Abstract
Protein glutathionylation is a redox post-translational modification occurring under oxidative stress conditions and playing a major role in cell regulation and signaling. This modification has been mainly studied in nonphotosynthetic organisms, whereas much less is known in photosynthetic organisms despite their important exposure to oxidative stress caused by changes in environmental conditions. We report a large scale proteomic analysis using biotinylated glutathione and streptavidin affinity chromatography that allowed identification of 225 glutathionylated proteins in the eukaryotic unicellular green alga Chlamydomonas reinhardtii. Moreover, 56 sites of glutathionylation were also identified after peptide affinity purification and tandem mass spectrometry. The targets identified belong to a wide range of biological processes and pathways, among which the Calvin-Benson cycle appears to be a major target. The glutathionylation of four enzymes of this cycle, phosphoribulokinase, glyceraldehyde-3-phosphate dehydrogenase, ribose-5-phosphate isomerase, and phosphoglycerate kinase was confirmed by Western blot and activity measurements. The results suggest that glutathionylation could constitute a major mechanism of regulation of the Calvin-Benson cycle under oxidative stress conditions.
Protein post-translational modifications (PTMs)1 play a pivotal role in cellular signaling (1). Recently, redox PTMs have emerged as important mechanisms of signaling and regulation in all organisms. Our increasing understanding of the molecular mechanism of cell signaling has revealed that reactive oxygen species (ROS) and reactive nitrogen species (RNS) act as signaling molecules to transfer extracellular or intracellular information and elicit specific responses. ROS and RNS have generally been considered to be toxic molecules that have to be continuously scavenged and efficiently detoxified. Plant cells exhibit a remarkable ability to cope with high rates of ROS/RNS production as a result of a complex scavenging system that includes either antioxidant molecules or enzymes (2). In photosynthetic organisms, ROS and RNS are continuously produced during normal aerobic metabolism but are also produced transiently in response to various types of endogenous or exogenous signals, such as biotic and abiotic stresses. This production activates specific signaling pathways, resulting in transcriptional, post-transcriptional and post-translational responses that will, in fine, allow adaptation to new environmental conditions. These past decades, redox modifications have emerged as central mechanisms in these processes, at the interface between ROS/RNS and the adaptative responses to environmental changes.
ROS/RNS signaling operates mainly through a set of PTMs of thiol residues on proteins (3). Indeed, cysteine residues can undergo different states of oxidation such as sulfenic, sulfinic, and sulfonic acids in addition to protein disulfide bridges (intra- or intermolecular), S-thiolation (mainly glutathionylation), or nitrosylation. Small disulfide oxidoreductases named thioredoxins (TRXs) and glutaredoxins play a prominent role in the control of most of these modifications that can affect the function of numerous proteins by modifying their activity, their subcellular localization, their stability, or their interactions with partner proteins.
Glutathione serves as one of the major cellular antioxidant redox buffers. It is a highly abundant tripeptide (γ-l-glutamyl-l-cysteinyl-l-glycine) present at millimolar concentrations in many subcellular compartments. Glutathione occurs mostly under the reduced form (GSH) because oxidized glutathione (GSSG) is continuously regenerated into GSH by glutathione reductase using NADPH as electron donor. Glutathione has numerous documented physiological functions. It is generally considered to constitute a redox buffer and also participates in the detoxification of ROS, heavy metals (through phytochelatins), and xenobiotics (through glutathione S-transferases) (2, 4). In addition to these functions, glutathione can form a mixed disulfide bridge between the thiol group of its cysteine and an accessible free thiol on a protein, a reaction termed protein S-glutathionylation. This PTM can protect specific cysteine residues from irreversible oxidation but can also modulate protein activities. The exact mechanism(s) leading to protein glutathionylation in vivo remain(s) unclear, whereas the reverse reaction, named deglutathionylation, is likely catalyzed by glutaredoxins, proteins belonging to the TRX family. Glutathionylation appears to play a major role in numerous fundamental cell processes and is implicated in a broad spectrum of human diseases including cancer, diabetes, and several neurodegenerative, cardiovascular, or pulmonary diseases (5, 6). Moreover, there is also a strong interplay between glutathionylation and other redox PTMs (7), especially nitrosylation as recently illustrated by the demonstration that the activity of endothelial nitric oxide synthase is regulated by glutathionylation (8).
To date, glutathionylation has been generally studied in nonphotosynthetic organisms where proteomic studies, mainly based on the use of [35S]cysteine labeling, have allowed identification of nearly 200 targets involved in diverse cell processes (5, 9–12). Very recently, a large scale analysis in Plasmodium falciparum identified 493 putative targets of glutathionylation (13). Although the number of studies on glutathionylation in plants remains limited (14), several plant enzymes have been shown to undergo glutathionylation including TRXs (15, 16), protein tyrosine phosphatase 1B (17), glyceraldehyde-3-phosphate dehydrogenase (18, 19), isocitrate lyase (20), galactono-γ-lactone dehydrogenase (21), glycine decarboxylase (22), peroxiredoxins (23, 24), and methionine sulfoxide reductase (25).
Only three proteomic studies aimed at analyzing the diversity of glutathionylated proteins in photosynthetic organisms have been reported. Two studies on Arabidopsis using biotinylated glutathione identified two (26) or 79 (27) glutathionylated proteins. In the unicellular eukaryotic green alga Chlamydomonas reinhardtii, a major photosynthetic model organism, [35S]cysteine labeling allowed 25 glutathionylated proteins to be identified (23).
Here we report a large scale proteomic analysis that allowed identification of 225 glutathionylated proteins in C. reinhardtii using biotinylated glutathione and streptavidin affinity chromatography. Moreover, the sites of glutathionylation (cysteine residues) were also identified after peptide affinity purification and tandem mass spectrometry. The identified targets belong to a wide range of biological processes and pathways, among which the Calvin-Benson cycle appears as a major target with 10 of 11 enzymes identified. The glutathionylation of four of these enzymes was confirmed by Western blot and activity measurements. Our results strongly suggest that glutathionylation could constitute a major mechanism of regulation of the Calvin-Benson cycle under oxidative stress conditions.
MATERIALS AND METHODS
Materials and Enzymes
Modified trypsin was obtained from Promega (Madison, WI). EZ-Link Sulfo-NHS-Biotin was from Perbio Science (Cramlington, UK). NAP-5 columns and 5,5′-dithiobis-2-nitrobenzoic acid were purchased from GE Healthcare and Pierce, respectively. Biotinylated glutathione ethyl ester (BioGEE) was obtained from Invitrogen. High performance liquid chromatography grade ethanol and acetic acid were purchased from VWR France, and all of the other reagents were from Sigma-Aldrich.
Chlamydomonas Cultures and Protein Extraction
The C. reinhardtii D66 cell wall-less strain was grown in Tris acetate phosphate (TAP) medium (28) at 25 °C under constant agitation and continuous light (100 μE·m−2·s−1). Two liters of Chlamydomonas culture at 5–7 × 106 cells/ml were harvested by centrifugation and resuspended in 2 ml of 30 mm Tris-HCl, pH 7.9, 1 mm EDTA, supplemented with 100 μm PMSF and a mixture of protease inhibitors (Complete, Mini, EDTA-free; Roche Applied Science). Total soluble proteins were extracted by two freeze/thaw cycles in liquid nitrogen, and protein concentrations were determined by the Bradford assay using bovine serum albumin as a standard (29).
Synthesis of BioGSSG
EZ-Link Sulfo-NHS-Biotin, a soluble biotinylation reagent, was used to couple biotin to the primary amino group of oxidized glutathione (GSSG) under mild alkaline conditions. The biotinylation reagent (50 μl, 64 mm) was added to GSSG (50 μl, 32 mm) in 100 mm potassium phosphate buffer, pH 8.0, and the mixture was left for 1 h at room temperature. After incubation, unreacted biotin was quenched by adding 35 μl of 0.6 m ammonium bicarbonate (NH4HCO3).
To assess the efficiency of the synthesis, reverse phase high performance liquid chromatography analysis using an Alliance system (Waters, UK) equipped with a diode array detector showed a major and a minor chromatographic peaks. Each peak was collected manually, and the identities of the products were determined by MALDI-TOF-MS (Voyager-DE STR; Perseptive Biosystems, Framingham, MA) and ESI-IT-MS/MS (Agilent Technologies, Santa Clara, CA). MS and MS/MS spectra (data not shown) revealed that the compound present in the major peak had a mass of 1065.4 Da corresponding to N,N′-biotinyl glutathione disulfide, named BioGSSG for brevity, formed by the addition of one biotin moiety on each amino group of γ-Glu residues and that the compound present in the minor peak had a mass of 839.3 Da corresponding to N-biotinyl glutathione disulfide (the addition of one biotin moiety on one of the two amino groups of γ-Glu residues).
Synthesis of BioGEE
BioGEE was either purchased directly from Invitrogen or synthesized from glutathione ethyl ester in the laboratory, as follows. EZ-Link Sulfo-NHS-Biotin, a soluble biotinylation reagent, was used to couple biotin to the primary amino group of glutathione ethyl ester. The biotinylation reagent (300 μl, 25 mm) was added to glutathione ethyl ester (300 μl, 25 mm) in 50 mm potassium phosphate buffer, pH 7.2, and the mixture was left to derivatize for 1 h at room temperature. After incubation, unreacted biotin was quenched by adding 82.5 μl of 0.6 m ammonium bicarbonate buffer. The concentration of BioGEE was determined spectrophotometrically with 5,5′-dithiobis-2-nitrobenzoic acid using a molar extinction coefficient at 412 nm of 14,150 m−1 cm−1 to calculate the free thiol content of BioGEE (30).
Detection of Glutathionylation in Vivo
A Chlamydomonas D66 cell culture was grown in 100 ml of TAP medium to 6–8 × 106 cells/ml. After centrifugation (4,000 × g, 3 min), the pellet was washed once with fresh TAP medium and resuspended to 8 × 106 cells/ml in TAP medium supplemented with BioGEE or BioGSSG (final concentration, 0.5–2 mm). After different incubation times (30 min to 2 h) to allow entry of biotinylated glutathione, 0.5 or 1 mm H2O2 or diamide was added for various times (from 5 to 60 min) with or without DMSO to force BioGSSG entry, either in the light or in the dark. After treatment, the medium was discarded by centrifugation, and cells were washed twice with TAP medium to remove the biotinylated glutathione not conjugated with proteins and resuspended in 30 mm Tris-HCl, pH 7.9. Total soluble proteins were then extracted by two freeze/thaw cycles in liquid nitrogen, and protein concentration was determined as described above. Protein extracts were loaded on gradient (4–12%) SDS-polyacrylamide gel and analyzed by Western blot using anti-biotin antibodies as described previously (20). As control, a protein extract was treated for 15 min with 20 mm reduced DTT before loading to assess the reversibility of the reaction.
Glutathionylation of Protein Extracts
Freshly prepared protein extracts (3.4 mg/ml) were incubated in 100 mm Tris-HCl, pH 7.9, in the presence of various concentrations of BioGSSG ranging from 0 to 5 mm. Control samples were incubated with 100 mm iodoacetamide (IAM) for 30 min in the dark prior to incubation in the presence of 2 mm BioGSSG. After 1 h of incubation, BioGSSG-treated samples were alkylated in the presence of 100 mm IAM or treated with 20 mm reduced DTT for 15 min to assess the reversibility of the reaction. All of the treatments were performed at room temperature.
SDS-PAGE and Western Blotting
BioGSSG-treated samples were loaded on SDS-polyacrylamide gel electrophoresis using the Bio-Rad Mini Protean system. Sixteen μl of samples in SDS sample buffer without any reducing agent were resolved by 10% SDS-PAGE. After electrophoresis, the proteins were transferred to nitrocellulose membranes using a Bio-Rad semidry blotter. Signals corresponding to glutathionylated proteins tagged with biotin were visualized by Western blot using anti-biotin antibodies, as described previously (20).
Purification of Glutathionylated Proteins
Freshly prepared protein extracts (12 mg/ml) were incubated with 2 mm BioGSSG for 1 h at room temperature in a final volume of 400 μl. A control experiment was performed without BioGSSG. After incubation, the excess of BioGSSG was removed by a single passage on G-25 gel filtration columns (HiTrap desalting, 5 ml; GE Healthcare). Desalted samples were then loaded on streptavidin-agarose columns (1 ml) pre-equilibrated with 30 mm Tris-HCl, pH 7.9, and 150 mm NaCl. Subsequently, the column was extensively washed with 10 ml of washing buffer (30 mm Tris-HCl, pH 7.9, 1 mm EDTA, 0.25% Triton X-100 plus 600 mm NaCl) and with 2 ml of washing buffer without NaCl. Finally, bound proteins were eluted with 20 mm DTT in 10 mm ammonium bicarbonate (NH4HCO3). The eluted proteins were concentrated by evaporation using a vacuum concentrator and separated on a reducing 10% SDS-PAGE gel and stained with Coomassie Brilliant Blue R-250. Visible protein bands were excised manually and then submitted to destaining and in-gel digestion with modified porcine trypsin using the automated system Digest Pro96 (Intavis AG, Bremen, Germany) as previously described (31). The peptides were dissolved in 18 μl of 3% acetonitrile (CH3CN) containing 0.1% v/v formic acid and analyzed by mass spectrometry.
Tandem Mass Spectrometry
NanoLC-CHIP-IT-MS/MS experiments were performed on an Agilent 1200 nanoflow LC system coupled to a 6330 Ion Trap equipped with the Chip Cube orthogonal ionization system (Agilent Technologies, Santa Clara, CA) as previously described (32). Briefly, peptide samples (4 μl) were loaded at a flow of 4 μl/min onto the enrichment column equilibrated in 3% CH3CN in 0.1% formic acid. Then the peptides were eluted toward the analytical column with a constant flow of 300 nl/min. The gradient profile consisted in two linear gradients (3–20% CH3CN in 0.1% formic acid over 3 min and 20–50% CH3CN in 0.1% formic acid over 12 min) followed by an isocratic step at 70% CH3CN in 0.1% formic acid for 5 min and an equilibration step with 3% CH3CN in 0.1% formic acid for 7 min. Five scans were averaged to obtain a full scan spectrum from which the four most abundant ions (singly charged ions excluded) were selected for fragmentation by collision-induced dissociation. For each fragmentation spectrum, four scans were averaged. All of the spectra were saved in profile mode, and other ion trap parameters were set as follows: capillary, −2000 V; skimmer, 30 V; Cap exit, 100 V; fragmentation amplitude, 1.30 V; smart parameter settings, on; drying gas, nitrogen at 300 °C with a flow rate of 4 liters/min. Helium was used as cooling and fragmentation gas at a pressure of 6 × 10−7 mbar.
Purification of Glutathionylated Peptides
After glutathionylation and G-25 gel filtration steps, the proteins were concentrated up to 8–10 mg/ml using Microcon devices YM-10 (Millipore). 65 μg of proteins were denatured with the Protein extraction Reagent-4 kit (Sigma) 10 min at room temperature and then alkylated with 15 mm IAM for 90 min in the dark. IAM concentration was lowered by three times dilution in 65 mm ammonium bicarbonate, and protein extract was loaded on immobilized trypsin column (Trypsin spin column from Sigma, prepared following the manufacturer's recommendations) for 30 min at room temperature. The peptides were eluted with 100 mm ammonium bicarbonate. Eluted peptides were loaded onto a 1-ml streptavidin-agarose column and eluted with DTT following the procedure described above. Eluted peptides were then concentrated up to 50 μl by evaporation using a vacuum concentrator and analyzed by tandem mass spectrometry as described above by injecting increasing amounts of peptides. Each analysis was performed in duplicate.
Data Analysis and Database Search
Raw MS/MS data were processed by Data Analysis software version 3.4 (Agilent Technologies, Santa Clara, CA). For protein and peptide identifications, files corresponding to control or glutathionylation conditions were concatenated into a single file for further use with Mascot MS/MS Ions Search and Phenyx.
For identifications with Mascot, the NCBInr database (release 20091202, 10,107,245 entries) was reduced to the Viridiplantae taxonomy. Mass accuracy tolerance was set to 100 ppm on the parent ion mass and 0.6 Da in MS/MS mode. One missed cleavage per peptide was allowed. No molecular weight or isoelectric point restriction were selected. Only high quality mass spectra (individual ion score above identity threshold i.e. 40 for a p value < 0.05) were taken into account to identify glutathionylated sites or glutathionylated proteins.
For identifications with Phenyx, a sequence database restricted to C. reinhardtii (15,006 entries) was uploaded in Fasta format from the Uniprot release 15.15 (March 2, 2010) database. The parameters selected for scoring model and parent charge were ESI ion trap and (two or three, trust = yes), respectively. Parent tolerance was set to 100 ppm, and methionine oxidation was considered as variable modification. Acceptance thresholds were set as follow: peptide length ≥ 6; peptide z-score ≥ 6; peptide p value ≤ 10−4 with a global AC score ≥ 6 (for identification of glutathionylated sites) or 12 (for identification of glutathionylated proteins). Because two sets of calculations have been carried out, the first round was more stringent (enzyme: trypsin KR_noP; missed cleavage ≤ 1; cleavage mode: normal) than the second (enzyme: trypsin KR; missed cleavage ≤ 2; cleavage mode: semi-tryptic).
For protein identifications with the two search engines, methionine oxidation and cysteine carboxyamidomethylation were taken into account as variable modifications. For identification of glutathionylated peptides, only methionine oxidation was selected as variable modification. Glutathionylated proteins were validated if they were identified by both programs with at least two different peptides meeting acceptance criteria, whereas glutathionylated cysteine-containing peptides proposed by both Mascot and Phenyx software were only considered as identified after manual validation of the corresponding MS/MS spectra.
Consensus Sequence Analysis
For each putative site of glutathionylation (SSG-modified sites) 10 residues on each side of the glutathionylated cysteine were selected. Frequencies of flanking residues were computed with motif-x software (33). For predictions of secondary structure and solvent accessibility, the NetSurfP server was employed (34).
Plasmid Construction for Expression of RPI and PGK in Escherichia coli
The cDNA clones encoding RPI (AV390085) and PGK (BP097682) from C. reinhardtii were obtained from the Kazusa DNA Research Institute (Chiba, Japan). The coding sequences were amplified by PCR, and specific restriction sites (underlined) were introduced at the 5′- and 3′-ends of the coding sequences using the following specific primers: 5′NcoI-RPI, CGCACCATGGCCGCGCCGGTCTCAA; 3′BamHI-RPI, GCACGGGATCCTTAGTGCTTCTTGGGGTTGGG; 5′NdeI-PGK, CATCGTCCATATGGCGGTGAAGAAGTCGGTTGG; and 3′BamHI-PGK, ATTGCGGGATCCTTACTTCTCGTCCAGGGCGG. After purification, the PCR products were digested by NcoI/NdeI and BamHI and cloned into the NcoI/NdeI and BamHI restriction sites of modified pET-3c-His (NdeI) or pET-3d-His (NcoI) vectors (Novagen) allowing expression of the proteins with a polyhistidine tag at the N terminus. All of these expression vectors were verified by DNA sequencing.
Protein Purification
Glyceraldehyde-3-phosphate dehydrogenase (A4-GAPDH) was purified from Chlamydomonas as previously described (35). Recombinant proteins (RPI and PGK) were produced using the pET vectors/BL21 expression system. Bacteria were grown in LB medium supplemented with 100 μg/ml ampicillin at 37 °C until the absorbance at 600 nm reached 0.6. The expression of RPI or PGK was then induced by transfer to 27 °C for 18 h with (RPI) or without (PGK) the addition of 200 μm isopropyl-β-d-thiogalactopyranoside. The cells were harvested by centrifugation, resuspended in 30 mm Tris-HCl, pH 7.9, 10 μg/ml PMSF, and broken using a French press (7 × 107 Pa), and cell debris was removed by centrifugation. The supernatants containing the soluble His-tagged proteins were loaded onto a Ni2+ HiTrap chelating resin (His-select® nickel affinity gel; Sigma) pre-equilibrated with 30 mm Tris-HCl, pH 7.9. The recombinant proteins were then purified using imidazole step gradients according to the manufacturer's instructions. The molecular mass and purity of the recombinant proteins were checked by SDS-PAGE after dialysis against 30 mm Tris-HCl, pH 7.9, 1 mm EDTA. The protein concentrations were determined spectrophotometrically using calculated molar extinction coefficient at 280 nm (14,690 m−1·cm−1 and 20,065 m−1·cm−1 for RPI and PGK, respectively). The resulting homogeneous proteins were stored at −20 °C.
Enzyme Assays
For phosphoribulokinase activity assays, total protein extracts from Chlamydomonas (0.16 mg/ml) were treated with 10 mm DTT, and the excess of reducing agent was removed by desalting in 100 mm Tris-HCl, pH 7.9, using NAP-5 columns. The eluates were then incubated in the presence of 5 mm GSSG, and PRK activity was monitored as described in Ref. 36. For the analysis of Chlamydomonas A4-GAPDH, incubations were performed as described previously (19). Briefly, the enzyme (2.5 μm) was incubated in 50 mm Tris-HCl, pH 7.9, and 0.2 mm NADP+ at room temperature in the presence of H2O2 and GSH at the indicated concentration. When indicated, BPGA (5 units/ml of yeast 3-phosphoglycerate kinase, 2 mm ATP, and 3 mm 3-phosphoglycerate) was incubated with the enzyme for 5 min prior to inactivation treatments. Reactivation was performed by adding 20 mm DTT to the treated samples. At the indicated times, the aliquots were withdrawn to assay enzyme activity monitored as described previously (19).
In Vitro Glutathionylation of RPI, PGK, and A4-GAPDH Using BioGSSG
Before BioGSSG treatments, all of the proteins were reduced with 10 mm DTT and desalted in 100 mm Tris-HCl, pH 7.9, using NAP-5 columns. The prereduced proteins were incubated in 100 mm Tris-HCl, pH 7.9, in the presence of 2 mm BioGSSG. Control samples were incubated with 100 mm IAM for 30 min in the dark prior to incubation in the presence of 2 mm BioGSSG. After 1 h of incubation, BioGSSG-treated samples were alkylated in the presence of 100 mm IAM or treated with 20 mm DTT for 30 min to assess the reversibility of the reaction. All of the treatments were performed at room temperature. Proteins were then loaded on nonreducing SDS-PAGE and analyzed by Western blotting using anti-biotin antibodies as described previously (20).
RESULTS
Detection of S-Glutathionylated Proteins in Chlamydomonas
Initially, we focused on the identification of proteins undergoing glutathionylation in vivo. For this purpose, protein glutathionylation was analyzed in Chlamydomonas cell cultures treated with different biotinylated forms of glutathione: BioGSSG or BioGEE, and under various culture conditions as described under “Materials and Methods.” Although some glutathionylated proteins have been occasionally detected during these experiments, the efficiency of the labeling was unsatisfactory, and this method was not further considered for identifying glutathionylated proteins.
Therefore the detection of S-glutathionylated proteins was performed with Chlamydomonas cell extracts treated with BioGSSG. This molecule promotes protein glutathionylation by itself via thiol-disulfide exchange reactions and allows visualization of glutathionylated proteins by immunoblotting using anti-biotin antibodies (Fig. 1, procedure 2). As shown in Fig. 2, numerous proteins were labeled after treatment with BioGSSG, and the signals increased following a dose-dependent profile relative to increasing concentrations of BioGSSG (0.1–5 mm) (Fig. 2). In the control sample (absence of BioGSSG) two prominent bands of ∼35 and ∼70 kDa were observed that likely represent endogenously biotinylated proteins.
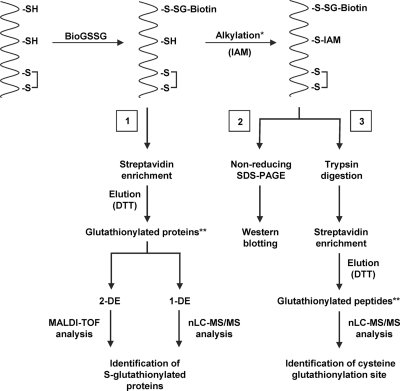
Fig. 1.
Schematic diagram of the methods employed to study protein glutathionylation in the present study. Procedure 1, work flow for identification of putative glutathionylated targets. Procedure 2, Western blot using anti-biotin antibodies. Procedure 3, work flow for identification of S-glutathionylation sites (cysteines) on proteins. *, alkylation during procedure 3 was performed under denaturating conditions. **, proteins and peptides initially glutathionylated with BioGSSG.
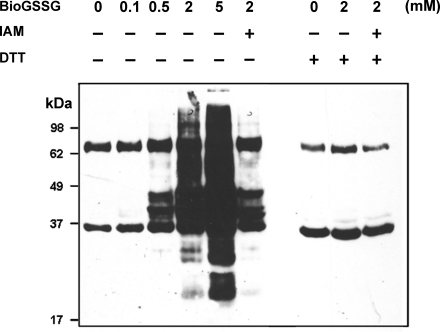
Fig. 2.
Western blot probed with anti-biotin antibodies to detect S-glutathionylated proteins in protein extract from Chlamydomonas following BioGSSG treatment. The figure shows a concentration-dependent increase in S-glutathionylation following a 1-h incubation in the presence of BioGSSG (0–5 mm). Two endogenous biotin-reactive bands at ∼35 and 70 kDa remain constant throughout and fortuitously serve as loading controls. A control sample was incubated with 100 mm IAM for 30 min in the dark followed by a 1-h incubation in the presence of 2 mm BioGSSG. The reversibility of the reaction was assessed by treating control, BioGSSG-treated, and IAM/BioGSSG-treated samples with 20 mm DTT for 30 min as indicated. The proteins were resolved by nonreducing SDS-PAGE and transferred to nitrocellulose for Western blotting with anti-biotin antibodies as described under “Materials and Methods.”
The extent of protein S-glutathionylation after treatment with 2 mm BioGSSG was strongly diminished by a pretreatment with the thiolate alkylating agent IAM (Fig. 2). This is consistent with the fact that IAM pretreatment blocks free accessible cysteines and confirms that the BioGSSG labeling is dependent on the presence of reactive cysteines on proteins.
Signals generated by BioGSSG at 2 mm were abolished by treatment with the reducing agent DTT, indicating that these signals indeed correspond to glutathionylated proteins (Fig. 2). In fact, the reducing treatment also removed the signals in the BioGSSG-treated sample preincubated with IAM. This indicates that the signals remaining even after IAM treatment likely correspond to unblocked cysteines rather than to aspecific reaction of BioGSSG with noncysteine amino acids. On the contrary, the two bands present in all samples, including in the control sample not treated with BioGSSG, did not disappear after DTT treatment. Therefore, these bands likely correspond to proteins carrying a biotin molecule as biotin decarboxylases, where biotin is covalently bound to the ε-amino group of lysine (37, 38).
Overall, these results demonstrate the efficiency and specificity of BioGSSG for detection of S-glutathionylated proteins in Chlamydomonas extracts. Because numerous bands could be detected, this prompted us to purify and identify the corresponding proteins.
Identification of Glutathionylated Proteins in Chlamydomonas Cell Extracts
Chlamydomonas cell extracts were treated for 1 h with BioGSSG, and taking advantage of the biotin moieties, S-glutathionylated proteins were purified using affinity chromatography on a streptavidin-agarose matrix (Fig. 1, procedure 1). Proteins bound to the matrix by association with the biotinylated glutathione tag were selectively released using DTT to cleave the mixed disulfides and then analyzed by two-dimensional gel electrophoresis (Fig. 3A).
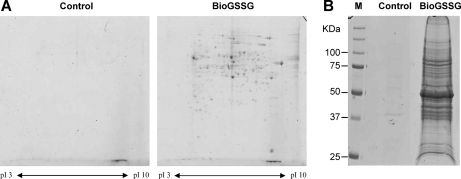
Fig. 3.
Coomassie-stained two- and one-dimensional gels of S-glutathionylated proteins purified by streptavidin-agarose affinity chromatography. A and B, Coomassie-stained S-glutathionylated proteins from Chlamydomonas protein extracts after two-dimensional (A) or one-dimensional electrophoresis (B). Proteins labeled with BioGSSG were purified as described under “Materials and Methods.” Control gel (A) and control lane (B) correspond to the protein extract submitted to the procedure of streptavidin-agarose affinity chromatography but omitting the initial BioGSSG treatment. For two-dimensional gels, the proteins were separated based on pI (3–10 linear gradient, left to right), and molecular mass (12% SDS-PAGE, top to bottom), whereas for one-dimensional gels, the proteins were separated based on molecular mass (12% SDS-PAGE).
In the eluate derived from BioGSSG treatment, ∼52 spots could be detected, after Coomassie Blue staining, in the pH range between 3 and 10. The visible spots were excised from the gel and digested with trypsin. The resulting peptides were subjected to MALDI-TOF mass spectrometry, allowing identification of 34 proteins by peptide mass fingerprinting (supplemental Table S1, proteins marked in the 2D column). Very few spots were visible in the control sample corresponding to the DTT eluate from a streptavidin column loaded with a protein extract not treated with BioGSSG. This indicates that the procedure allows identification of glutathionylated proteins with high specificity.
To increase the diversity of identified glutathionylated proteins, BioGSSG-treated extracts were affinity-purified as above, and the eluate was separated by one-dimensional gel electrophoresis and subjected to nanoLC-MS/MS (Fig. 3B). As observed previously, very few proteins were detected in the control sample, whereas the eluate derived from BioGSSG treatment contained numerous proteins. All of the protein bands were analyzed by nanoLC-MS/MS, allowing identification of 208 proteins (supplemental Table S1). Each assignment was based on data submitted to two different search engines for two distinct peptides for which a high degree of certainty was achieved (p < 0.05). A similar analysis performed on the control sample identified few peptides belonging to only four very abundant Chlamydomonas proteins (Rubisco, elongation factor 1α, tubulin α, and enolase). These four proteins were also identified in the treated sample but with numerous peptides and high sequence coverage. This indicates that they are likely retained on the column after BioGSSG treatment because they are glutathionylated and that their presence as traces in the control sample likely reflects their abundance. Among all of the proteins identified in the BioGSSG-treated sample, only two lack cysteines, but the sequences of these predicted proteins are probably partial because they are both translated from genome sequences containing several gaps. The full-length sequence of these proteins may therefore contain one or several cysteine residues.
Among the 208 proteins identified by nanoLC-MS/MS, 31 of 34 proteins identified by two-dimensional gel electrophoresis and MALDI-TOF MS could be recovered. Overall, the two approaches employed (MALDI-TOF and nanoLC-MS/MS) allowed identification of 211 potential targets of protein glutathionylation in C. reinhardtii.
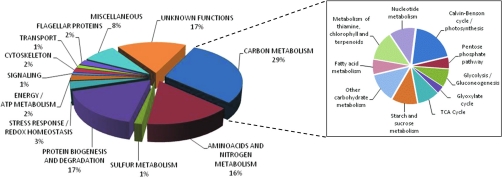
Most of the proteins identified were never found previously as potential candidates for glutathionylation in plant cells, including a number of miscellaneous and hypothetical proteins. The 211 identified proteins are involved in numerous cell processes and metabolic pathways (Fig. 4). Many proteins participate in amino acid and nitrogen metabolism (17%) and protein biogenesis and degradation (17%), but the most prominent category is carbon metabolism (29%), which accounts for nearly one-third of the proteins identified. This category can be further divided in subcategories, thereby revealing that the process containing the highest number of putative targets of glutathionylation is photosynthesis and especially CO2 assimilation (Fig. 4). Indeed, 10 of 11 enzymes of the Calvin-Benson cycle were identified (supplemental Table S1). Overall, these results suggest that, as in mammals, numerous proteins can undergo glutathionylation the green alga C. reinhardtii. This strengthens the potential importance of glutathionylation in plants and its role in the redox control of numerous cell processes and metabolic pathways.
Fig. 4.
Functional categories of S-glutathionylated proteins identified using BioGSSG in Chlamydomonas cell extract. The pie chart represents the functional categories of S-glutathionylated proteins identified in C. reinhardtii cell extracts after BioGSSG treatment. The prominent category, carbon metabolism, is further detailed in the right panel.
Identification of S-Glutathionylation Sites
Concomitantly to the identification of proteins undergoing S-glutathionylation, the procedure described above was adapted to enable unbiased specification of SSG-cysteine residues on proteins. Because the BioGSSG-based method allowed capture and identification of S-glutathionylated proteins, we reasoned that it could similarly be used to capture SSG-cysteine containing peptides in tryptic digests of S-glutathionylated proteins. Then for peptides with a suitable mass to charge ratio, it should be possible to use tandem mass spectrometry to elucidate the peptide sequence and identify both the protein of origin and the site of S-glutathionylation. To this end, the scheme depicted in Fig. 1 (procedure 3) was applied to isolate and analyze S-glutathionylated peptides by nanoLC-MS/MS.
Chlamydomonas extracts were treated for 1 h with BioGSSG followed by alkylation with IAM under denaturating conditions to block completely all remaining free cysteine thiols as done previously for affinity purification of S-glutathionylated proteins. At this stage, instead of capturing intact S-glutathionylated proteins on streptavidin-agarose, the proteins were subjected to complete trypsinolysis. Glutathionylated peptides were affinity-purified on streptavidin agarose, followed by selective cleavage of the disulfide bond linking the peptide cysteine and its biotinylated glutathione tag (using 20 mm DTT). This purification procedure based on selective peptide capture allows unequivocal identification of the target cysteine, not only when one cysteine is present in the eluted peptide (unmodified cysteine in the eluate) but also when the peptide contains several cysteines. Indeed, because of the IAM alkylation before trypsinolysis, when a peptide containing several cysteine residues is eluted from the column, the cysteine originally glutathionylated is unmodified, whereas the other cysteines are carbamidomethylated. Moreover, because the elution of peptides is performed under gentle conditions and follows extensive washing steps, one would not predict significant contamination by peptides that bind streptavidin-agarose nonspecifically as observed in the elution profile of control samples not treated with BioGSSG (Fig. 3).
The eluted peptides were analyzed by nanoLC-MS/MS, and a total of 71 different peptides were identified. A total of 12 peptides were excluded: 2 peptides identified in the control, 4 false positive peptides without cysteine, and 5 that could not be validated because of poor fragmentation spectra. The remaining 59 peptides correspond to 41 different proteins (Table I). Many peptides belong to proteins already identified above under native conditions, but several sites were also identified in new targets. This method allowed identification of 56 putative glutathionylated cysteines: 39 sites in 27 proteins among the 211 putative targets previously identified and 17 sites in 14 new putative target proteins (Table I). Interestingly, many of these new potential targets of glutathionylation correspond to proteins belonging to high molecular weight complexes, such as ribosomal proteins. This suggests that some of these glutathionylation sites might be shielded under native conditions and therefore not retained on streptavidin column. By contrast, these sites could have been exposed after trypsinolysis, allowing the purification and identification of the corresponding peptides. If we include these new proteins, a total of 225 potential glutathionylated proteins were identified using BioGSSG. Analysis of the primary amino acid sequence surrounding the 56 putative sites of glutathionylation did not reveal any consensus sequence motifs (data not shown). Further analysis of the secondary structure immediately surrounding modified cysteines indicated that SSG sites have a random distribution within α-helical, β-strand, or coil structures (data not shown). The structural determinants underlying the receptive nature of these glutathionylation sites are therefore most probably dependent on the microenvironment of the cysteine residue within the three-dimensional structure of the protein.
Table I. Glutathionylated peptides identified in C. reinhardtii.
RefSeq, accession number in NCBI database. The glutathionylated cysteine-containing peptide column shows the primary sequences of glutathionylated peptides; the positions of modified cysteines in the full-length sequences of the corresponding proteins are indicated as exponents. Column # shows protein identified as glutathionylated by BioGSSG (this work). Column S shows proteins identified as in vivo targets of S-thiolation in C. reinhardtii by [35S]cysteine labeling (23). Column T shows proteins identified as thioredoxin targets in C. reinhardtii (55).
| Protein name | RefSeq | # | S | T | Glutathionylated cysteine-containing peptide |
|---|---|---|---|---|---|
| Carbon metabolism (17 proteins) | |||||
| Calvin-Benson cycle/photosynthesis | |||||
| Apoferredoxin 1 | XP_001692808 | × | K.TPSGDKTIEC48PADTYILDAAEEAGLDLPYSC69R.A | ||
| Fructose-1,6-bisphosphatase | XP_001690872 | × | K.GTIDGELATVISSVSLAC109K.Q | ||
| Plastocyanin | XP_001702952 | A.GEYGYYC130EPHQGAGMVGK.I | |||
| K.LTAAGEYGYYC130EPHQGAGMVGK.I | |||||
| Phosphoglycerate kinase | XP_001699523 | × | × | K.GC411ITIIGGGDSVAAVEQAGVAEK.M | |
| K.VDDC158IGPEVEK.A | |||||
| Phosphoribulokinase | XP_001694038 | × | × | K.TVVIGLAADSGC47GK.S | |
| K.MFDPVYLFDEGSTISWIPC274GR.K | |||||
| Ribulose bisphosphate carboxylase large chain | NP_958405 | × | × | N.ATAGTC247EEMMKR.A | |
| R.GLLGC172TIKPK.L | |||||
| R.VALEAC427TQAR.N | |||||
| Transketolase | XP_001701881 | × | K.SGHPGMPMGC84APMGYVLWNEVMK.Y | ||
| Gluconeogenesis, glycolysis | |||||
| Enolase | XP_001702971 | × | × | K.KVEEVLNLC23VK.E | |
| K.LTTENIC357QVVGDDILVTNPVR.V | |||||
| Fructose-1,6-bisphosphate aldolase | XP_001700659 | × | × | R.GILAMDESNATC58GK.R | |
| R.GILAMDESNATC58GKR.L | |||||
| K.VMFEGILLKPAMVTPGADC256K.N | |||||
| K.VMFEGILLKPAMVTPGADC256KNK.A | |||||
| Phosphoenolpyruvate carboxykinase, splice variant | XP_001694964 | × | K.NVVLLAC405DAFGALPPVSR.L | ||
| Glyoxylate cycle | |||||
| Isocitrate lyase | XP_001695331 | × | × | × | R.TAEGFYC247VR.G |
| Malate synthase | XP_001695632 | × | K.C79APPAPGLVDR.R | ||
| TCA cycle | |||||
| Aconitate hydratase, mitochondrial | XP_001689702 | × | × | K.AGLIGSC400TNSSYEDMAR.A | |
| Starch and sucrose metabolism | |||||
| ADP-glucose pyrophosphorylase large subunit | XP_001693447 | × | R.LIDVPMSNC112INSGISK.I | ||
| Phosphorylase, plastidial | XP_001700091 | R.LAAC171FLDSMATLDLPGWGYGIR.Y | |||
| Metabolism of thiamine, chlorophyll, and terpenoids | |||||
| Full-length thiazole biosynthetic enzyme | XP_001698672 | × | × | K.DMDEFAESDVVIVGAGSAGLAC106AFELGR.I | |
| Magnesium-chelatase subunit chlI, chloroplastic | XP_001691232 | × | × | K.KIPMVDLPLGATEDRVC184GTIDIEK.A | |
| Amino acids and nitrogen metabolism (3 proteins) | |||||
| Acetohydroxy acid isomeroreductase | XP_001702649 | × | × | K.GHPFSEIC439NESIIEAVDSLNPYMHAR.G | |
| Adenosylhomocysteinase | XP_001693339 | × | K.DIAEADFGRLEIDLAEAEMPGLMAC41R.S | ||
| R.LEIDLAEAEMPGLMAC41R.S | |||||
| K.SKFDNVYGC243R.H | |||||
| Isopropylmalate dehydratase, large subunit | XP_001702135 | × | R.MNEPEVC444VSTTNR.N | ||
| Protein biogenesis and degradation (11 proteins) | |||||
| Protein biosynthesis, translation | |||||
| Eukaryotic translation elongation factor 1 α1 | XP_001696568 | × | × | K.EHLSIVIC13GHVDSGK.S | |
| K.NMISGAAQADVC112LLMVPADGNFTTAIQK.G | |||||
| R.VEQGVVKPGDEVIFLPTHTTANPC303TGK.V | |||||
| Elongation factor 2 | XP_001703215 | × | × | R.LC728EPVYLVEIQAPEQALGGIYSTLNTK.R | |
| Elongation factor Tu | XP_001696344 | × | × | × | R.HYAHVDC82PGHADYVK.N |
| H.VDC82PGHADYVK.N | |||||
| Ribosomal protein L4 | XP_001694804 | R.AGQGAFGNMC96R.G | |||
| Ribosomal protein L9 | XP_001700532 | R.SAALINQSC165HVR.V | |||
| Ribosomal protein L21 | XP_001698024 | R.C101REEFLTR.R | |||
| Ribosomal protein S5 | XP_001695400 | K.TIAEC161LADELVNAAK.G | |||
| Ribosomal protein S6 | XP_001695102 | R.GC102IVSPDLAVLNLVIVK.K | |||
| 40 S ribosomal protein S12 | XP_001698669 | R.KVVGC115SC117AVITDYGEETAGLSMLQEYLK.S | |||
| Folding/molecular chaperones | |||||
| Chaperonin 60B2 | XP_001692504 | × | R.C537SLENAVSVAK.T | ||
| R.MLAEYENC249R.I | |||||
| Degradation | |||||
| E3 ubiquitin ligase | XP_001690964 | K.GLLDLTC115QTVAQMIK.G | |||
| Energy/ATP metabolism (1 protein) | |||||
| Vacuolar ATP synthase subunit E | XP_001692936 | × | × | R.INC200SNTLDDR.L | |
| Cytoskeleton (3 proteins) | |||||
| Actin | XP_001699068 | × | × | K.EKLC219YVALDFEQEMATALSSSALEK.T | |
| Tubulin α1/α2 chain | XP_001691876 | × | R.AVC376MISNSTAIGEIFSR.L | ||
| XP_001703110 | R.C65IFLDLEPTVVDEVR.T | ||||
| Tubulin β1/β2 chain | XP_001693997 | × | R.EIVHIQGGQC12GNQIGAK.F | ||
| XP_001694072 | K.NMMC301AADPR.H | ||||
| Miscellaneous (2 proteins) | |||||
| Inosine-5′-monophosphate dehydrogenase | XP_001699045 | R.GFTSVC138VTDTGALGGK.L | |||
| Low CO2 inducible protein LCIB | XP_001698344 | × | K.C320YTVVNGLK.T | ||
| Unknown functions (4 proteins) | |||||
| Predicted protein | XP_001696868 | K.AAELEAVGSAVC109AK.K | |||
| K.SC248LGIVDNTGK.L | |||||
| Predicted protein | XP_001694293 | R.VEVQDTSGGC30GAMYR.I | |||
| Predicted protein | XP_001698018 | × | R.NPDKC38AALAAEGAK.V | ||
| Predicted protein | XP_001691572 | K.AGGVVVEAAC154VIELPFLK.G |
Calvin-Benson Cycle Enzymes as Major Targets of S-Glutathionylation
The Calvin-Benson cycle appears to be a major target of glutathionylation in Chlamydomonas with 10 of 11 enzymes of this pathway identified using BioGSSG (supplemental Table S1). Only triose phosphate isomerase has not been found but was identified as a target of glutathionylation in an earlier study in Arabidopsis (26). These results are consistent with the previous identification in Chlamydomonas, using an in vivo proteomic approach based on [35S]cysteine radiolabeling (23), of two Calvin-Benson cycle enzymes: RPI and PGK. They are also consistent with the fact that all Calvin-Benson cycle enzymes were previously suggested to be redox-regulated (39).
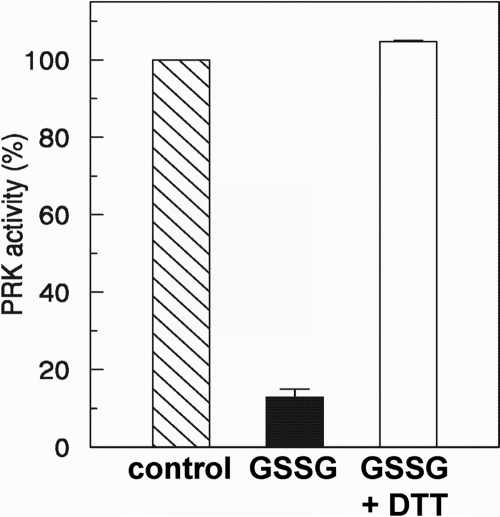
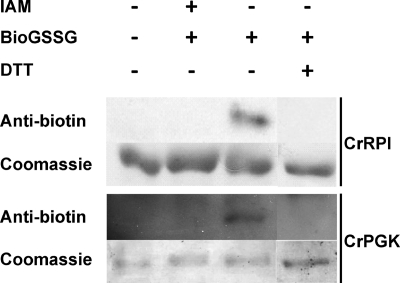
The glutathionylation of several Calvin-Benson cycle enzymes was further investigated at the biochemical level. We focused on four enzymes, namely PRK, PGK, RPI, and A4-GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Because PRK activity is restricted to the Calvin-Benson cycle enzyme, the effect of glutathionylation treatments on the enzyme activity was analyzed in total protein extracts of Chlamydomonas. After incubation with 5 mm GSSG, a classical in vitro glutathionylation test, PRK activity was found to decrease to ∼15% of its initial activity (Fig. 5). This inactivation was reversed in the presence of DTT as expected for a modification by glutathionylation. To analyze the glutathionylation of PGK and RPI in vitro, we cloned the corresponding cDNAs and expressed the recombinant proteins in E. coli. Both enzymes were purified to homogeneity by nickel affinity chromatography. To confirm their glutathionylation, the reduced enzymes were treated with 2 mm BioGSSG for 1 h, and the presence of glutathione adducts was analyzed by Western blot using anti-biotin antibodies. For both proteins, glutathionylation was detected after BioGSSG treatment, and the signal disappeared in the presence of DTT (Fig. 6). Moreover, no signal was detected when the protein was pretreated with IAM, a specific cysteine alkylating agents, suggesting that the absence of free cysteines prevents biotin labeling via glutathionylation. These results confirm that purified PGK and RPI undergo glutathionylation in the presence of BioGSSG in vitro.
Fig. 5.
Effect of GSSG on PRK activity. Protein extract of Chlamydomonas cell culture was treated with 5 mm GSSG (black bar) and enzyme activity was determined according to Ref. 36. The PRK activity in untreated control extract was set at 100% (hatched bar). For restoring PRK activity, GSSG-treated extract was desalted in 100 mm Tris-HCl, pH 7.9, using NAP-5 columns and then treated with 10 mm DTT for 15 min (white bar). The data are represented as the mean percentages of maximal activity ± S.D. (n = 3).
Fig. 6.
Analysis of recombinant PGK and RPI glutathionylation with BioGSSG. Recombinant PGK and RPI were incubated for 1 h in the presence of 2 mm BioGSSG with or without prior incubation with 100 mm IAM. The proteins were resolved by nonreducing SDS-PAGE and transferred to nitrocellulose for anti-biotin immunoblotting. The reversibility of the reaction was assessed by treatment with 20 mm DTT for 30 min as indicated. The Coomassie staining of the gel shows equal loading in each lane.
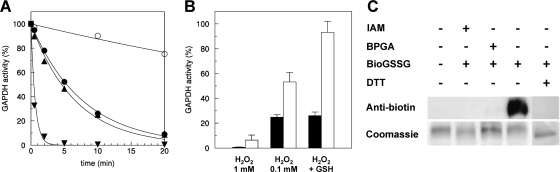
A4-GAPDH catalyzes the only reductive step of the Calvin-Benson cycle. Glycolytic and photosynthetic GAPDH enzymes contain a strictly conserved catalytic cysteine and are highly sensitive to oxidants (18, 19, 40, 41). Consistently, incubation of Chlamydomonas A4-GAPDH in the presence of 1 mm H2O2 led to a complete loss of activity within 2 min (Fig. 7A). This inactivation could not be reversed by DTT treatment (Fig. 7B), suggesting that H2O2 induced irreversible oxidation, most likely affecting the sulfhydryl group of the active site cysteine. Irreversibly oxidized cysteine thiols are typically converted to sulfinic (-SO2H) or sulfonic acids (-SO3H) (42). The inactivation of the enzyme was slower in the presence of a 10-fold lower H2O2 concentration (0.1 mm) (Fig. 7A), but in this case DTT could restore ∼50% of the initial activity (Fig. 7B). This partial recovery, not observed after treatment with 1 mm H2O2, indicated that part of the A4-GAPDH molecules were reversibly oxidized, probably to sulfenic acid (-SOH) by primary oxidation of the catalytic cysteine. The involvement of this residue is further supported by the fact that the enzyme substrate, BPGA, which forms a covalent intermediate with the catalytic cysteine, protects A4-GAPDH from H2O2-dependent oxidation (Fig. 7A). Moreover, by contrast with the partial recovery after treatment with 0.1 mm H2O2, an almost complete recovery of the initial activity after DTT treatment was observed for A4-GAPDH samples treated with 0.1 mm H2O2 and 0.5 mm GSH (Fig. 7B). This suggested that, in the presence of GSH, the mechanism of inactivation is different and most likely involves glutathionylation. This was confirmed by in vitro BioGSSG treatments of purified A4-GAPDH and immunoblotting with anti-biotin antibodies. A strong signal was detected after BioGSSG treatment and was reversed by DTT and blocked by prior alkylation with IAM. Moreover, BPGA also prevented glutathionylation of A4-GAPDH, thereby confirming that the catalytic cysteine is most probably the target of glutathionylation. All of the these data clearly demonstrate that glutathionylation inhibits the activity of Chlamydomonas A4-GAPDH but also protects it from irreversible oxidative inactivation. These results are consistent with those reported for Arabidopsis GAPDH isoforms (18, 19) and suggest that the mechanisms of redox regulation by glutathionylation of this Calvin-Benson cycle enzyme are conserved between green algae and higher plants. Higher plants contain two types of GAPDH: the A2B2 form, which is TRX-dependent, and the A4 form, which is not regulated by TRX but undergoes glutathionylation. By contrast, algae such as C. reinhardtii only contain the A4-GAPDH form. Therefore our results confirm that the only chloroplastic GAPDH isoform present in Chlamydomonas can also be redox-regulated by glutathionylation.
Fig. 7.
Analysis of recombinant A4-GAPDH oxidation and glutathionylation. A, kinetics of inactivation of A4-GAPDH by different oxidants. Reduced A4-GAPDH (2.5 μm) was incubated with 1 mm H2O2 (▾) or 0.1 mm H2O2 (▴) or 0.1 mm H2O2 plus 0.5 mm GSH (●) or 0.1 mm H2O2 plus 0.5 mm GSH in the presence of BPGA (○). Aliquots were withdrawn at the indicated times, and the remaining NADPH-dependent activity was determined. Activity is given as the percentage of the initial activity. B, reversal of A4-GAPDH inactivation by DTT. Reduced A4-GAPDH was incubated for 10 min in the presence of different oxidants as indicated (black bars). The reversibility of A4-GAPDH inactivation was assessed by incubation in the presence of 20 mm DTT (white bars). The data are represented as mean percentage of maximal activity ± S.D. (n = 3–5). C, immunoblot analysis of A4-GAPDH glutathionylation with BioGSSG. Reduced A4-GAPDH was incubated for 1 h in the presence of 2 mm BioGSSG with or without prior incubation with 100 mm IAM or BPGA. The proteins were resolved by nonreducing SDS-PAGE and transferred to nitrocellulose for Western blotting with anti-biotin antibodies. The reversibility of the reaction was assessed by treatment with 20 mm DTT for 30 min as indicated. The Coomassie staining of the gel shows equal loading in each lane.
DISCUSSION
Our proteomic analysis based on the use of BioGSSG has allowed identification of 225 putative glutathionylated proteins and 56 potential sites of modification. This is the most extensive study on protein glutathionylation in a photosynthetic organism and confirms that this modification is likely to constitute, as in mammals, an important mechanism of cell regulation and signaling, e.g. under oxidative stress conditions. Our results confirm the utility of BioGSSG for analysis of protein glutathionylation (43, 44). The proteins identified participate in various metabolic pathways and cell processes. Almost one-third of the proteins participate in carbon metabolism including enzymes involved in the Calvin-Benson and TCA cycles, glycolysis/gluconeogenesis, starch, and sucrose metabolism or fatty acid metabolism. These results are consistent with studies in yeast and mammals reporting that numerous glycolytic or TCA cycle enzymes are regulated by glutathionylation (45–50). No protein involved in the regulation of gene expression, such as transcription factors, was identified as glutathionylated in Chlamydomonas. This may reflect the low abundance of these enzymes, although this seems unlikely because of the sensitivity of the method employed and the selective enrichment of glutathionylated proteins by streptavidin affinity chromatography. Another possibility is that these enzymes were identified but are found among the 17% of proteins with unknown functions. Indeed, the mechanisms of regulation of gene expression in Chlamydomonas are much less known than in organisms where redox-sensitive transcription factors have been identified such as OxyR in E. coli (51), Yap1 in yeast (52), or NF-κB or AP-1 in mammals (5, 53, 54). Finally, another possibility could be that the redox-dependent regulation of protein abundance by glutathionylation mainly operates at the translational or post-translational level, as illustrated by the fact that 17% of identified proteins are involved in protein biogenesis and degradation.
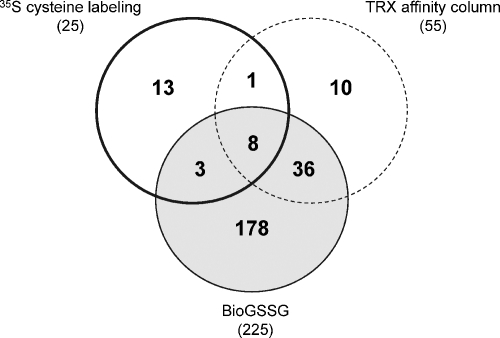
Nevertheless, many of the 225 proteins identified were previously found to be redox-regulated in Chlamydomonas (Table I and Fig. 8). Among the 25 proteins identified as S-thiolated in vivo using [35S]cysteine labeling (23), 11 were found to be glutathionylated with BioGSSG. This suggests that although BioGSSG labeling was performed on Chlamydomonas extracts, the proteins and sites of modification identified may be relevant in vivo. Some proteins may not be detected with BioGSSG because this molecule triggers glutathionylation only through thiol-disulfide exchange, whereas other mechanisms may occur in vivo. Similarly, 41 among the 55 proteins retained on a TRX affinity column (55) were also identified with BioGSSG. This is consistent with the lack of specificity of monocysteinic TRX columns that can retain proteins harboring different types of oxidative modifications, including protein disulfide bonds and glutathionylation (39). A detailed biochemical characterization of Chlamydomonas isocitrate lyase, a protein binding to TRX columns, revealed that the enzyme can be inactivated by glutathionylation of its catalytic cysteine but does not appear to be regulated by TRX (20). Alternatively, it is also possible that some of the identified proteins might be subject to a double regulation by both TRX and glutathionylation. For example, PRK activity is increased by TRXf-dependent reduction of a disulfide bond in the light (56), whereas the enzyme was shown to be inactivated by a glutathionylation treatment in the present study. Similarly, Chlamydomonas PRX1 was shown to be reduced by TRX (57) and to undergo glutathionylation in vitro (23). For each of these enzymes, further work will be required to confirm the double regulation and to determine whether these mechanisms of redox regulation are independent or interconnected. Finally, 178 of the 225 proteins identified with BioGSSG correspond to new potential targets, not previously suggested to be redox-regulated in Chlamydomonas. These results strengthen the potential importance of glutathionylation in photosynthetic organisms for regulation of numerous biological processes and pathways.
Fig. 8.
Analysis of redox-regulated proteins identified in Chlamydomonas. The Venn diagram shows the distribution of Chlamydomonas proteins identified as putative target of S-glutathionylation (BioGSSG), TRX (monocysteinic TRX affinity column (55)), and S-thiolation (35S-cysteine radiolabeling (23)).
The method employed to determine glutathionylation sites appears to be reliable, because many of the sites identified correspond to cysteines known to undergo glutathionylation or oxidative modifications such as Cys217 in human actin (58), Cys385 in porcine aconitase (59), Cys98 in yeast PGK (60), Cys247 in Chlamydomonas isocitrate lyase (20, 23), and Cys172 in Chlamydomonas Rubisco (61). A search for motifs in the primary sequence surrounding the 56 sites identified did not reveal any consensus sequence. Similarly, no motif was identified for S-nitrosylated cysteines (62). Multiple factors appear to contribute to the sensitivity of a given cysteine residue to S-glutathionylation including pKa, accessibility, reactivity, and its microenvironment, especially surrounding amino acids (63). A combination of these factors is likely required because protein cysteine residues exhibit a striking differential susceptibility to glutathionylation (9). In the case of human TRX and plant f-type TRXs from diverse species, the glutathionylated cysteine is accessible but also surrounded by a patch of positive charges that could facilitate interaction with the negatively charged glutathione molecule (16, 64). Several other plant TRX isoforms that also contain accessible cysteines but lack surrounding positive charges are apparently not susceptible to glutathionylation (16). Anyway, the identification of glutathionylation sites together with the identity of the modified protein provides important information that will greatly facilitate future studies on the functional role of this modification for the different targets identified.
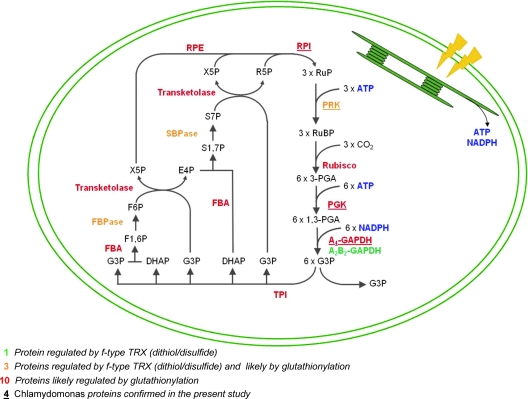
The Calvin-Benson cycle is regulated by multiple redox post-translational modifications (Fig. 9). Four enzymes of this cycle (fructose-1,6-bisphosphatase, sedoeptulose-1,7-bisphosphatase, A2B2-GAPDH, and PRK) are well established targets of TRX in chloroplasts. These enzymes have a low activity in the dark and are activated upon illumination by photosynthetically reduced TRX (39, 65). Although chloroplasts contain several isoforms of TRX (f, m, x, y, and z), regulation of Calvin-Benson cycle enzymes is preferentially or exclusively operated by f-type TRXs. Moreover, all Calvin-Benson cycle enzymes from diverse organisms were retained on TRX affinity columns (39, 66–68), suggesting that they might all be redox-regulated. The Calvin-Benson cycle is also regulated either directly or indirectly by glutathionylation. The indirect regulation operates through glutathionylation of TRXf because this modification results in an impaired ability of TRXf to activate its target enzymes in the light (16). Several Calvin-Benson cycle enzymes were reported to be modified directly by glutathionylation including Arabidopsis A4-GAPDH (19), fructose-1,6-bisphosphate aldolase, and triose phosphate isomerase (26). Moreover, early studies demonstrated that GSSG could inhibit Calvin cycle enzymes, although at this time GSSG was thought to mediate dark deactivation (69). In the present study, all of the Calvin-Benson cycle enzymes except triose phosphate isomerase were identified as putatively glutathionylated, and four enzymes (PGK, PRK, RPI, and GAPDH) were confirmed. Overall, glutathionylation appears to decrease the activity of Calvin-Benson cycle enzymes. This suggests that under conditions of oxidative stress leading to protein glutathionylation in the chloroplast, the turnover of the Calvin-Benson cycle could be decreased. Such a regulation could have several physiological roles. First, this negative regulation in the presence of ROS could allow a redistribution of reducing power within chloroplasts by decreasing NADPH consumption by the Calvin cycle. This would increase NADPH availability for ROS detoxifying enzymes such as glutathione reductase or monodehydroascorbate reductase. Moreover, this slowing down could also allow a redistribution of electrons to other detoxifying enzymes like peroxiredoxins, glutathione peroxidases, or ascorbate peroxidases with the possible implication of TRX isoforms whose activity is not apparently regulated by glutathionylation (16, 39). This type of regulation may allow a fine tuning of cell metabolism under stress conditions. The idea that enzymes of the Calvin-Benson cycle could be down-regulated in response to oxidative stress to enhance survival by enabling the plant to redirect energy resources to antioxidant defense was recognized early on in proteomic studies of TRX targets (68). Indeed, carbon dioxide assimilation is among the first chloroplast processes affected when plants experience stress. The Calvin cycle typically slows down with the onset of stress, well before photosynthetic electron flow and ATP synthesis (70, 71). Early results provided evidence that several enzymes of the Calvin cycle such as transketolase or ribulose-3-phosphate-epimerase are sensitive to H2O2 and are sites of H2O2-dependent inhibition (72). The identification of these enzymes as glutathionylation targets in the present study may provide a link with these initial studies because H2O2 oxidation is an efficient mediator of protein glutathionylation.
Fig. 9.
The Calvin-Benson cycle is a major target of redox regulation. All of the enzymes of the Calvin-Benson cycle are potentially regulated by TRXf-dependent reduction of a disulfide bond and/or by glutathionylation, as indicated. The Calvin-Benson cycle is represented in a schematic chloroplast. Light energy is used by thylakoids to generate ATP and NADPH consumed by the Calvin-Benson cycle for CO2 fixation. Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase; TPI, triose phosphate isomerase; FBA, fructose-1,6-bisphosphate aldolase; FBPase, fructose-1,6-bisphosphatase; SBPase, sedoheptulose-1,7-bisphosphatase; RPE, ribulose phosphate-3-epimerase; RuBP, ribulose-1,5-bisphosphate; 3-PGA, 3-phosphoglycerate; G3P, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; F1,6P, fructose-1,6-bisphosphate; F6P, fructose-6-phosphate; X5P, xylulose-5-phosphate; E4P, erythrose-4-phosphate; S1,7P, sedoheptulose-1,7-bisphosphate; S7P, sedoheptulose-7-phosphate; R5P, ribose-5-phosphate; RuP, ribulose-5-phosphate.
However, current models rather suggest that overreduction of Photosystem I caused by a decreased availability of electron acceptors would tend to increase ROS production. Thus, an alternative hypothesis would be to consider that the slowing down of the Calvin cycle would increase ROS production and thereby reinforce the initial oxidative signal. Clearly, further studies are required to determine the different factors controlling the distribution of reducing equivalents between the different Photosystem I electron acceptors. Finally, all of these data suggest that the Calvin-Benson cycle is tightly regulated by multiple redox post-translational modifications including glutathionylation. The underlying molecular mechanisms will have to be examined thoroughly because they might be crucial for fine tuning of photosynthetic efficiency and carbon fixation under varying environmental conditions.
Footnotes
* This work was supported in part by Agence Nationale de la Recherche Grant 08-BLAN-0153 GLUTAPHOTO (to M. B., S. D. L., H. G., C. M., P. D., and C. C. C.) and by a PRIN 2008 grant (to M. Z.) from the Ministero dell'Istruzione, dell'Università e della Ricerca of Italy. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PTM
- post-translational modification
- BioGEE
- N-biotinyl glutathione ethyl-ester
- BioGSSG
- N,N′-biotinyl glutathione disulfide
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GSH
- reduced glutathione
- GSSG
- glutathione disulfide
- IAM
- iodoacetamide
- LC
- liquid chromatography
- PGK
- phosphoglycerate kinase
- PRK
- phosphoribulokinase
- RNS
- reactive nitrogen species
- ROS
- reactive oxygen species
- RPI
- ribose-5-phosphate isomerase
- TRX
- thioredoxin
- TAP
- Tris acetate phosphate
- BPGA
- 1,3-bisphosphoglycerate.
REFERENCES
- 1. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 2. Foyer C. H., Noctor G. (2009) Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11, 861–905 [DOI] [PubMed] [Google Scholar]
- 3. Hancock J. T. (2009) The role of redox mechanisms in cell signalling. Mol. Biotechnol. 43, 162–166 [DOI] [PubMed] [Google Scholar]
- 4. Rouhier N., Lemaire S. D., Jacquot J. P. (2008) The role of glutathione in photosynthetic organisms: Emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166 [DOI] [PubMed] [Google Scholar]
- 5. Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D. (2008) Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 10, 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiong Y., Uys J. D., Tew K. D., Townsend D. M. (2011) S-Glutathionylation: From molecular mechanisms to health outcomes. Antioxid. Redox Signal. 15, 233–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martínez-Ruiz A., Lamas S. (2007) Signalling by NO-induced protein S-nitrosylation and S-glutathionylation: Convergences and divergences. Cardiovasc. Res. 75, 220–228 [DOI] [PubMed] [Google Scholar]
- 8. Chen C. A., Wang T. Y., Varadharaj S., Reyes L. A., Hemann C., Talukder M. A., Chen Y. R., Druhan L. J., Zweier J. L. (2010) S-Glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature 468, 1115–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dalle-Donne I., Rossi R., Colombo G., Giustarini D., Milzani A. (2009) Protein S-glutathionylation: A regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96 [DOI] [PubMed] [Google Scholar]
- 10. Ghezzi P., Di Simplicio P. (2007) Glutathionylation pathways in drug response. Curr. Opin. Pharmacol. 7, 398–403 [DOI] [PubMed] [Google Scholar]
- 11. Michelet L., Zaffagnini M., Massot V., Keryer E., Vanacker H., Miginiac-Maslow M., Issakidis-Bourguet E., Lemaire S. D. (2006) Thioredoxins, glutaredoxins, and glutathionylation: New crosstalks to explore. Photosynth. Res. 89, 225–245 [DOI] [PubMed] [Google Scholar]
- 12. Shelton M. D., Mieyal J. J. (2008) Regulation by reversible S-glutathionylation: Molecular targets implicated in inflammatory diseases. Mol. Cells 25, 332–346 [PMC free article] [PubMed] [Google Scholar]
- 13. Kehr S., Jortzik E., Delahunty C., Yates J. R., Rahlfs S., Becker K. (2011) Protein S-glutathionylation in malaria parasites. Antioxid. Redox Signal. 15, 2855–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zaffagnini M., Bedhomme M., Marchand C. H., Couturier J., Gao X. H., Rouhier N., Trost P., Lemaire S. D. (2012) Glutaredoxin S12: Unique properties for redox signaling. Antioxid. Redox Signal. 16, 17–32 [DOI] [PubMed] [Google Scholar]
- 15. Gelhaye E., Rouhier N., Gérard J., Jolivet Y., Gualberto J., Navrot N., Ohlsson P. I., Wingsle G., Hirasawa M., Knaff D. B., Wang H., Dizengremel P., Meyer Y., Jacquot J. P. (2004) A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc. Natl. Acad. Sci. U.S.A. 101, 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Michelet L., Zaffagnini M., Marchand C., Collin V., Decottignies P., Tsan P., Lancelin J. M., Trost P., Miginiac-Maslow M., Noctor G., Lemaire S. D. (2005) Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc. Natl. Acad. Sci. U.S.A. 102, 16478–16483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dixon D. P., Fordham-Skelton A. P., Edwards R. (2005) Redox regulation of a soybean tyrosine-specific protein phosphatase. Biochemistry 44, 7696–7703 [DOI] [PubMed] [Google Scholar]
- 18. Holtgrefe S., Gohlke J., Starmann J., Druce S., Klocke S., Altmann B., Wojtera J., Lindermayr C., Scheibe R. (2008) Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant 133, 211–228 [DOI] [PubMed] [Google Scholar]
- 19. Zaffagnini M., Michelet L., Marchand C., Sparla F., Decottignies P., Le Maréchal P., Miginiac-Maslow M., Noctor G., Trost P., Lemaire S. D. (2007) The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J. 274, 212–226 [DOI] [PubMed] [Google Scholar]
- 20. Bedhomme M., Zaffagnini M., Marchand C. H., Gao X. H., Moslonka-Lefebvre M., Michelet L., Decottignies P., Lemaire S. D. (2009) Regulation by glutathionylation of isocitrate lyase from Chlamydomonas reinhardtii. J. Biol. Chem. 284, 36282–36291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leferink N. G., van Duijn E., Barendregt A., Heck A. J., van Berkel W. J. (2009) Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C. Plant Physiol. 150, 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Palmieri M. C., Lindermayr C., Bauwe H., Steinhauser C., Durner J. (2010) Regulation of plant glycine decarboxylase by S-nitrosylation and glutathionylation. Plant Physiol. 152, 1514–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michelet L., Zaffagnini M., Vanacker H., Le Maréchal P., Marchand C., Schroda M., Lemaire S. D., Decottignies P. (2008) In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J. Biol. Chem. 283, 21571–21578 [DOI] [PubMed] [Google Scholar]
- 24. Noguera-Mazon V., Lemoine J., Walker O., Rouhier N., Salvador A., Jacquot J. P., Lancelin J. M., Krimm I. (2006) Glutathionylation induces the dissociation of 1-Cys d-peroxiredoxin non-covalent homodimer. J. Biol. Chem. 281, 31736–31742 [DOI] [PubMed] [Google Scholar]
- 25. Tarrago L., Laugier E., Zaffagnini M., Marchand C., Le Maréchal P., Rouhier N., Lemaire S. D., Rey P. (2009) Regeneration mechanisms of Arabidopsis thaliana methionine sulfoxide reductases B by glutaredoxins and thioredoxins. J. Biol. Chem. 284, 18963–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito H., Iwabuchi M., Ogawa K. (2003) The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: Detection using biotinylated glutathione. Plant Cell Physiol. 44, 655–660 [DOI] [PubMed] [Google Scholar]
- 27. Dixon D. P., Skipsey M., Grundy N. M., Edwards R. (2005) Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 138, 2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rochaix J. D., Mayfield S. P., Goldschmidt-Clermont M., Erickson J. (1988) Plant molecular biology: A practical approach, IRL, Oxford [Google Scholar]
- 29. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 30. Ellman G. L. (1959) Tissue sulfhydryl groups. Arch. Biochem. Biophys 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 31. Gillet S., Decottignies P., Chardonnet S., Le Maréchal P. (2006) Cadmium response and redoxin targets in Chlamydomonas reinhardtii: A proteomic approach. Photosynth. Res. 89, 201–211 [DOI] [PubMed] [Google Scholar]
- 32. Pasternak T. P., Prinsen E., Ayaydin F., Miskolczi P., Potters G., Asard H., Van Onckelen H. A., Dudits D., Fehér A. (2002) The role of auxin, pH, and stress in the activation of embryogenic cell division in leaf protoplast-derived cells of alfalfa. Plant Physiol. 129, 1807–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 34. Petersen B., Petersen T. N., Andersen P., Nielsen M., Lundegaard C. (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Graciet E., Lebreton S., Camadro J. M., Gontero B. (2003) Characterization of native and recombinant A4 glyceraldehyde 3-phosphate dehydrogenase: Kinetic evidence for confromation changes upon association with the small protein CP12. Eur. J. Biochem. 270, 129–136 [DOI] [PubMed] [Google Scholar]
- 36. Gontero B., Cárdenas M. L., Ricard J. (1988) A functional five-enzyme complex of chloroplasts involved in the Calvin cycle. Eur. J. Biochem. 173, 437–443 [DOI] [PubMed] [Google Scholar]
- 37. Chapman-Smith A., Cronan J. E., Jr. (1999) The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem. Sci 24, 359–363 [DOI] [PubMed] [Google Scholar]
- 38. Healy S., McDonald M. K., Wu X., Yue W. W., Kochan G., Oppermann U., Gravel R. A. (2010) Structural impact of human and Escherichia coli biotin carboxyl carrier proteins on biotin attachment. Biochemistry 49, 4687–4694 [DOI] [PubMed] [Google Scholar]
- 39. Lemaire S. D., Michelet L., Zaffagnini M., Massot V., Issakidis-Bourguet E. (2007) Thioredoxins in chloroplasts. Curr. Genet. 51, 343–365 [DOI] [PubMed] [Google Scholar]
- 40. Grant C. M., Quinn K. A., Dawes I. W. (1999) Differential protein S-thiolation of glyceraldehyde-3-phosphate dehydrogenase isoenzymes influences sensitivity to oxidative stress. Mol. Cell. Biol. 19, 2650–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Talfournier F., Colloc'h N., Mornon J. P., Branlant G. (1998) Comparative study of the catalytic domain of phosphorylating glyceraldehyde-3-phosphate dehydrogenases from bacteria and archaea via essential cysteine probes and site-directed mutagenesis. Eur. J. Biochem. 252, 447–457 [DOI] [PubMed] [Google Scholar]
- 42. Poole L. B., Karplus P. A., Claiborne A. (2004) Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 43. Brennan J. P., Miller J. I., Fuller W., Wait R., Begum S., Dunn M. J., Eaton P. (2006) The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol. Cell. Proteomics 5, 215–225 [DOI] [PubMed] [Google Scholar]
- 44. Lind C., Gerdes R., Hamnell Y., Schuppe-Koistinen I., von Löwenhielm H. B., Holmgren A., Cotgreave I. A. (2002) Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch. Biochem. Biophys. 406, 229–240 [DOI] [PubMed] [Google Scholar]
- 45. Applegate M. A., Humphries K. M., Szweda L. I. (2008) Reversible inhibition of α-ketoglutarate dehydrogenase by hydrogen peroxide: Glutathionylation and protection of lipoic acid. Biochemistry 47, 473–478 [DOI] [PubMed] [Google Scholar]
- 46. Fratelli M., Demol H., Puype M., Casagrande S., Villa P., Eberini I., Vandekerckhove J., Gianazza E., Ghezzi P. (2003) Identification of proteins undergoing glutathionylation in oxidatively stressed hepatocytes and hepatoma cells. Proteomics 3, 1154–1161 [DOI] [PubMed] [Google Scholar]
- 47. Kil I. S., Park J. W. (2005) Regulation of mitochondrial NADP+-dependent isocitrate dehydrogenase activity by glutathionylation. J. Biol. Chem. 280, 10846–10854 [DOI] [PubMed] [Google Scholar]
- 48. Mohr S., Hallak H., de Boitte A., Lapetina E. G., Brüne B. (1999) Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 274, 9427–9430 [DOI] [PubMed] [Google Scholar]
- 49. Shenton D., Grant C. M. (2003) Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 374, 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shin S. W., Oh C. J., Kil I. S., Park J. W. (2009) Glutathionylation regulates cytosolic NADP+-dependent isocitrate dehydrogenase activity. Free Radic. Res. 43, 409–416 [DOI] [PubMed] [Google Scholar]
- 51. Kim S. O., Merchant K., Nudelman R., Beyer W. F., Jr., Keng T., DeAngelo J., Hausladen A., Stamler J. S. (2002) OxyR: A molecular code for redox-related signaling. Cell 109, 383–396 [DOI] [PubMed] [Google Scholar]
- 52. Toledano M. B., Delaunay A., Monceau L., Tacnet F. (2004) Microbial H2O2 sensors as archetypical redox signaling modules. Trends Biochem. Sci. 29, 351–357 [DOI] [PubMed] [Google Scholar]
- 53. Pineda-Molina E., Klatt P., Vázquez J., Marina A., García de Lacoba M., Pérez-Sala D., Lamas S. (2001) Glutathionylation of the p50 subunit of NF-κB: A mechanism for redox-induced inhibition of DNA binding. Biochemistry 40, 14134–14142 [DOI] [PubMed] [Google Scholar]
- 54. Reynaert N. L., van der Vliet A., Guala A. S., McGovern T., Hristova M., Pantano C., Heintz N. H., Heim J., Ho Y. S., Matthews D. E., Wouters E. F., Janssen-Heininger Y. M. (2006) Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory κB kinase β. Proc. Natl. Acad. Sci. U.S.A. 103, 13086–13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lemaire S. D., Guillon B., Le Maréchal P., Keryer E., Miginiac-Maslow M., Decottignies P. (2004) New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 101, 7475–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marri L., Zaffagnini M., Collin V., Issakidis-Bourguet E., Lemaire S. D., Pupillo P., Sparla F., Miginiac-Maslow M., Trost P. (2009) Prompt and easy activation by specific thioredoxins of calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol. Plant 2, 259–269 [DOI] [PubMed] [Google Scholar]
- 57. Goyer A., Haslekås C., Miginiac-Maslow M., Klein U., Le Marechal P., Jacquot J. P., Decottignies P. (2002) Isolation and characterization of a thioredoxin-dependent peroxidase from Chlamydomonas reinhardtii. Eur. J. Biochem. 269, 272–282 [DOI] [PubMed] [Google Scholar]
- 58. Hamnell-Pamment Y., Lind C., Palmberg C., Bergman T., Cotgreave I. A. (2005) Determination of site-specificity of S-glutathionylated cellular proteins. Biochem. Biophys. Res. Commun. 332, 362–369 [DOI] [PubMed] [Google Scholar]
- 59. Han D., Canali R., Garcia J., Aguilera R., Gallaher T. K., Cadenas E. (2005) Sites and mechanisms of aconitase inactivation by peroxynitrite: Modulation by citrate and glutathione. Biochemistry 44, 11986–11996 [DOI] [PubMed] [Google Scholar]
- 60. McDonagh B., Ogueta S., Lasarte G., Padilla C. A., Bárcena J. A. (2009) Shotgun redox proteomics identifies specifically modified cysteines in key metabolic enzymes under oxidative stress in Saccharomyces cerevisiae. J. Proteomics 72, 677–689 [DOI] [PubMed] [Google Scholar]
- 61. Moreno J., Spreitzer R. J. (1999) C172S substitution in the chloroplast-encoded large subunit affects stability and stress-induced turnover of ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Biol. Chem. 274, 26789–26793 [DOI] [PubMed] [Google Scholar]
- 62. Marino S. M., Gladyshev V. N. (2010) Structural analysis of cysteine S-nitrosylation: A modified acid-based motif and the emerging role of trans-nitrosylation. J. Mol. Biol. 395, 844–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dalle-Donne I., Milzani A., Gagliano N., Colombo R., Giustarini D., Rossi R. (2008) Molecular mechanisms and potential clinical significance of S-glutathionylation. Antioxid. Redox Signal. 10, 445–473 [DOI] [PubMed] [Google Scholar]
- 64. Casagrande S., Bonetto V., Fratelli M., Gianazza E., Eberini I., Massignan T., Salmona M., Chang G., Holmgren A., Ghezzi P. (2002) Glutathionylation of human thioredoxin: A possible crosstalk between the glutathione and thioredoxin systems. Proc. Natl. Acad. Sci. U.S.A. 99, 9745–9749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schürmann P., Buchanan B. B. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 10, 1235–1274 [DOI] [PubMed] [Google Scholar]
- 66. Lindahl M., Mata-Cabana A., Kieselbach T. (2011) The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid. Redox Signal. 14, 2581–2642 [DOI] [PubMed] [Google Scholar]
- 67. Montrichard F., Alkhalfioui F., Yano H., Vensel W. H., Hurkman W. J., Buchanan B. B. (2009) Thioredoxin targets in plants: The first 30 years. J. Proteomics 72, 452–474 [DOI] [PubMed] [Google Scholar]
- 68. Balmer Y., Koller A., del Val G., Manieri W., Schürmann P., Buchanan B. B. (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. U.S.A. 100, 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wolosiuk R. A., Buchanan B. B. (1977) Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature 266, 565–567 [Google Scholar]
- 70. Lawlor D. W., Cornic G. (2002) Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ. 25, 275–294 [DOI] [PubMed] [Google Scholar]
- 71. Weis E. (1981) Reversible heat-inactivation of the calvin cycle: A possible mechanism of the temperature regulation of photosynthesis. Planta 151, 33–39 [DOI] [PubMed] [Google Scholar]
- 72. Kaiser W. (1976) The effect of hydrogen peroxide on CO2 fixation of isolated intact chloroplasts. Biochim. Biophys. Acta 440, 476–482 [DOI] [PubMed] [Google Scholar]