Abstract
Ankylosing Spondylitis (AS) is a common inflammatory rheumatic disease with a predilection for the axial skeleton, affecting 0.2% of the population. Current diagnostic criteria rely on a composite of clinical and radiological changes, with a mean time to diagnosis of 5 to 10 years. In this study we employed nano liquid-chromatography mass spectrometry analysis to detect and quantify proteins and small compounds including endogenous peptides and metabolites in serum from 18 AS patients and nine healthy individuals. We identified a total of 316 proteins in serum, of which 22 showed significant up- or down-regulation (p < 0.05) in AS patients. Receiver operating characteristic analysis of combined levels of serum amyloid P component and inter-α-trypsin inhibitor heavy chain 1 revealed high diagnostic value for Ankylosing Spondylitis (area under the curve = 0.98). We also depleted individual sera of proteins to analyze endogenous peptides and metabolic compounds. We detected more than 7000 molecular features in patients and healthy individuals. Quantitative MS analysis revealed compound profiles that correlate with the clinical assessment of disease activity. One molecular feature identified as a Vitamin D3 metabolite—(23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone—was down-regulated in AS. The ratio of this vitamin D metabolite versus vitamin D binding protein serum levels was also altered in AS as compared with controls. These changes may contribute to pathological skeletal changes in AS. Our study is the first example of an integration of proteomic and metabolomic techniques to find new biomarker candidates for the diagnosis of Ankylosing Spondylitis.
Ankylosing Spondylitis (AS)1 is a common inflammatory disease affecting more than two million people in the United States (1–3) and is associated with significant morbidity and mortality. AS affects the axial skeleton, frequently targeting the sacroiliac joint in the pelvis, but can also involve peripheral joints and is associated with both uveitis and inflammatory bowel disease. Patients suffer symptoms of back pain and arthritis without clear diagnosis for many years (4). The etiology of Ankylosing Spondylitis is unknown but is thought to be immune-mediated and has a strong genetic association with the class I human Leukocyte antigen allotype HLA-B27 (5). Additional genes including the endopeptidase ERAP1 and the IL23R receptor (IL23R) have also been associated with Ankylosing Spondylitis (3, 6–8). Structural and functional studies of endoplasmatic reticulum aminopeptidase (ERAP1) have recently demonstrated altered N-terminal peptide trimming for the AS-associated ERAP1 variant K528R, supporting a key role for Major histocompatibility complex I-associated antigen processing in AS pathogenesis (9, 10). Even though these studies have contributed significantly to the understanding of AS, the diagnosis is still challenging and is commonly based on a composite of clinical features and sacroiliac joint radiological changes, with a mean time to diagnosis of 5 to 10 years. With effective and potentially disease-modifying treatments such as tumor necrosis factor (TNF) inhibitors becoming widely available (11), the diagnostic delay becomes the critical rate-limiting factor for the mobility and quality of life of AS patients.
Blood markers such as autoantibodies against citrullinated proteins or Rheumatoid factor are used for the early diagnosis of Rheumatoid Arthritis with high specificity and selectivity (12, 13). However, potential markers for the diagnosis of AS such as C-reactive protein, IgG, or IgA have not archived sufficient diagnostic sensitivity or specificity (14, 15). Recently an array of autoantibodies in plasma targeting skeletal and connective tissue exhibited a specificity of 60% for AS compared with healthy controls or Rheumatoid Arthritis patients (16, 17). There is also preliminary data suggesting that gene-expression profiling may prove diagnostically useful (18). In addition, several proteins including the metalloprotease MMP-3 (19), CA1 (20), talin-1, and integrin-linked kinase 1 have been reported to be aberrantly expressed in the context of AS (21). The pathology of AS is complex, and it has been suggested that the alteration of bone growth in AS may be uncoupled from the inflammatory process, which can be separately monitored by a differential set of biomarkers (22). Also, TNF inhibitors given in established disease may not reduce the abnormal bone formation in the spine (23). The diagnostic delay and the variability of findings in previous biomarker studies already imply that an array of markers rather than a single compound will be more promising for an accurate diagnosis of AS (15, 24).
In the present discovery study, we show that the mass profile of proteins and small compounds in the serum of individuals can be used to differentiate between AS subjects and healthy individuals. Since the early diagnosis of AS can have an impact on the treatment of subjects with AS, we also present a set of molecular features that can be used to differentiate the AS patients according to the severity of symptoms, represented by the Bath Ankylosing Spondylitis Disease Activity Index (BASDAI). We also demonstrate a down-regulation of the Vitamin D metabolite (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone in our metabolomic approach. Although its abundance in serum was in concordance with levels of Vitamin D binding protein in healthy individuals, this was not the case in AS subjects. Interestingly 25- hydroxyl Vitamin D and its metabolites are involved in the regulation of bone formation (25), providing a potential link to AS-associated osteoporosis. Our integrative approach represents the first study which combines the analysis of proteins and small compounds such as metabolites to define an array of diagnostic serum markers for AS.
EXPERIMENTAL PROCEDURES
Patients Sample Collection
Patients with definite AS (modified New York criteria) were recruited from the Ankylosing Spondylitis Clinic Oxford (UK) under the appropriate ethical permission Oxfordshire REC 06/Q1606/139. These samples were used for method development. Patient samples reported here (Table I), were obtained under the appropriate ethical permission Queensland (AU) HREC/05/QPAH/221. All participants gave informed, written consent.
Table I. Clinical data of the patients and controls used in this study. Clinical data were acquired on date of sampling. CRP is [mg/L], ESR is [mm/h], ALP is [IU/L], Ca [mmol/L], PO4 is [mmol/L] and Alb is [g/L].
| Number | Gender | Age | AS/Control | TNF treatment | Steroids | BASDAI | CRP | ESR | ALP | Ca (Alb corr) | PO4 | Alb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 66 | M | 47 | AS | TNF | None | 3.4 | 20 | 8 | 98 | 2.25 | 1.06 | 38 |
| 72 | M | 37 | AS | None | None | 2 | 21 | 8 | 68 | 2.29 | 1.05 | 37 |
| 73 | M | 35 | AS | None | None | 7.2 | 83 | 84 | 100 | 2.47 | 0.89 | 36 |
| 77 | M | 38 | Control | |||||||||
| 78 | M | 43 | Control | |||||||||
| 83a | M | 61 | AS | previous TNF | None | 7.3 | 19 | 54 | 97 | 2.33 | 1.51 | 38 |
| 92 | M | 41 | Control | |||||||||
| 93 | M | 30 | AS | None | None | 6.1 | 11 | 36 | 79 | 2.29 | 1.54 | 40 |
| 95 | M | 38 | AS | None | 25 mg/day | 9.5 | 149 | 34 | 69 | 2.15 | 0.94 | 43 |
| 96 | M | 18 | AS | NA | None | 7.6 | 98 | 85 | 84 | 2.37 | 1.21 | 35 |
| 99 | M | 35 | AS | None | None | 5.4 | 30 | 39 | 99 | 2.3 | 1.32 | 37 |
| 103 | M | 25 | AS | None | None | 8.8 | 9.1 | 51 | 104 | 2.2 | 1.32 | 41 |
| 104 | F | 37 | AS | None | None | 6.6 | 20 | 68 | 77 | 2.3 | 1.05 | 39 |
| 107 | F | 26 | Control | None | ||||||||
| 108 | M | 33 | Control | None | ||||||||
| 110 | M | 31 | AS | None | None | 8.4 | 11 | 25 | 92 | 2.37 | 1.46 | 43 |
| 115 | M | 63 | AS | None | None | 6.3 | 8.8 | 85 | 40 | 2.38 | 1.45 | 31 |
| 116 | M | 64 | Control | |||||||||
| 120 | M | 32 | Control | |||||||||
| 126 | M | 67 | AS | None | None | 5.6 | 34 | 52 | 86 | 2.28 | 1.06 | 37 |
| 128 | M | 41 | AS | None | None | 8 | 7.6 | 17 | 101 | 2.27 | 1.09 | 42 |
| 130 | M | 45 | AS | None | 6.1 | 14 | 22 | 137 | 2.41 | 1.35 | 42 | |
| 131 | M | 39 | AS | None | 8.6 | 7.4 | 28 | 70 | 2.35 | 1.05 | 35 | |
| 132 | M | 52 | Control | |||||||||
| 136 | M | 35 | AS | None | 5.9 | 14 | 21 | 97 | 2.48 | 1.51 | 35 | |
| 140 | M | 35 | AS | None | 8.9 | 31 | 39 | 105 | 2.45 | 1.52 | 35 | |
| 141 | M | 39 | Control |
a Used in metabolomic study only.
Blood was collected from AS patients and age-gender matched controls (supplemental Table S1) into 8 ml Vacuette Z Serum Sep Clot Activator tubes (Greiner, Kremsmünster, Austria), left to clot at room temperature for 2 h then spun at 3000 × g for 15 min. Serum was aliquoted and stored at −80 °C until further use.
Metabolomic Workflow
Serum of patients and healthy individuals was centrifuged at 16000 × g for 15 min and filtered through a 0.45-μm syringe filter. One hundred microliters of serum were precipitated twice, using chloroform and methanol as described (26). The protein-free aqueous phase was ultra-filtered with a MWCO of 5000 Da and the flow through was dried and resuspended in 2% acetonitrile, 0.1% formic acid. Samples were randomized and subjected to a nLC-MS/MS workflow (Agilent Chip/6250 QTOF-MS) in triplicates using a Ultra High Capacity chip (Agilent, G4240–65010) packed with Zorbax 80SB-C18 5 μm (25 mm 500 nl enrichment and 150 mm × 75 μm separation). Chromatographic separation was achieved by a 10 min gradient (5–40% Acetonitrile) followed by 40–98% Acetonitrile in 3 min at a flow rate of 600 nl/min. Data was acquired in positive ionization mode at a 1.37 spectra/s between m/z 50–1700 with a fragmentor voltage of 100 V. Tandem MS (MS/MS) spectra were acquired with an isolation width of 1.3 m/z at 0.91 spectra/s. For data analysis we used Progenesis LC-MS software version 3.1.4003.30577 (http://www.nonlinear.com/). Subjects were grouped by disease state and disease activity (BASDAI). Abundance profiles of compounds were searched manually for significant differences according to the grouping and ANOVA p ≤ 0.01. Isotopic and chromatographic profile of regulated compounds was reviewed manually and compounds exhibiting poorly defined profiles were excluded from further analysis. Principal component analysis (PCA) was carried out to validate clustering of patients according to their grouping either with the curated set of regulated compounds or with a selection of molecular features with the highest separation power between the groups.
Proteomics and Mass Spectrometry Workflow
One hundred microliters of centrifuged and filtered serum was depleted from IgGs and albumin according to Fu et al. (27) with minor changes. In brief, 200 μl of 0.15 m NaCl was added before the incubation with 200 μl Protein G agarose beads (Invitrogen) for 1 h at room temperature. The supernatant was combined with 200 μl of 100 mm NaCl, 10 mm Tris, pH7.4 washing solution. The solution was brought to 4 °C before adding 396 μl of cold 95% ethanol to selectively precipitate nonalbumin proteins. The samples were pelleted at 16,000 × g and the pellet washed with ice-cold 42% Ethanol. Proteins were resuspended in 6 m Urea, reduced with dithiotreitol and alkylated with iodacetamide before proteolysis with Trypsin. Tryptic digests were desalted and subjected to LC-MS/MS (Waters, nAcquity, 75 μm × 250 mm, 1.7 μm particle size) analysis using a Thermo LTQ Orbitrap Velos (30,000 Resolution, Top 20, collision-induced dissociation) workflow and a gradient of 1–40% acetonitrile in 48 min at a flow rate of 250 nl/min.
Samples were randomized and analyzed in triplicates. Peptides were detected and quantified with Progenesis LC-MS software (version 3.1.4003.30577) using default settings (no deconvolution/deisotoping, 200 most intense MS/MS peaks). A merged peaklist generated by Progenesis LC-MS was searched against the IPI human database (version.3.80, 86719 entries) using Mascot (http://www.matrixscience.com/) version 2.3.01, allowing one missed cleavage and 20ppm/0.5 Da mass deviations in MS and MS/MS. Carbamidomethylation of cysteine was a fixed modification. Oxidation of methionine and deamidation of asparagine and glutamine were used as variable modifications. Using a significance threshold of p < 0.05, a total of 158,054 out of 251,351 spectra could be matched to the IPI human database. 3469 decoy hits were identified, indicating a false discovery rate of 2.21%. Protein identifications were grouped into 316 groups based on the highest scoring protein in each group (summarized in supplemental Table S2). For label-free protein quantitation Mascot results were imported into Progenesis LC-MS. Similar proteins were grouped and only non-conflicting features were used for quantitation. Raw abundances were exported and used for LIMMA analysis in R (28).
Statistical Analysis
Statistical analysis of quantified proteins was performed using R (version 2.12.1) (29). The Linear Models for Microarray (LIMMA) package was used to identify significant differences in protein expression between AS and control samples (28). Raw protein abundance values from Progenesis LC-MS were first normalized using quantile normalization, and technical replicates aggregated by taking the median normalized value for each protein in each sample. Twenty-two proteins indicated by LIMMA to be differentially regulated in AS patients versus controls (p value < 0.05, Benjamini Hochberg correction applied) were then used to construct a logistic regression predictive model. Bootstrap forward-backward selection was performed using the boot.stepAIC R package to select proteins for inclusion in the final model. Generalized predictive performance of the final model was estimated with the bootstrap .632+ method (30) using the ModelGood package for R. Bootstrap procedures were chosen because of the small sample size, which prohibited the use of held-out test data or nested cross-validation methods.
RESULTS
Identification of Multiple Serum Proteins Differentially Regulated in AS
Serum samples were obtained from 18 AS patients and nine age-gender matched healthy controls (Table I and supplemental Table S1). IgG- and albumin-depleted serum samples were precipitated, digested with trypsin and analyzed in triplicates using an Orbitrap Velos (Thermo) mass spectrometer. Following to the workflow in Fig. 1, 316 protein groups were identified by Mascot. Two hundred and one proteins were identified with at least two unique peptides and quantified using a label-free approach (supplemental Table S3). Applying LIMMA data (31) we identified 22 proteins which were significantly up- or down- regulated in AS patients compared with healthy individuals (Table II). Among the up-regulated proteins were several acute phase proteins such as C reactive protein (CRP), complement proteins, APCS and Serpin3A. Fig. 2A shows that CRP levels measured semi-quantitatively by MS/MS correlated extremely closely with those obtained by ELISA-based routine biochemical analysis at the time of sampling (R2 = 0.97, p < 0.001).
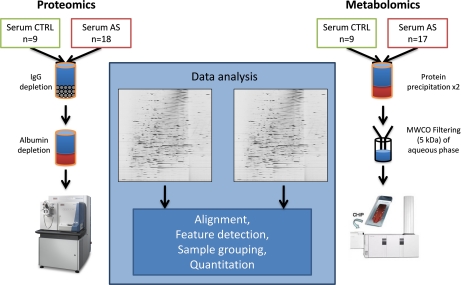
Fig. 1.
Workflow for accessing proteins and small molecules in serum. The proteomic workflow comprises a biochemical depletion of IgGs and albumin followed by tryptic digest and nLC-MS analysis with a LTQ Oribitrap Velos (Thermo) instrument in triplicate. For the metabolomic workflow, proteins in sera of AS patients and healthy individuals were precipitated with chloroform and methanol two times. The aqueous phase was filtered through a molecular cutoff filter (5 kDa). The flow-through was analyzed for small molecules by nLC-MS with a QTOF 6250 (Agilent) in triplicate. In both workflows the data was analyzed with Progenesis LCMS (nonlinear Dynamics).
Table II. Proteins significantly up- or down-regulated in AS compared to controls. Individual sera were IgG depleted and albumin was biochemically removed. Proteins were digested and analysed by mass spectrometry. We identified 22 proteins which were significantly regulated between AS and control using LC-Progenesis software (p < 0.05 for significance of regulation).
| ±log2(AS/CTRL) | p value | Accession [IPI] | Gene | Protein | Expected level in acute phase |
|---|---|---|---|---|---|
| 2.239 | 0.001 | IPI00022389 | CRP | CRP Isoform 1 of C-reactive protein | up[52] |
| 0.799 | 0.005 | IPI00022395 | C9 | C9 Complement component C9 | up[52] |
| 0.653 | 0.014 | IPI00006543 | CFHR5 | CFHR5 Complement factor H-related 5 | up[54] |
| 0.465 | 0.016 | IPI00022463 | TF | TF Serotransferrin | down[52] |
| 0.419 | 0.014 | IPI00019591 | CFB | CFB cDNAFLI55673, highly similar to Complement factor B | up[52] |
| 0.406 | 0.014 | IPI00022391 | APCS | APCS Serum amyloid P-component | up[40] |
| 0.405 | 0.035 | IPI00550991 | SERPINA3 | SERPINA3 cDNAFLI35730fis | up[52] |
| −0.310 | 0.047 | IPI00026314 | GSN | GSN Isoform 1 of Gelsolin | down[54] |
| −0.371 | 0.014 | IPI00218732 | PON1 | PON1 Serum paraoxonase/arylesterase 1 | down[55] |
| −0.374 | 0.041 | IPI00305461 | ITIH2 | ITIH2 Inter-alpha (Globulin) inhibitor H2, isoform CRA_a | up[52] |
| −0.376 | 0.016 | IPI00027482 | SERPINA6 | SERPINA6 Corticosteroid-binding globulin | down[56] |
| −0.394 | 0.035 | IPI00292530 | ITIH1 | ITIH1 Inter-alpha-trypsin inhibitor heavy chain H1 | up[52] |
| −0.441 | 0.036 | IPI00022229 | APOB | APOB Apolipoprotein B-100 | - |
| −0.483 | 0.015 | IPI00022431 | AHSG | AHSG cDNA FLI55606, highly similar to Alpha-2-HS-glycoprotein | down[58] |
| −0.524 | 0.035 | IPI00020996 | IGFALS | IGFALS Insulin-like growth factor-binding protein complex acid labile subunit | - |
| −0.531 | 0.008 | IPI00032179 | SERPINC1 | SERPINC1 Antithrombin-III | down[57] |
| −0.531 | 0.010 | IPI00296099 | THBS1 | THBS1 Thrombospondin-1 | - |
| −0.672 | 0.014 | IPI00016381 | RAB27A | RAB27A Isoform Long of Ras-related protein Rab-27A | - |
| −0.677 | 0.014 | IPI00795528 | C21orf59 | C21orf59 Protein | - |
| −0.677 | 0.015 | IPI00022432 | TTR | TTR Transthyretin | down[52] |
| −0.767 | 0.014 | IPI00301689 | PPFIBP2 | PPFIBP2 Liprin-beta-2 | - |
| −0.821 | 0.001 | IPI00328609 | SERPINA4 | SERPINA4 Kallistatin | down[59] |
Fig. 2.
MS-based quantitation of CRP correlates strictly with ELISA. MS-based quantitation of APCS and ITIH2 has diagnostic utility for Ankylosing Spondylitis. A, CRP concentration as measured by ELISA was plotted against median signal intensities of CRP derived peptides measured by MS exhibiting a close correlation between MS and ELISA readout. B, ROC analysis shows the diagnostic utility of combined quantitation of APCS and ITIH2. C, Signal intensities of APCS and ITIH2 exhibit orthogonality in the detection of AS. In patients where APCS is not up-regulated we observe a down-regulation of ITIH2 and vice versa. 1) represents two overlaying “no AS” data points.
To determine if a diagnosis of AS could be achieved using quantitative assays for a small number of proteins, we developed a predictive logistic regression model from the obtained MS data. Because of the small sample size precluding the use of held-out test data or nested cross validation, we used bootstrap procedures to create and assess this model (see also experimental procedures). Bootstrap forward-backward selection resulted in a final logistic regression model comprising the proteins amyloid P-component serum protein (APCS) and inter-α-trypsin inhibitor 2 (ITIH2). Using normalized abundances of only these two proteins for classification we achieved highly specific and sensitive detection of AS. Using the bootstrap .632+ procedure to estimate the generalized performance of this classifier we observed an area under the Receiver operating characteristic (ROC) curve of 0.98 (Fig. 2B). Although individually APCS and ITIH2 do not separate AS and control patients as strongly as other proteins, their orthogonality facilitates efficient classification when combined (Fig. 2C).
Quantitation of Small Molecular Compounds in Serum Can Discriminate AS from Healthy Controls
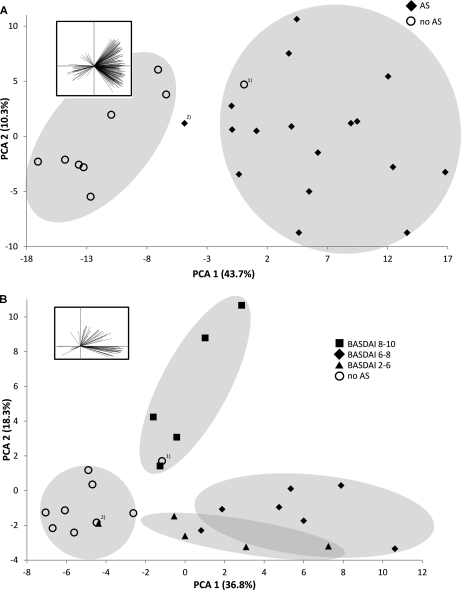
To enable the detection of small water-soluble molecules in serum, we denatured and precipitated serum proteins by chloroform/methanol precipitation (26) followed by an ultrafiltration of the aqueous phase using a 5 kDa molecular cutoff. The filtrate was dried and resuspended in 2% acetonitrile, 0.1% formic acid followed by Chip MS analysis (Agilent 6520) in analytical triplicates. The data were acquired in MS mode to generate as many data points for quantitation as possible and then analyzed using Progenesis LC-MS. The software detected 7813 features that exhibit at least 3 isotopic peaks with a charge state of [M+H]+, [M+2H]2+, or [M+3H]3+ in the compiled dataset. The subjects were grouped according to disease activity as measured by the Bath Ankylosing Spondylitis Disease Activity Index (BASDAI). The small molecules exhibiting a significant differential abundance between the subject groups, based on ANOVA scores (p < 0.01) were reviewed for a defined isotopic and chromatographic profile. We found 215 molecular features that are significantly regulated between AS subjects and healthy controls. The PCA of these compounds enabled a clear separation of the individuals according into healthy or AS (Fig. 3A). However, the assessment of 215 variables in a diagnostic test is challenging, so we repeated the PCA using the 10 molecular features with the highest separating power only (largest negative and positive component loadings of PC1) (supplemental Fig. S1). Using only ten molecular features resulted in a clear separation between healthy individuals and AS subjects. In an effort to further distinguish between AS subjects with different disease activity, a different set of 65 molecular features that enables a separation of at least 2 BASDAI groups was selected (ANOVA p ≤ 0.01). PCA of these compounds permits separation and clustering of patients according to their BASDAI group (Fig. 3 B). Although we observed some overlap between the AS patient groups with BASDAIs of 2–6 and 6–8, this technique clearly separated patients with the most severe disease (BASDAI 8–10). The same set of compounds can still be used to distinguish between AS subjects and healthy individuals in general. Tables of the molecular features detected can be found in the supplemental materials (supplemental Tables S4 and S5).
Fig. 3.
Principal Component Analysis of selected molecular features distinguishes healthy individuals from AS patients and correlates with disease activity. A, LC-MS analysis revealed 215 molecular features with defined chromatographic and isotopic profiles exhibiting significant differences in abundance between AS and controls (ANOVA p < 0.01). Principal Component Analysis (PCA) with this set allows separation of AS from healthy control subjects. Ten molecular features selected for their highest separation power (PC1 loadings, as visualized in the inserted diagram), are sufficient to separate AS from controls (PCA in supplemental Fig. S1). B, A set of 65 molecular features was used for a PCA (ANOVA p ≤ 0.01) after classifying the AS patients into BASDAI groups, which indicate the severity of their symptoms (PC loadings visualized in diagram insert). Patients with most severe symptoms could be separated from the low and medium disease activity groups. Again the control group could be separated from the cases with the exception of one individual (2) with the lowest BASDAI in the cohort and one control individual (1) with the highest age in the control group.
The Vitamin D Metabolite (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone Is Down-regulated in AS
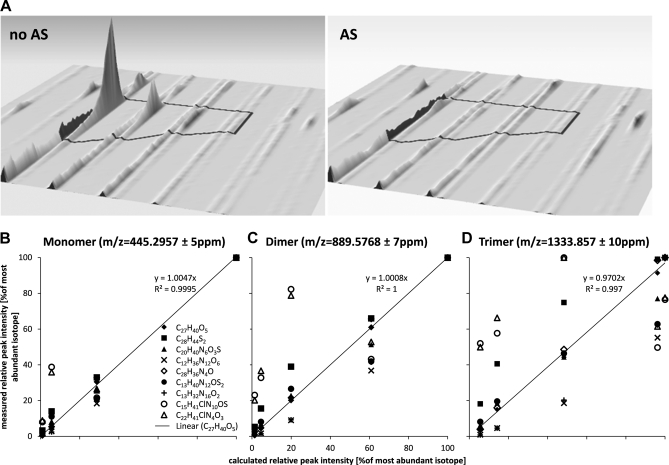
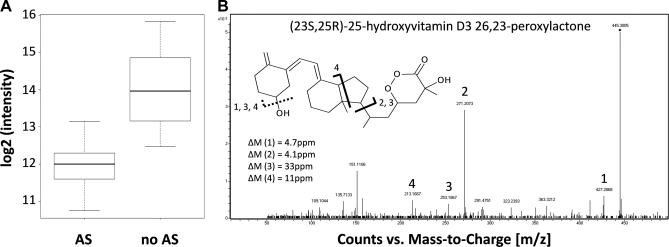
In our MS-based screening experiment for regulated profiles of molecular features we detected a singly charged ion with an m/z of 445.2957, clearly resolved from the internal lock mass of polydimethylcyclosiloxane (PCM) at 445.120024 (Fig. 4A). Besides the [M+H]+ ion we also detected [2M+H]+ at 889.5768 (Δm = 7 ppm) and [3M+H]+ at 1333.857 (Δm = 10 ppm) (data not shown). Nine different chemical formulae were found to match the detected mass within a mass error of ± 5 ppm using Agilent's Masshunter software (version B.03.01). However, only the formula C27H40O5 and its di-/trimer matched the isotopic distribution of the observed ions (Figs. 4B, 4C, and 4D). A search against the Metlin database (32) revealed six different compounds with the chemical formula C27H40O5 (Δm = 1.9 ppm), of which five are isomers of 1,25-Dihydroxyvitamin D3–26,23-lactone and one is (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone. m/z 445.2957 was hardly detectable in 14 AS patients and was found at low intensity in the remaining three (supplemental Fig. S2). Five of the nine healthy individuals exhibited strong signal intensities in serum whereas the compound showed low intensity in the serum of the other four individuals (summarized in box-plot Fig. 5A).
Fig. 4.
Detection of a molecular feature at m/z 445. 2957 and matching of the measured isotopic patterns to compound candidates with the same mass (± 5ppm). A, A singly charged compound was detected at m/z of 445.2957, well separated from the lock-mass of PCM (M+H+ = 445.120024 Da). The left signal represents the strongest signal in the control group whereas the right signal represents the strongest signal in the AS group. The mass of the detected compound can be matched to nine different chemical formulae. The elemental composition of each theoretical compound generates a specific isotopic profile because of the natural abundance of heavy isotopes of each element. The only compound matching the detected isotopic profiles of (B) the monomer at m/z 445.2957, (C) the dimer at m/z 889.5768 and (D) the trimer at m/z 1333.857 is a molecule with the monomeric formula C27H40O5.
Fig. 5.
Differential regulation of a compound at m/z 445. 2957 and its identification as the Vitamin D3 metabolite (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone. A, The compound with a precursor ion mass m/z of 445.2957 is down-regulated in AS patients (ANOVA p ≤ 5.44E-15). B, The MS/MS spectrum of the compound exhibits characteristic fragment ions for (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone. Fragment 1) represents the precursor after loss of water, 2) and 3) the secosteroid nucleus before and after water-loss, and 4) the conjugated ring system after water-loss.
MS/MS analysis of m/z 445.2957 permitted the identification of this compound as (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone (Fig. 5B); however, the exact configuration of the compound cannot be determined by the methods used here. The diagnostic fragment ions at m/z 271.2073 (M+-side chain, Δm = 6 ppm) and 253.1867 (271.2073 - H2O, Δm = 33 ppm), representing the secosteroid nucleus, allow the distinction between this vitamin D derivative and 1,25-Dihydrovitamin D3–26,23-lactone or its isomers, in which these fragment ions are not observed (see also (33)). Also, these fragment ions indicate the presence of the other four oxygen atoms in the side chain, consistent with the molecular structure of (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone with a precursor mass of 445.2957 Da. We also measured the concentration of Vitamin D binding protein (DBP) in the individual sera and found no significant correlation between (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone and DBP, although a pattern was more apparent in controls as compared with AS (healthy controls: p = 0.078, AS patients: p = 0.679, data not shown).
DISCUSSION
In this study we report MS profiling of proteins and small compounds from the sera of AS subjects following depletion of IgGs and Albumin (27). Three hundred and sixteen serum proteins were detected, with 201 identified using two or more unique peptides. We believe this list will be extremely powerful in future clinical studies, since all of the proteins identified should be quantifiable using ELISA-based methods.
We show that 22 proteins are up- or down-regulated in AS (listed in Table II, p < 0.05). Most have roles in innate immunity and/or the acute phase inflammatory response. Our data thus strongly support an innate inflammatory pathogenesis in AS, and identify key pathways for further study. These data are complementary to the recent genetic and gene expression data obtained in AS (5, 18). We applied LIMMA analysis to quantify 22 proteins that are up- or down-regulated in AS patients (p < 0.05). We observed an expected up-regulation of C-reactive protein (CRP) in AS patients, with the mass spectrometry-derived data closely and linearly correlated with ELISA-based measurements of CRP, confirming the validity of our quantitative MS approach. Besides CRP, a set of six other proteins were up-regulated in serum from AS subjects that, except for the iron transporter Serotransferrin, have been reported to increase in blood during an acute phase response. The concentration of serotransferrin typically decreases in the context of an inflammatory response. Interestingly however, Low et al. (34) have reported an up-regulation of serotransferrin in juvenile idiopathic arthritis. The activation, proliferation and maturation of most immune cells is iron dependent, so that high levels of serotransferrin could directly contribute to the pathogenesis of AS. Among the down-regulated proteins in AS we observed two components of the inter-α-trypsin inhibitor complex (ITIH1 and ITIH2). Both proteins have previously been associated with multiple disorders through their interaction with the cell surface antigen CD44 (35). ITIH1 and ITIH2 are part of the SHAP-HA multi-protein-complex with the mucopolysaccharide hyaluronan, a determinant of synovial fluid viscosity and modulator of TNF signaling (36). Interestingly SHAP-HA was found in high levels in both sera and synovial fluid from rheumatoid arthritis but not osteoarthritis patients (37). The down-regulation of components of the SHAP-HA complex here observed in Ankylosing Spondylitis may prove useful diagnostically and could additionally play a role in the pathogenesis of AS. Furthermore we detected a down-regulation of the negative acute phase proteins transthyretin and antithrombin-3. Kallistatin, a TNF-α antagonist that inhibits vascular inflammation (38), appears to be up-regulated in plasma and inflammatory joints of Rheumatoid Arthritis patients (39). Thus several of the “acute phase” proteins in Table II are regulated differently in AS compared with other chronic inflammatory conditions. We propose that the measurement of these proteins may enable the distinction of Ankylosing Spondylitis from other inflammatory rheumatic diseases.
We conducted a bootstrap ROC analysis on a generated logistic regression model in order to determine the combination of proteins with the strongest diagnostic value. This method enables ROC analysis to be performed even with the limited sample numbers used in our study. We found the greatest diagnostic utility in combining measurement of the up-regulated protein APCS (Serum amyloid P-component, also SAP) with the down-regulated inter-α-trypsin inhibitor subunit ITIH2. APCS has 51% sequence homology with CRP and has been identified as acute phase protein which is up-regulated during inflammation in mice (40). However Bijl et al. (41) showed that APCS does not exhibit the kinetics of a typical acute phase protein, with decreasing levels in the early stage of inflammation and elevated levels in sustained inflammation. Others have reported that APCS is slightly elevated in Rheumatoid Arthritis and AS patients but not systemic lupus erythematosus (42). We propose that the combined determination of APCS and ITIH2 levels in patients can contribute to the diagnosis of Ankylosing Spondylitis with high sensitivity and specificity. However, we recognize the limitations inherent in assessing classifier performance on small sample sizes regardless of method. Our results nevertheless suggest a promising classification model worthy of further investigation in different clinical groups.
As part of an integrative approach to profile changes in serum from AS subjects, we complemented the proteomic profiling with a metabolomic screen. The metabolomic workflow was designed to be fast, cost-effective and easily reproducible in a clinical and analytical laboratory. The analysis of the metabolomic samples was conducted with reversed phase chromatography and a quadrupole time-of-flight (Q-Tof) mass spectrometer in positive ionization mode. The limitation of this configuration is that only compounds that are water-soluble, bind to a hydrophobic stationary phase and ionize well in positive mode in electro-spray are detected. Our results show that even with these technical limitations we can profile a highly complex but well defined subset of small molecular features in serum without a need for prefractionation. We detected molecular features that could separate healthy individuals from AS subjects in a principal component analysis. A refinement of this analysis to the 10 molecular features with the largest absolute loadings of PC1 was still able to differentiate between the two groups, a trait that may be further explored in a clinical setting. Furthermore, we found a set of 65 molecular features that exhibited a clustering of subjects according to disease activity based on the BASDAI. Even though we did not identify these molecular features, our data suggests that they could be used to diagnose Ankylosing Spondylitis, even when patients suffer from less severe symptoms. Confident identifications for the differential molecular features are desirable, but the identification of hundreds of metabolites and small compounds is challenging.
Nevertheless we successfully identified (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone as a compound down-regulated in AS by accurate mass, isotopic pattern and characteristic MS/MS fragmentation. At least 37 chemically characterized vitamin D metabolites are known, and 25(OH)D3 26,23-peroxylactone appears to be produced from 25(OH)D3 in the kidney and secreted into the serum (43). 25(OH)D3 26,23-peroxylactone can convert nonenzymatically into 25(OH) D3 26,23-lactone, which appears to antagonize differentiation of leukemia cell types (44). The hormonally active form of Vitamin D3 (1,25-dihydroxyvitamin D3) is itself an intermediate in the production of the lactone similar to the peroxylactone derivative identified here (45). The biological and physiological functions of the 25(OH)D3–26,23-peroxylactone are currently not fully explored, but thought to be intermediates of lactone biosynthesis (46). Lower levels of 25(OH)D3–26,23-peroxylactone in sera of AS subjects may therefore indicate an altered Vitamin D3 metabolism. Under normal conditions more than 99% of Vitamin D3 and its metabolites are bound to DBP, saturating the protein only up to 2% (47). As expected we observed a linear trend in healthy individuals when we compared the ELISA readouts for DBP with the relative abundance of (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone. However, in AS patients DBP did not correlate with (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone (p < 0.078 in controls, p > 0.679 in AS patients, data not shown). Interestingly Vitamin D3 and its metabolites have a direct influence on bone remodelling. A lower abundance of (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone in AS patients could point to generally lower Vitamin D3 abundance, reduced production of one of its intermediates upstream of (23S,25R)-25-hydroxyvitamin D3 26,23-peroxylactone or differential affinity of DBP or the Vitamin D3 receptor to Vitamin D3 in AS patients due to AS-specific polymorphisms. Besides the immune-modulatory effects of Vitamin D3 in suppressing activated T-cells and proliferation (48), the compound has been associated with AS and osteoporosis in various studies (49, 50). Lower levels of Vitamin D3 may lead to an activation of osteoclasts, increased bone resorption and reduced trans-cellular calcium transport and absorption. Furthermore, it has been reported that high levels of pro-inflammatory cytokines such as TNF-α or Interleukin-1 can modulate bone turnover in chronic inflammation (51). All these effects converge on a decreased bone mineral density and unbalanced bone turnover in AS patients. Our results suggest that Vitamin D3 and its metabolites play a significant role in the pathogenesis of AS.
The relevance of the candidate molecules APCS and ITIH2, the vitamin D3 derivative and other distinct molecular features in discriminating between AS and healthy control subjects remains to be validated in larger independent and double-blinded patient cohorts. Discovery based studies as described here have their limitations as far as the number of sample processing and analysis are concerned. Even with the simple workflows used in this study, a moderate number of individual samples prepared in a different fashion for both metabolite and proteome analyses with various LC-MS workflows performed in triplicates required significant instrumental and human resources. We addressed this limitation in part by the application of bootstrap methods to generate and predict the generalized classification performance of our AS/control prediction models leading to increased confidence in the utility of these models given the small-sample size.
In summary, quantitative MS analysis of proteins and small compounds in the serum of AS and healthy control patients provides an integrative view that accelerates the discovery of biomarker candidates useful for the diagnosis and disease monitoring of AS, and additionally identifies biochemical pathways such as vitamin D metabolism to be of potential importance in AS pathogenesis.
Acknowledgments
The Mascot in-house server used for the analysis of mass spectrometry data is supported by the Computational Biology Research Group (CBRG, Oxford, UK). The data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash: bdQICBWSdaXmnFcK0ZoEvcKrousbWcmtr9NX0qz7NfBFh6dKwQKGtDerY9IqIYf1Cr0iXLvLqaSJO1wG8RkC7qlRVQ0AAAAAAAq5jg== The hash may be used to prove exactly what files were published as part of this manuscript's data set, and the hash may also be used to check that the data has not changed since publication.
Footnotes
* This study was supported by the Biomedical Research Centre (NIHR), Oxford, UK. D.C.T. was part supported by ERC FP7 grant 233240 to C.J. Schofield and P.J. Ratcliffe, University of Oxford, UK. This work was funded in part by a program grant from the National Health and Medical Research Council (Australia) reference 569938. G.T. was supported by a Lions Senior Medical Research Fellowship and M.A.B was funded by a National Health and Medical Research Council (Australia) Principal Research Fellowship. R.F. is supported by an Action Medical Research Grant to P.B. and to B.M.K. (University of Oxford, UK).
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S5.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S5.
1 The abbreviations used are:
- AS
- Ankylosing Spondylitis
- nLC-MS
- nano liquid-chromatography mass spectrometry
- ROC
- Receiver operating characteristic
- DBP
- vitamin D binding protein
- ERAP
- endoplasmatic reticulum aminopeptidase
- TNF
- tumor necrosis factor
- IgG
- Immunoglobulin G
- IgA
- Immunoglobulin A
- BASDAI
- Bath Ankylosing Spondylitis disease activity index
- PCA
- Principal component analysis
- LIMMA
- Linear models of microarray data
- CRP
- C-reactive protein
- ITIH2
- Inter-α-trypsin-inhibitor heavy chain H2
- ANOVA
- Analysis of variance
- ppm
- parts per million
- D3
- Vitamin D3.
REFERENCES
- 1. Elyan M., Khan M. A. (2006) Diagnosing ankylosing spondylitis. J. Rheumatol. 78, 12–23 [PubMed] [Google Scholar]
- 2. Rostom S., Dougados M., Gossec L. (2010) New tools for diagnosing spondyloarthropathy. Joint Bone Spine 77, 108–114 [DOI] [PubMed] [Google Scholar]
- 3. Thomas G. P., Brown M. A. (2010) Genomics of ankylosing spondylitis. Discov. Med. 10, 263–271 [PubMed] [Google Scholar]
- 4. Dougados M., Baeten D. (2011) Spondyloarthritis. Lancet 377, 2127–2137 [DOI] [PubMed] [Google Scholar]
- 5. Thomas G. P., Brown M. A. (2010) Genetics and genomics of ankylosing spondylitis. Immunol. Rev. 233, 162–180 [DOI] [PubMed] [Google Scholar]
- 6. Evans D. M., Spencer C. C., Pointon J. J., Su Z., Harvey D., Kochan G., Opperman U., Dilthey A., Pirinen M., Stone M. A., Appleton L., Moutsianis L., Leslie S., Wordsworth T., Kenna T. J., Karaderi T., Thomas G. P., Ward M. M., Weisman M. H., Farrar C., Bradbury L. A., Danoy P., Inman R. D., Maksymowych W., Gladman D., Rahman P., Morgan A., Marzo-Ortega H., Bowness P., Gaffney K., Gaston J. S., Smith M., Bruges-Armas J., Couto A. R., Sorrentino R., Paladini F., Ferreira M. A., Xu H., Liu Y., Jiang L., Lopez-Larrea C., Diaz-Pena R., Lopez-Vazquez A., Zayats T., Band G., Bellenguez C., Blackburn H., Blackwell J. M., Bramon E., Bumpstead S. J., Casas J. P., Corvin A., Craddock N., Deloukas P., Dronov S., Duncanson A., Edkins S., Freeman C., Gillman M., Gray E., Gwilliam R., Hammond N., Hunt S. E., Jankowski J., Jayakumar A., Langford C., Liddle J., Markus H. S., Mathew C. G., McCann O. T., McCarthy M. I., Palmer C. N., Peltonen L., Plomin R., Potter S. C., Rautanen A., Ravindrarajah R., Ricketts M., Samani N., Sawcer S. J., Strange A., Trembath R. C., Viswanathan A. C., Waller M., Weston P., Whittaker P., Widaa S., Wood N. W., McVean G., Reveille J. D., Wordsworth B. P., Brown M. A., Donnelly P. (2011) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harvey D., Pointon J. J., Evans D. M., Karaderi T., Farrar C., Appleton L. H., Sturrock R. D., Stone M. A., Oppermann U., Brown M. A., Wordsworth B. P. (2009) Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum .Mol. Genet. 18, 4204–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karaderi T., Harvey D., Farrar C., Appleton L. H., Stone M. A., Sturrock R. D., Brown M. A., Wordsworth P., Pointon J. J. (2009) Association between the interleukin 23 receptor and ankylosing spondylitis is confirmed by a new UK case-control study and meta-analysis of published series. Rheumatology 48, 386–389 [DOI] [PubMed] [Google Scholar]
- 9. Kochan G., Krojer T., Harvey D., Fischer R., Chen L., Vollmar M., von Delft F., Kavanagh K. L., Brown M. A., Bowness P., Wordsworth P., Kessler B. M., Oppermann U. (2011) Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. U.S.A. 108, 7745–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nguyen T. T., Chang S. C., Evnouchidou I., York I. A., Zikos C., Rock K. L., Goldberg A. L., Stratikos E., Stern L. J. (2011) Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat. Struct. Mol. Biol. 18, 604–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Heijde D., Sieper J., Maksymowych W. P., Dougados M., Burgos-Vargas R., Landewé R., Rudwaleit M., Braun J. (2011) 2010 Update of the international ASAS recommendations for the use of anti-TNF agents in patients with axial spondyloarthritis. Ann Rheum. Dis. 70, 905–908 [DOI] [PubMed] [Google Scholar]
- 12. Braun J., Sieper J. (2009) Classification criteria for rheumatoid arthritis and ankylosing spondylitis. Clin. Exp. Rheumatol. 27, S68–73 [PubMed] [Google Scholar]
- 13. Tilleman K., Van Steendam K., Cantaert T., De Keyser F., Elewaut D., Deforce D. (2008) Synovial detection and autoantibody reactivity of processed citrullinated isoforms of vimentin in inflammatory arthritides. Rheumatology 47, 597–604 [DOI] [PubMed] [Google Scholar]
- 14. Benhamou M., Gossec L., Dougados M. (2010) Clinical relevance of C-reactive protein in ankylosing spondylitis and evaluation of the NSAIDs/coxibs' treatment effect on C-reactive protein. Rheumatology 49, 536–541 [DOI] [PubMed] [Google Scholar]
- 15. Chandra P. E., Sokolove J., Hipp B. G., Lindstrom T. M., Elder J. T., Reveille J. D., Eberl H., Klause U., Robinson W. H. (2011) Novel multiplex technology for diagnostic characterization of rheumatoid arthritis. Arthritis Res. Ther. 13, R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. López-Longo F. J., Rodriguez-Mahou M., Sánchez-Ramón S., Estecha A., Balsera M., Plaza R., Fernández-Cruz E., Pèrez L. C. (2006) Anti-cyclic citrullinated peptide versus anti-Sa antibodies in diagnosis of rheumatoid arthritis in an outpatient clinic for connective tissue disease and spondyloarthritis. J. Rheumatol. 33, 1476–1481 [PubMed] [Google Scholar]
- 17. Wright C., Sibani S., Trudgian D., Fischer R., Kessler B., Labaer J., Bowness P. (2010) Detection of multiple autoantibodies in patients with ankylosing spondylitis using nucleic acid programmable protein arrays. Mol. Cell. Proteomics. 2, M9.003841-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duan R., Leo P., Bradbury L., Brown M. A., Thomas G. (2010) Gene expression profiling reveals a downregulation in immune-associated genes in patients with AS. Ann. Rheum. Dis. 69, 1724–1729 [DOI] [PubMed] [Google Scholar]
- 19. Arends S., van der Veer E., Groen H., Houtman P. M., Jansen T. L., Leijsma M. K., Bijzet J., Limburg P. C., Kallenberg C. G., Spoorenberg A., Brouwer E. (2011) Serum MMP-3 Level as a Biomarker for Monitoring and Predicting Response to Etanercept Treatment in Ankylosing Spondylitis. J. Rheumatol. 38, 1644–1650 [DOI] [PubMed] [Google Scholar]
- 20. Chang X., Han J., Zhao Y., Yan X., Sun S., Cui Y. (2010) Increased expression of carbonic anhydrase I in the synovium of patients with ankylosing spondylitis. BMC Musculoskelet. Disord. 11, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li T., Zheng B., Huang Z., Lu H., Lin Q., Liao Z., Lin Z., Zhao L., Wang X., Gu J. (2010) Over-expression of talin 1 and integrin-linked kinase in PBMCs of patients with ankylosing spondylitis: a proteomic study. Clin. Exp. Rheumatol. 28, 828–835 [PubMed] [Google Scholar]
- 22. Pedersen S. J., Sørensen I. J., Garnero P., Johansen J. S., Madsen O. R., Tvede N., Hansen M. S., Thamsborg G., Andersen L. S., Majgaard O., Loft A. G., Erlendsson J., Asmussen K., Jurik A. G., Moller J., Hasselquist M., Mikkelsen D., Skjodt T., Lambert R., Hansen A., Østergaard M. (2011) ASDAS, BASDAI and different treatment responses and their relation to biomarkers of inflammation, cartilage and bone turnover in patients with axial spondyloarthritis treated with TNF{alpha} inhibitors. Ann. Rheum. Dis. 70, 1375–1381 [DOI] [PubMed] [Google Scholar]
- 23. Pedersen S. J., Chiowchanwisawakit P., Lambert R. G., Østergaard M., Maksymowych W. P. (2011) Resolution of inflammation following treatment of ankylosing spondylitis is associated with new bone formation. J. Rheumatol. 38, 1349–1354 [DOI] [PubMed] [Google Scholar]
- 24. Tam L. S., Gu J., Yu D. (2010) Pathogenesis of ankylosing spondylitis. Nat. Rev. Rheumatol. 6, 399–405 [DOI] [PubMed] [Google Scholar]
- 25. Farach-Carson M. C. (2001) Bioactive analogs that simulate subsets of biological activities of 1alpha,25(OH)(2)D(3) in osteoblasts. Steroids 66, 357–361 [DOI] [PubMed] [Google Scholar]
- 26. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 27. Fu Q., Bovenkamp D. E., Van Eyk J. E. (2007) A rapid, economical, and reproducible method for human serum delipidation and albumin and IgG removal for proteomic analysis. Methods Mol. Biol. 357, 365–371 [DOI] [PubMed] [Google Scholar]
- 28. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y., Szustakowski J., Schinke M. (2009) Bioinformatics analysis of microarray data. Methods Mol. Biol. 573, 259–284 [DOI] [PubMed] [Google Scholar]
- 30. Tibshirani R. (1997) The lasso method for variable selection in the Cox model. Stat. Med. 16, 385–395 [DOI] [PubMed] [Google Scholar]
- 31. Smyth G. K., Yang Y. H., Speed T. (2003) Statistical issues in cDNA microarray data analysis. Methods Mol. Biol. 224, 111–136 [DOI] [PubMed] [Google Scholar]
- 32. Smith C. A., O'Maille G., Want E. J., Qin C., Trauger S. A., Brandon T. R., Custodio D. E., Abagyan R., Siuzdak G. (2005) METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751 [DOI] [PubMed] [Google Scholar]
- 33. Ishizuka S., Ishimoto S., Norman A. W. (1984) Isolation and identification of 1 alpha,25-dihydroxy-24-oxovitamin D3, 1 alpha,25-dihydroxyvitamin D3 26,23-lactone, and 1 alpha,24(S),25-trihydroxyvitamin D3: in vivo metabolites of 1 alpha,25-dihydroxyvitamin D3. Biochemistry 23, 1473–1478 [DOI] [PubMed] [Google Scholar]
- 34. Low J. M., Chauhan A. K., Gibson D. S., Zhu M., Chen S., Rooney M. E., Ombrello M. J., Moore T. L. (2009) Proteomic analysis of circulating immune complexes in juvenile idiopathic arthritis reveals disease-associated proteins. Proteomics Clin. Appl. 3, 829–840 [DOI] [PubMed] [Google Scholar]
- 35. Zhuo L., Kanamori A., Kannagi R., Itano N., Wu J., Hamaguchi M., Ishiguro N., Kimata K. (2006) SHAP potentiates the CD44-mediated leukocyte adhesion to the hyaluronan substratum. J. Biol. Chem. 281, 20303–20314 [DOI] [PubMed] [Google Scholar]
- 36. Termeer C., Benedix F., Sleeman J., Fieber C., Voith U., Ahrens T., Miyake K., Freudenberg M., Galanos C., Simon J. C. (2002) Oligosaccharides of Hyaluronan activate dendritic cells via toll-like receptor 4. J. Exp. Med. 195, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kida D., Yoneda M., Miyaura S., Ishimaru T., Yoshida Y., Ito T., Ishiguro N., Iwata H., Kimata K. (1999) The SHAP-HA complex in sera from patients with rheumatoid arthritis and osteoarthritis. J Rheumatol 26, 1230–1238 [PubMed] [Google Scholar]
- 38. Yin H., Gao L., Shen B., Chao L., Chao J. (2010) Kallistatin inhibits vascular inflammation by antagonizing tumor necrosis factor-alpha-induced nuclear factor kappaB activation. Hypertension 56, 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang C. R., Chen S. Y., Shiau A. L., Wu C. L., Jou I. M., Chao L., Chao J. (2007) Upregulation of kallistatin expression in rheumatoid joints. J. Rheumatol. 34, 2171–2176 [PubMed] [Google Scholar]
- 40. Pepys M. B., Baltz M., Gomer K., Davies A. J., Doenhoff M. (1979) Serum amyloid P-component is an acute-phase reactant in the mouse. Nature 278, 259–261 [DOI] [PubMed] [Google Scholar]
- 41. Bijl M., Bootsma H., Van Der Geld Y., Limburg P. C., Kallenberg C. G., Van Rijswijk M. H. (2004) Serum amyloid P component levels are not decreased in patients with systemic lupus erythematosus and do not rise during an acute phase reaction. Ann. Rheum. Dis. 63, 831–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strachan A. F., Johnson P. M. (1982) Protein SAP (serum amyloid P-component) in Waldenstrom's macroglobulinaemia, multiple myeloma and rheumatic diseases. J. Clin. Lab. Immunol. 8, 153–156 [PubMed] [Google Scholar]
- 43. Ishizuka S., Ishimoto S., Norman A. W. (1982) Isolation and identification of 25-hydroxyvitamin D3–26,23-peroxylactone. A novel in vivo metabolite of vitamin D3. J. Biol. Chem. 257, 14708–14713 [PubMed] [Google Scholar]
- 44. Miura D., Manabe K., Gao Q., Norman A. W., Ishizuka S. (1999) 1alpha,25-dihydroxyvitamin D(3)-26,23-lactone analogs antagonize differentiation of human leukemia cells (HL-60 cells) but not of human acute promyelocytic leukemia cells (NB4 cells). FEBS Lett. 460, 297–302 [DOI] [PubMed] [Google Scholar]
- 45. Tanaka Y., DeLuca H. F., Schnoes H. K., Ikekawa N., Eguchi T. (1981) 23,25-Dihydroxyvitamin D3: a natural precursor in the biosynthesis of 25-hydroxyvitamin D3–26,23-lactone. Proc. Natl. Acad. Sci. U.S.A. 78, 4805–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ikekawa N. (1987) Structures and biological activities of vitamin D metabolites and their analogs. Med. Res. Rev. 7, 333–366 [DOI] [PubMed] [Google Scholar]
- 47. Chun R. F., Lauridsen A. L., Suon L., Zella L. A., Pike J. W., Modlin R. L., Martineau A. R., Wilkinson R. J., Adams J., Hewison M. (2010) Vitamin D-binding protein directs monocyte responses to 25-hydroxy- and 1,25-dihydroxyvitamin D. J. Clin. Endocrinol. Metab. 95, 3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams J. S., Hewison M. (2008) Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat. Clin. Pract. Endocrinol. Metab. 4, 80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lange U., Teichmann J., Strunk J., Müller-Ladner U., Schmidt K. L. (2005) Association of 1.25 vitamin D3 deficiency, disease activity and low bone mass in ankylosing spondylitis. Osteoporos Int 16, 1999–2004 [DOI] [PubMed] [Google Scholar]
- 50. Lange U., Jung O., Teichmann J., Neeck G. (2001) Relationship between disease activity and serum levels of vitamin D metabolites and parathyroid hormone in ankylosing spondylitis. Osteoporos Int. 12, 1031–1035 [DOI] [PubMed] [Google Scholar]
- 51. Polzer K., Joosten L., Gasser J., Distler J. H., Ruiz G., Baum W., Redlich K., Bobacz K., Smolen J. S., van den Berg W., Schett G., Zwerina J. (2010) Interleukin-1 is essential for systemic inflammatory bone loss. Ann. Rheum. Dis. 69, 284–290 [DOI] [PubMed] [Google Scholar]
- 52. Gabay C., Kushner I. (1999) Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340, 448–454 [DOI] [PubMed] [Google Scholar]
- 53. Whicher J. T., Barnes M. P., Brown A., Cooper M. J., Read R., Walters G., Williamson R. C. (1982) Complement activation and complement control proteins in acute pancreatitis. Gut 23, 944–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Osborn T. M., Verdrengh M., Stossel T. P., Tarkowski A., Bokarewa M. (2008) Decreased levels of the gelsolin plasma isoform in patients with rheumatoid arthritis. Arthritis Res. Ther 10, R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ayub A., Mackness M. I., Arrol S., Mackness B., Patel J., Durrington P. N. (1999) Serum paraoxonase after myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 19, 330–335 [DOI] [PubMed] [Google Scholar]
- 56. Séralini G. E. (1991) A new role for corticosteroid binding globulin (CBG), member of SERPIN superfamily. C R Seances Soc Biol. Fil. 185, 500–509 [PubMed] [Google Scholar]
- 57. Niessen R. W., Lamping R. J., Jansen P. M., Prins M. H., Peters M., Taylor F. B., Jr., de Vijlder J. J., ten Cate J. W., Hack C. E., Sturk A. (1997) Antithrombin acts as a negative acute phase protein as established with studies on HepG2 cells and in baboons. Thromb. Haemos. 78, 1088–1092 [PubMed] [Google Scholar]
- 58. Gangneux C., Daveau M., Hiron M., Derambure C., Papaconstantinou J., Salier J. P. (2003) The inflammation-induced down-regulation of plasma Fetuin-A (alpha2HS-Glycoprotein) in liver results from the loss of interaction between long C/EBP isoforms at two neighbouring binding sites. Nucleic Acids Res. 31, 5957–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chai K. X., Chen V. C., Ni A., Lindpaintner K., Rubattu S., Chao L., Chao J. (1997) Molecular cloning and expression of rat kallistatin gene. Biochim. Biophys. Acta 1353, 277–286 [DOI] [PubMed] [Google Scholar]