Google Scholar]
- 38. McKenzie N. L., Nodwell J. R. (2007) Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J. Bacteriol. 189, 5284–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yepes A., Rico S., Rodríguez-García A., Santamaría R. I., Díaz M. (2011) Novel two-component systems implied in antibiotic production in Streptomyces coelicolor. PloS One 6, e19980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ryding N. J., Anderson T. B., Champness W. C. (2002) Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 184, 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hunt A. C., Servín-González L., Kelemen G. H., Buttner M. J. (2005) The bldC developmental locus of Streptomyces coelicolor encodes a member of a family of small DNA-binding proteins related to the DNA-binding domains of the MerR family. J. Bacteriol. 187, 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ryu Y. G., Butler M. J., Chater K. F., Lee K. J. (2006) Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl. Environ. Microbiol. 72, 7132–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Diacovich L., Peirú S., Kurth D., Rodríguez E., Podestá F., Khosla C., Gramajo H. (2002) Kinetic and structural analysis of a new group of Acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J. Biol. Chem. 277, 31228–31236 [DOI] [PubMed] [Google Scholar]
- 44. Rodríguez E., Banchio C., Diacovich L., Bibb M. J., Gramajo H. (2001) Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3(2). Appl. Environ. Microbiol. 67, 4166–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L., Tian X., Wang J., Yang H., Fan K., Xu G., Yang K., Tan H. (2009) Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. U.S.A. 106, 8617–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]