Abstract
Analysis of cellular signaling networks typically involves targeted measurements of phosphorylated protein intermediates. However, phosphoproteomic analyses usually require affinity enrichment of phosphopeptides and can be complicated by artifactual changes in phosphorylation caused by uncontrolled preanalytical variables, particularly in the analysis of tissue specimens. We asked whether changes in protein expression, which are more stable and easily analyzed, could reflect network stimulation and inhibition. We employed this approach to analyze stimulation and inhibition of the epidermal growth factor receptor (EGFR) by EGF and selective EGFR inhibitors. Shotgun analysis of proteomes from proliferating A431 cells, EGF-stimulated cells, and cells co-treated with the EGFR inhibitors cetuximab or gefitinib identified groups of differentially expressed proteins. Comparisons of these protein groups identified 13 proteins whose EGF-induced expression changes were reversed by both EGFR inhibitors. Targeted multiple reaction monitoring analysis verified differential expression of 12 of these proteins, which comprise a candidate EGFR inhibition signature. We then tested these 12 proteins by multiple reaction monitoring analysis in three other models: 1) a comparison of DiFi (EGFR inhibitor-sensitive) and HCT116 (EGFR-insensitive) cell lines, 2) in formalin-fixed, paraffin-embedded mouse xenograft DiFi and HCT116 tumors, and 3) in tissue biopsies from a patient with the gastric hyperproliferative disorder Ménétrier's disease who was treated with cetuximab. Of the proteins in the candidate signature, a core group, including c-Jun, Jagged-1, and Claudin 4, were decreased by EGFR inhibitors in all three models. Although the goal of these studies was not to validate a clinically useful EGFR inhibition signature, the results confirm the hypothesis that clinically used EGFR inhibitors generate characteristic protein expression changes. This work further outlines a prototypical approach to derive and test protein expression signatures for drug action on signaling networks.
Signaling networks and pathways regulate essential cellular functions. Activities of pathways are controlled by post-translational modification of key pathway intermediates, such as signaling receptors and their downstream effectors, which undergo reversible phosphorylation. Immunoblot methods are most commonly used to monitor protein phosphorylation changes, but this approach is limited by the availability and specificity of antibody reagents. Mass spectrometry-based proteomic approaches aimed at the detection of phosphorylation modifications have proven useful in the investigation of cellular signaling events (1–3) and have been shown to identify protein phosphorylation changes in response to drug treatments (4). Phosphoproteome analysis methods typically require affinity enrichment of phosphorylated proteins or peptides to detect low abundance phosphorylated forms (1, 3–5). The transient nature of phosphorylation modifications also presents the challenge of preserving phosphorylation status during sample preparation. Most work in phosphoproteomics has been done in cell culture models, which offer the advantages of controlled experimental conditions, relatively easy sample workup, and scalability to enable analysis of low abundance phosphoproteins. Phosphoproteomic analysis of tissue specimens is complicated by sample heterogeneity, limiting amounts of available tissue, and low abundance of modified peptides. In addition, acquisition practices for biopsies and surgical resections do not permit rigorous control of preanalytical variables, such as ischemic time and temperature, which trigger stress responses that may obscure the status of network intermediates in vivo (6–8). Given these considerations, more robust approaches to measure signaling networks are needed to overcome the shortcomings of direct phosphoproteome analyses.
One of the most extensively studied signaling pathways is driven by the epidermal growth factor receptor (EGFR),1 a receptor tyrosine kinase that influences a broad range of signaling events and biological processes. Upon ligand binding, EGFR dimerizes with itself or with other ErbB proteins and the receptor is autophosphorylated at multiple residues (9). Signal transduction occurs by recruitment of adaptor proteins and activation of downstream kinases in the mitogen-activated protein kinase, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways (10). EGFR activation plays a critical role in many human cancers, and several anticancer drugs directed at this receptor tyrosine kinase are in clinical use (11). EGFR mutations can modify responsiveness to EGFR-inhibiting drugs and are associated with acquired resistance to inhibitors (12, 13).
Despite the broad importance of EGFR as a therapeutic target, prediction and assessment of therapeutic responses to EGFR inhibitors present a significant clinical problem (14). Negative predictors of response include mutations in KRAS, which constitutively activate mitogen-activated protein kinase signaling and block cellular response to EGFR inhibiting drugs (12, 15). Similarly, mutations in PIK3CA also confer resistance to EGFR inhibition with cetuximab (16, 17). Protein and phosphoprotein analyses in tumor tissues by reverse phase protein array methods have identified putative signatures for EGFR inhibitor responses (18–21). Studies in cell models using global phosphoproteomics and targeted analysis of EGF pathway phosphoprotein intermediates have provided the most comprehensive analyses of EGFR-driven signaling networks (1, 22–24). Despite the rapid growth of information about EGFR signaling networks, identification of robust molecular markers linking network status and therapeutic response remains an open challenge. Indeed, skin rash remains one of the most effective early indicators of clinical response to EGFR inhibitors (25).
We asked whether changes in global protein expression levels could produce distinct protein signatures indicative of a cellular response to EGFR modulation. To address this issue, we employed a model system in A431 cells using EGF and two clinically used EGFR inhibitors, gefitinib and cetuximab, to manipulate the EGFR signaling axis. We analyzed differentially treated A431 cells with a standardized shotgun proteomics platform that combines peptide isoelectric focusing and LC-MS/MS (26, 27). Comparison of these data sets indicated proteins that differed significantly in expression between treatment conditions and constitute potential stimulation and inhibition signatures. A set of proteins whose expression was changed by EGF and reversed by both gefitinib and cetuximab comprised a candidate “EGFR inhibition signature,” which we further verified by multiple reaction monitoring (MRM) analyses. We then tested this EGFR inhibition signature in three other models: 1) a comparison of DiFi (EGFR inhibitor-sensitive) and HCT116 (EGFR-resistant) cell lines, 2) in formalin-fixed, paraffin-embedded (FFPE) mouse xenograft DiFi and HCT116 tumors, and 3) in frozen tissue biopsies from a patient with the gastric hyperproliferative disorder Ménétrier's disease who was treated with cetuximab. The results verified a multiprotein EGFR inhibition signature in all three models and illustrate the application of protein expression changes as surrogate measures of signaling network activation and inhibition.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Iodoacetamide, ammonium bicarbonate, sodium molybdate, β-glycerophosphate, sodium molybdate, sodium orthovanadate, 4-(2-aminoethyl)benzenesulfonyl fluoride, aprotinin, leupeptin, betastatin, pepstatin A, dimethyl sulfoxoide, and sodium dodecyl sulfate (all >99% purity) were purchased from Sigma. Dithiothreitol was from Bio-Rad; 2,2,2-trifluoroethanol (TFE) was from Acros (Geel, Belgium). C-terminal isotopically labeled β-actin peptide (GYSFTTTAE*R) containing U-13C6, U-15N4-arginine was obtained from New England Peptide at 99% isotopic purity and 95% chemical purity. Fetal bovine serum (FBS) was from Atlas Biologicals (Fort Collins, CO). Mass spectrometry grade trypsin (Trypsin Gold) was purchased from Promega (Madison, WI). HPLC grade water and ACN were from Mallinckrodt Baker (Phillipsburg, NJ). Phosphate-buffered saline, Dulbecco's modified Eagle's medium, improved minimal essential medium, McCoy's 5A medium, NuPAGE® MOPS, and MES SDS running buffer, NuPAGE® lithium dodecyl sulfate sample buffer, NuPAGE® 10% BisTris gels, and polyvinylidene difluoride membrane were from Invitrogen. EGF, EGFR, pY1173, pY998, Jagged-1, c-Jun, caspase 3, and poly(ADP-ribose) polymerase antibodies (antibodies 8916LF, 4267s, 4407s, 2641, 2620, 9165, 9542, and 9662, respectively) were purchased from Cell Signaling Technologies (Danvers, MA). Claudin 4 and β-actin antibody (catalog numbers ab53156 and ab8224, respectively) were from AbCam (Cambridge, MA). 4G10 antibody (catalog number 05-1050X) was from Millipore (Billerica, MA). AlexaFluor® 680-conjugated fluorescent secondary antibodies were obtained from Molecular Probes (Eugene, OR). TFA, formic acid, urea, and tris-carboxyethylphosphine were purchased from Thermo Fisher Scientific (Waltham, MA). Gefitinib and cetuximab were gracious gifts from Dr. Carlos Arteaga and Dr. Robert Coffey, respectively, both at Vanderbilt University. Sub-X was from Surgipath (Richmond, IL).
Tissue Specimens
Frozen gastric epithelial biopsy specimens were obtained from our previous study (28), which was a single-arm clinical trial to explore the effectiveness of a 4-week course of cetuximab in patients with clinically and histologically confirmed Ménétrier disease. This prospective, open label trial was approved by the Vanderbilt University Medical Center Institutional Review Board, and all of the participants provided written informed consent. DiFi- and HCT116-derived mouse xenograft tumor samples were from our previous study (29), and tissues were obtained as archival FFPE sections.
Cell Culture
A431 human epithelial carcinoma cell lines were purchased from ATCC (Manassas, VA). A431 cells were maintained in improved minimal essential medium supplemented with 10% FBS. DiFi human colorectal carcinoma cells were a gift from Dr. Bruce Boman (Creighton University School of Medicine) and were maintained in Dulbecco's modified Eagle's medium and supplemented with 10% FBS (30). HCT116 human colorectal carcinoma cells were from ATCC and were maintained in McCoy's 5A medium also supplemented with 10% FBS. All of the cell lines were cultured at 37 °C in 5% CO2. Proliferating cells were grown to 70–75% confluency before collection, whereas all treated cells were grown to ∼60–65% confluency before incubation overnight in serum-free medium. These serum-starved cells were then either treated with 30 nm EGF only for 4 h (EGF-stimulated) or preincubated with 500 nm gefitinib or 10 μg/ml cetuximab (unless otherwise noted) for 30 min before treatment with 30 nm EGF for 4 h in the continued presence of the inhibitors. All of the cells were harvested on ice using cold magnesium- and calcium-free phosphate-buffered saline and supplemented with a phosphatase inhibitor mixture (1.0 mm sodium orthovanadate, 1.0 mm sodium molybdate, 1.0 mm sodium fluoride, and 10 mm of β-glycerophosphate). The cells were pelleted by centrifugation at 500 × g at 4 °C, and pellets were flash frozen in liquid nitrogen.
Western Blot Analysis
The cell pellets were resuspended and lysed in a modified radioimmune precipitation assay buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Igepal, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) supplemented with phosphatase inhibitor mixture (see above) and protease inhibitor mixture (0.5 μm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 10 mm aprotinin, 1.0 mm leupeptin, 5.0 μm bestatin, and 1.0 μm pepstatin). The lysates were chilled for 20 min on ice before sonication with five 1-s pulses at 30 watts and 20% output. The lysate was centrifuged at 13,000 × g, and the total protein concentration of the supernatant was determined using the bicinchoninic acid assay (Pierce) with bovine serum albumin as the protein standard. Sample loads were normalized for total protein concentration before reducing with dithiothreitol and adding NuPAGE® lithium dodecyl sulfate sample buffer and then were boiled for 7 min at 90 °C.
Serial frozen tissue biopsy specimens from a Ménétrier's disease patient were provided for this study as lysates generated using the TFE and ammonium bicarbonate lysis protocol described below. Prior to Western blot analysis, aliquots of the lysates were lyophilized and resuspended in HPLC water three times to remove residual TFE before resuspending in radioimmune precipitation assay buffer. All of the denatured samples were then resolved using NuPAGE® 10% BisTris gels at 160 V for ∼65 min in either MOPS or MES SDS running buffer (Invitrogen). Proteins were electrophoretically transferred from the gel to a polyvinylidene difluoride membrane for 3 h at 35 V at 4 °C.
The membranes were probed using primary antibodies to EGFR, pY1173, pY998, Jagged-1, c-Jun, Claudin 4, 4G10 (for global phosphotyrosine analysis), or β-actin overnight at 4 °C. AlexaFluor® 680-labeled goat anti-rabbit and goat anti-mouse secondary antibodies were used to detect the corresponding primary antibodies. Immunoreactive proteins were visualized using the OdysseyTM infrared imaging system and Odyssey software as described by the manufacturer (Li-Cor, Lincoln, NE).
Cell and Tissue Preparation for MS Analyses
A431, DiFi, and HCT116 cell pellets were resuspended and lysed in 100 μl of ammonium bicarbonate (100 mm, pH 8.0) and 100 μl of TFE and then incubated at 60 °C for 60 min at 1000 rpm on an Eppendorf Thermomixer before sonicating at 30 watts and 20% output for ten 1-s pulses. Protein concentration was assessed using the bicinchoninic acid assay, and equivalent sample aliquots were reduced with 40 mm tris-carboxyethylphosphine and 100 mm dithiothreitol at 60 °C for 30 min at 1000 rpm on the Eppendorf Thermomixer and then incubated at ambient temperature in the dark with 200 mm iodoacetamide. The samples were diluted with 50 mm ammonium bicarbonate, pH 8.0 (to 10% TFE) before adding Trypsin Gold at a 1:50 (w/w) ratio and incubating overnight at 37 °C with shaking. Aliquots (200 μg) of digested samples were lyophilized. The samples were resuspended in 1 ml of water, vortex-mixed vigorously, and desalted with Sep-Pak® 100 mg, C-18 columns (Waters) on a vacuum manifold. After washing with water, the peptides were eluted with 80% ACN and evaporated to dryness in vacuo. Digests from A431 lysate for shotgun analyses were then subjected to IEF as described below. Digests for MRM analysis were reconstituted in 0.1% (v/v) aqueous formic acid to a final concentration of 0.5 μg/μl, and the isotopically labeled β-actin peptide reference standard was spiked in at a concentration of 20 fmol/μl.
DiFi and HCT116 derived mouse tumor xenograft samples were acquired as FFPE tumor blocks. Three 30-μm slices were cut from each tumor block and placed into a single 1.5-ml Eppendorf tube. These were processed as we described previously (27). Rehydrated mouse xenograft samples and Ménétrier disease lysates were prepared for MRM analysis as described above for cultured cells, except that in the initial protein solubilization step, the tissue samples were sonicated three times at 30 watts and 20% output continuously for 20 s both before and after 60 min of incubation in 50 mm ammonium bicarbonate with 50% TFE.
Isoelectric Focusing of Peptides
Tryptic peptide mixtures corresponding to 200 μg of protein were resuspended in 155 μl of 6 m urea. The samples were loaded into sample loading wells of the ZOOM IPGRunner cassette (Invitrogen). ZOOM strips (10 cm) with immobilized pH gradient gel from 3.5–4.7 were placed into cassette and loaded with samples at room temperature for 1 h. Cassette was placed into a ZOOM IPGRunner cell and run for 15 min at 175 V, ramped from 175 to 2000 V over 45 min, and held at 2000 V for 105 min. The gel strips were then cut into 15 equal 4-mm segments, which were each placed in separate wells of a 96-well ELISA plate. The peptides were eluted from the strips as follows: 200 μl of 0.1% formic acid (FA) in water for 15 min; 200 μl of 50% ACN, 0.1% FA for 15 min; and 200 μl of 100% ACN, 0.1% FA for 15 min. Extracted peptides for each fraction were pooled, evaporated in vacuo, resuspended in 1 ml of 0.1% TFA and desalted over a 96-well, C18 Oasis hydrophilic-lipophilic balance 30-μm (μElution) plate (Waters Corp.). After washing with HPLC water and stepwise elution (200 μl of 30% CAN, 0.1% TFA; 200 μl of 70% ACN, 0.1% TFA; and 200 μl of 100% ACN, 0.1% TFA), the eluates for each IPG strip fraction were pooled, evaporated in vacuo, resuspended in 100 μl of 0.1% (v/v) FA in water, and placed in sample vials for LC-MS/MS analysis.
Reverse Phase LC-MS/MS
LC-MS/MS analyses were performed on an LTQ-XL mass spectrometer from Thermo Fisher Scientific equipped with an Eksigent nanoLC 1D plus pump and Eksigent autosampler (Dublin, CA). The peptides were resolved on 100 μm × 11 cm fused-silica capillary column (Polymicro Technologies, LLC Phoenix, AZ) packed with 5 μm, 300 Å Jupiter C-18 resin (Phenomenex, Torrance, CA) with an in-line solid phase extraction column (pre-column, 100 μm × 6 cm) packed with the same C-18 resin (using a frit generated with liquid silicate Kasil) similar to that previously described (31). LC was carried out at ambient temperature over 85 min using a gradient mixture of 0.1% (v/v) FA in water (solvent A) and 0.1% (v/v) FA in ACN (solvent B). A 10-min load period using 100% solvent A at 1 μl/min was followed by an elution gradient (600 nl/min) from 2 to 25% solvent B in 30 min, 25 to 90% solvent B over 15 min, and held at 90% solvent B for 17 min before returning to 2% solvent B to equilibrate column. Peptides eluting from the capillary tip were introduced into the LTQ source in microelectrospray mode with a capillary voltage of ∼2 kV. A full scan was obtained for eluting peptides in the range of 400–2000 atomic mass units followed by five data-dependent MS/MS scans of the most intense ions. MS/MS spectra were recorded using dynamic exclusion of previously analyzed precursors for 60 s with a repeat of 1 and a repeat duration of 1. MS/MS spectra were generated by collision-induced dissociation of the peptide ions at normalized collision energy of 35% to generate a series of b- and y-ions as major fragments.
MRM Analyses
MRM analyses were performed on a TSQ Vantage triple quadrupole mass spectrometer from Thermo Fisher Scientific equipped with an Eksigent NanoLC-ultra one-dimensional Plus pump (Dublin, CA) and a capillary column and precolumn similar to that described above. The mobile phase consisted of the gradient mixture of solvent A and solvent B used for LC-MS/MS analysis. Sample solutions (2 μl) containing 0.5 ng/μl peptide mixture (based on protein concentration) were loaded for 15 min onto the column using 100% solvent A at 1 μl/min followed by a gradient elution (400 nl/min) from 3 to 20% solvent B in 7 min, 20 to 60% solvent B over 35 min, 60 to 95% solvent B in 6 min, and held at 95% for 11 min before returning to 3% solvent B. MRM analyses of target peptides and β-actin peptides (isotope labeled and endogenous) were performed using a 1300 V electrospray voltage, 210 °C capillary temperature, and −5 V skimmer offset. Both Q1 and Q3 were set at unit resolution FWHM 0.7 Da and collision gas (helium) pressure in Q2 was held at 1.5 mTorr. Scan width was 0.004 m/z, and scan time was 10 ms for all analyses. Collision energy for each peptide was calculated using the open source software Skyline (32). Instrument quality control assessment was done as described previously (33).
Quantitative analyses were done by the method we described recently (33) using U-13C6, U-15N4-arginine β-actin peptide (GYSFTTTAE*R) as the reference standard. At least four MRM transitions were monitored for target peptides and the standard. Signature peptides for each protein measured were required to be between 7 and 25 amino acids long and were selected based on uniqueness and chemical stability. Although priority was given to peptides that were previously identified in the shotgun data set with high MS/MS spectral quality, additional peptides were selected by in silico digestion. Peptides containing cysteine or methionine residues were not excluded, and cysteines were present as carboxyamidomethylated derivatives following treatment with iodoacetamide during sample workup. Peptide uniqueness was confirmed by BLAST searching sequences against the UniProt database. Skyline software was used to extract and integrate transition peak areas for each target peptide. Dot product scores (≥0.85) and predicted retention times computed by Skyline were used as criteria to confirm peak area identifications (32). Summed peak areas for target peptide transitions were divided by the summed peak area for the reference standard peptide transitions to give normalized peak area (NPA), and coefficients of variation (CVs) were calculated across replicates for each treatment. For MRM analyses in cell culture experiments, three replicate cell cultures were analyzed for each cell culture and treatment. For DiFi cell xenografts, four replicate samples were analyzed, and five replicate HCT116 xenografts were analyzed; for Ménétrier disease, three separate gastric tissue biopsy samples were analyzed.
Data Analysis
The “ScanSifter” algorithm v2.0.4 read MS/MS spectra stored as centroided peak lists from Thermo RAW files and transcoded them to mzData files (34). Only MS/MS scans were written to the mzData files; MS scans were excluded. If 90% of the intensity of a MS/MS spectrum appeared at a lower m/z than that of the precursor ion, a single precursor charge was assumed; otherwise the spectrum was processed under both double and triple precursor charge assumptions. MS/MS spectra were assigned to peptides from the IPI Human database version 3.56 (May 5, 2009; 76591 entries) by the MyriMatch algorithm, version 1.6.33 (35). The sequence database was doubled to contain each sequence in both normal and reversed orientations, enabling a false discovery rate (FDR) estimation. MyriMatch was configured to expect all cysteines to bear carboxamidomethyl modifications and to allow for the possibility of oxidation on methionines. Candidate peptides were required to feature trypsin cleavages or protein termini at both ends, although any number of missed cleavages was permitted. A precursor error of 1.25 m/z was allowed, but fragment ions were required to match within 0.5 m/z. The IDPicker algorithm v2.6.1 (36) filtered the identifications for each reverse phase liquid chromatography run to include the largest set for which a 5% peptide level identification FDR could be maintained, as described by Qian et al. (37). Indistinguishable proteins were recognized and grouped, and parsimony rules were applied to generate a minimal list of proteins (protein groups) that explained all of the peptides that passed our entry criteria (36). This approach uses bipartite graph analysis to derive a minimal list of protein identifications with shared clusters of peptides. These identifications were pooled for each IEF sample set (15 fractions). Proteins were required to have at least two different peptide sequences observed within an IEF sample set. FDR for peptide identifications were computed by the formula (38): FDR = (2 × reverse)/(forward + reverse). The algorithm reported the number of spectra and number of distinct sequences observed for each protein and protein group in each sample set.
Statistical Analyses
Comparisons of protein abundance from shotgun data sets were made with spectral count data (39) using QuasiTel, a statistical software modeling package (40). QuasiTel uses a quasi-likelihood method based on Poisson distribution—commonly used for count data (41)—and applies a regression model to compare spectral count data. This statistical model was used to perform pair-wise comparisons between two treatment conditions (six replicates/condition), which generated a single combined inventory of protein identifications (comparison data set); the model also uses F-tests to compute p values and the FDR method to correct for multiple hypothesis comparisons of identified proteins (42). Thresholds were set for p values (≤0.20), total spectral counts (11), and spectral count log2 rate ratios (fold changes ≥ 2) generated by this model and were used as criteria to filter comparison data sets. Significance of measured differences for target proteins of all MRM analytes was determined with a two-tailed unpaired t test using Prism 5.0 (GraphPad Software, San Diego, CA).
RESULTS
EGFR Modulation in A431 Cells
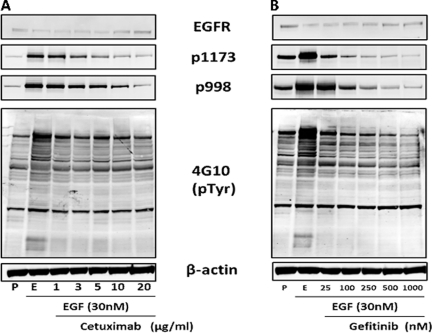
To establish the A431 cell model, we first established conditions for activation and inhibition of EGFR signaling. We measured the phosphorylation status of the EGF receptor as well as global tyrosine phosphorylation by Western blot analysis (Fig. 1). Cells proliferating in the presence of serum served as baseline controls, whereas serum-starved cells treated with EGF served as the benchmark for EGFR activation. After treatment with 30 nm EGF for 4 h, EGFR phosphorylation at residues Tyr-998 and Tyr-1173 increased dramatically, as did global phosphotyrosine levels indicating activation, as measured with the antiphosphotyrosine antibody 4G10. EGFR protein expression inversely varied with activation, which reflects enhanced receptor internalization and down-regulation upon activation (43–46). Increasing concentrations of both cetuximab and gefitinib lead to a decrease of EGFR phosphorylation at Tyr-1173 and Tyr-998 compared with EGF only stimulated cells. At the concentrations used, the tyrosine kinase inhibitor gefitinib produced a more profound inhibition of EGFR phosphorylation than did the ligand-binding domain inhibitor cetuximab. The highest concentration of cetuximab (20 μg/ml) only reduced EGFR phosphorylation to basal levels (Fig. 1A), whereas gefitinib at 250 nm reduced both Tyr-1173 and Tyr-998 phosphorylation well below basal levels (Fig. 1B). Total tyrosine phosphorylation detected in EGF-stimulated cells showed an increase over signals detected in proliferating cells, and although both inhibitors reduced detected levels of total tyrosine phosphorylation, only gefitinib was capable of reducing tyrosine phosphorylation below basal levels.
Fig. 1.
Activation and inhibition of EGFR in A431 cells. A431 cells were serum-starved overnight before being treated with either 30 nm EGF (lanes E) for 4 h or co-treated first with cetuximab (A) or gefitinib (B) at the indicated concentrations for 30 min prior to 4 h of treatment with EGF. Proliferating cells (lanes P) were not serum-starved and serve as a reference control. Immunoblots were performed for total EGFR, sites of EGFR phosphorylation at Tyr(P)-1173 and Tyr(P)-998, and total tyrosine phosphorylation was detected with the 4G10 antibody. β-Actin was used as a loading control.
With the exception of Tyr(P)-998 in cetuximab-treated cells, minimal differences were noted between EGFR and total tyrosine phosphorylation status for the two highest concentrations of each inhibitor (10 and 20 μg/ml for cetuximab or 500 and 1000 nm for gefitinib). Based on these results, a single concentration each for cetuximab (10 μg/ml) and gefitinib (500 nm) was selected for subsequent experiments. At the selected concentrations, both inhibitors significantly and reproducibly inhibited EGF receptor signaling. Although changes in phosphorylation were noted after 4 h, EGF treatment did not induce apoptosis (supplemental Fig. S1). A slight rounding of A431 cells in culture was noted for EGF-treated cells, in agreement with previous reports (47, 48), Cells co-treated with inhibitor and EGF retained an unaltered morphology. Neither EGF nor EGFR inhibitors produced significant detachment of cells from the culture dish.
Global Protein Expression Analyses
We performed shotgun proteome analyses of the four A431 treatment conditions described above: proliferating cells, EGF-treated, EGF- and gefitinib-treated, or EGF- and cetuximab-treated. The cells were collected and lysed, tryptic digests were prepared and resolved by IEF, and the IEF fractions were analyzed by data-dependent LC-MS/MS. A summary of the global protein analysis is shown in Table I. The values displayed are the averaged data from three separate cultures and two process replicates of each culture per treatment condition. Similar values for confident spectral identifiers (numbers of spectra matched to database sequences at 5% FDR) and protein groups (numbers of indistinguishable protein identifications supported by the identified peptides), and protein group CVs under 5% demonstrate high reproducibility of the analyses across biological replicates. A complete list of protein identifications is provided in supplemental Table S1.
Table I. LC-MS/MS data summary.
The table lists confident identifications (number of spectra successfully matched to peptides), protein groups, and %CV of replicate analysis for each of the treatment conditions. Protein group numbers are averages of six replicates (three separate cultures and two process replicates of each) per treatment (30 nm EGF, 10 μg/ml cetuximab, 500 nm gefitinib). A 5% peptide level FDR and minimum of two distinct peptides were required for confident protein identification. A protein group is defined by a set of proteins that cannot be distinguished based on the peptide identifications. A single protein is chosen to represent each protein group.
| Proliferating | EGF stimulated | Gefitinib + EGF | Cetuximab + EGF | |
|---|---|---|---|---|
| Confident peptide-spectrum matches | 32567 | 36156 | 34105 | 35322 |
| Protein groups | 5203 | 5294 | 5347 | 5250 |
| Protein group CV (%) | 4.4 | 4.8 | 4.9 | 4.5 |
Data Set Comparisons, Data Filtering, and Derivation of a Candidate EGFR Inhibition Signature
Next, we compared protein expression data sets from the four A431 cell conditions to identify proteins whose levels were changed by EGFR activation and for which the changes were reversed by the inhibitors. Protein expression was compared on the basis of spectral counts using QuasiTel, a quasi-likelihood modeling software package (40). QuasiTel performed pair-wise comparison between two treatment/biological conditions based on protein spectral counts and variance across replicate analyses and computed p values and rate ratios (fold changes) for detected proteins.
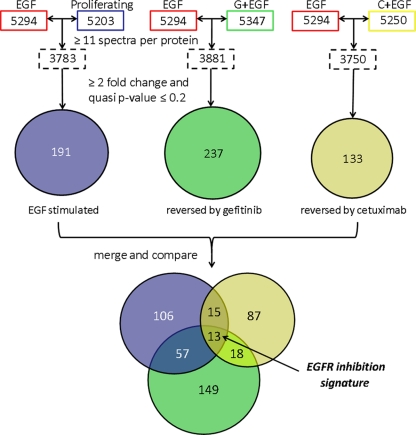
Pair-wise comparisons were made between 1) proliferating and EGF-stimulated cells, 2) EGF-stimulated and EGF/gefitinib-treated cells, and 3) EGF-stimulated and EGF/cetuximab-treated cells (Fig. 2, colored boxes). To control the protein level FDR, comparisons were limited to proteins with ≥11 spectral counts per protein across replicates. Accordingly, the three comparisons included 3783, 3881, and 3750 proteins, which corresponded to protein level FDRs of 3.3, 3.1, and 3.3% respectively. The colored circles in Fig. 2 represent proteins differentially expressed (both up and down) between treatments with a fold change of ≥2.0 and p values of ≤0.20. The blue circle (191 proteins) represents proteins differentially expressed between EGF-stimulated cells and nontreated, proliferating cells. The green circles (237 proteins) and yellow circles (133 proteins) signify proteins whose EGF-stimulated changes were reversed by gefitinib and cetuximab inhibition, respectively. Supplemental Tables S2–S4 provide a complete list of proteins in each of these groups. The group reversed by cetuximab is only approximately half as large as the group reversed by gefitinib, which is consistent with the lesser degree of EGFR inhibition produced by cetuximab in our model (Fig. 2), as well as with possible off target effects of gefitinib because of its inhibition of multiple kinases (49, 50).
Fig. 2.
Filtering and comparison of A431 proteome data sets. Colored rectangles show protein identifications from global proteomic analyses of the four treatment groups. Dashed rectangles show number of proteins in comparison data sets that were filtered to have ≥11 spectral counts/protein across replicates. Comparisons of protein spectral counts between treatment groups were performed by quasi-likelihood analysis, and a p value and spectral count rate ratio (fold change) were generated. The colored circles represent lists of proteins differentially expressed (both up and down) between treatments with a fold change of ≥2.0 and quasi p values of ≤0.20. The resulting protein groups represent proteins differentially expressed in response to EGF (blue), EGF-induced protein changes reversed by gefitinib (green), and EGF-induced protein changes reversed by cetuximab (yellow). The Venn diagram comparison indicates proteins whose expression changes are shared by the different experimental conditions. The central overlap indicates the “EGFR inhibition signature,” which refers to EGF-stimulated protein expression changes reversed by both inhibitors.
The Venn diagram (Fig. 2) compares these three groups of proteins to explore the common protein expression changes associated with EGF stimulation and EGFR inhibition. The three groups comprised 445 proteins, of which 13 proteins were shared between all three groups. Another 90 proteins were shared between two groups, and most (342 proteins) belonged to only a single group. The center overlap contained 13 proteins whose expression was significantly changed by EGF treatment, and these changes were reversed by both inhibitors. We will refer to this group hereafter as the “EGFR inhibition signature.” The 13 proteins are: CCDC50 (coiled-coiled domain containing 50), CDKN1A (cyclin-dependent kinase inhibitor 1A, p21), CLDN4 (claudin 4), COBLL1 (cordon-bleu-like protein 1), JAG1 (Jagged-1), JUN and JUND (proto-oncogenes c-Jun and JunD, respectively), PRIM1 (DNA primase subunit 1), RBM15 (RNA-binding motif protein 15), TOMM20 (translocase of outer mitochondrial membrane 20), TRAPPC3 (trafficking protein particle complex 3), TRIM 32 (tripartite motif containing protein 32), and GFPT2 (glutamine-fructose-6-phosphatetransaminase 2).
The Venn diagram also indicates protein subgroups with expression patterns similar to those in the EGFR inhibition signature. For example, EGF-induced expression changes were reversed only by cetuximab for 15 proteins and only by gefitinib for 57 proteins. Another 18 proteins were reversed by both cetuximab and gefitinib, but these proteins were not significantly elevated by EGF treatment at the selected cut-offs. The proteins found outside of any overlap showed significant differences between EGF stimulation and either nontreated (proliferating) or inhibitor-treated cells; however, these protein differentials were unique to individual treatments and comparisons. Supplemental Table S5 lists the proteins found in each section of the Venn diagram.
These protein groupings, including the EGFR inhibition signature, depend on the selected thresholds for fold change and p values. More lenient thresholds may increase detection of false-positive differences, whereas more stringent criteria may decrease detection of true differences. Manipulation of thresholds for p value and fold change can shift some proteins from one classification to another in the Venn diagram (supplemental Fig. S2).
MRM Verification of EGFR Inhibition Signature in A431 Cells
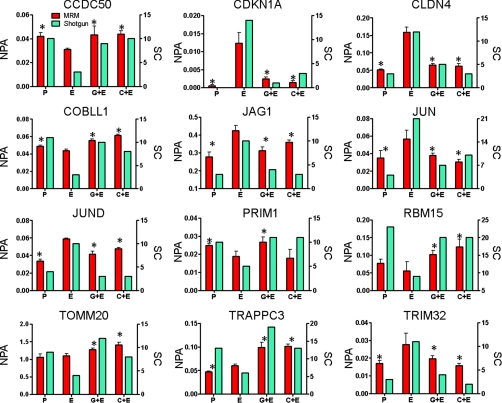
To verify expression differences for the 13 proteins in the EGFR inhibition signature, we performed MRM analyses using the labeled reference peptide method that we described recently (33) to quantitate target peptides with reference to a labeled β-actin peptide standard. A431 cells were treated as described for global proteome comparisons, but no IEF fractionation was performed. The data generated by MRM analyses are displayed together with spectral count data from the shotgun analyses (Fig. 3). MRM data were from analyses of three separate cultures for each treatment, and the NPA for a single, proteotypic peptide is shown for each protein. The spectral count data (Fig. 3, teal bars) display the total number of spectra identified for the corresponding protein across six replicate shotgun analyses (three separate cultures and two process replicates of each culture). The peptide sequences, monitored transitions, peptide and normalized peak areas, and CVs for the MRM data in Fig. 3 can be found in supplemental Table S6. CVs for NPA values calculated measured peptides across the treatment conditions for all but two of the peptides were below 30%. For 39 of these, the CVs were less than 18%. MRM analyses for a second, unique peptide for 10 of the 12 EGFR inhibition signature proteins produced similar results (supplemental Fig. S3).
Fig. 3.
Normalized MRM data and spectral count correlation of EGFR inhibition signature proteins. Each panel shows both MRM data (red bar) and spectral count data (teal bar) for each protein of interest across four A431 cell treatment conditions: proliferating cells (P), EGF-treated (E), EGF and gefitinib (G+E), and EGF and cetuximab (C+E). The left y axis is the NPA, which is the total MRM transition peak area for the target peptide divided by the peak area for the β-actin-labeled reference peptide. MRM data are representative of one unique peptide for each target protein across three separate cultures. Spectral counts (SC) are the total number of spectra identified for each protein across replicate analyses. *, significant difference compared with EGF treated cells as determined by Student's two-tailed, unpaired t test.
MRM analyses detected peptide transitions corresponding to 12 of the 13 EGFR inhibition signature proteins. None of the peptides monitored for GFPT2 (glutamine-fructose-6-phosphatetransaminase 2) produced detectable signals. This protein was not considered further. For the remaining 12 proteins, MRM analyses verified almost all of the expression differences detected by shotgun analyses (Fig. 3). EGF-induced protein expression compared with proliferating cells was significantly different by MRM in 10 of the 12 proteins, and the trends in the others were consistent with spectral count differences, except for TOMM20 and RBM15. Significant reversal of EGF-induced expression was verified by MRM for both inhibitors for all 12 proteins, except for the effect of cetuximab on PRIM1, which fell short of statistical significance, although the trend was consistent with the spectral count comparison. All of the protein expression differentials measured by spectral counts and MRM were consistent in direction, whereas the magnitude of the measured differences varied between the two analysis platforms. This reflects the greater precision of MRM measurements over the dynamic range of protein expression analyzed.
Confirmation of EGFR Inhibition Signature Protein Changes in DiFi and HCT116 Cell Lines
Our next studies analyzed changes in EGFR inhibition signature proteins in cells that differ in response to EGFR inhibitors. We used MRM analyses to measure the 12 EGFR inhibition signature proteins in DiFi and HCT116 colorectal cancer cell lines. DiFi cells demonstrate both gene amplification and protein overexpression of the EGF receptor and also harbor an adenomatous polyposis coli gene mutation frequently found in hereditary colorectal cancers (30, 51). DiFi cells thus would be expected to show similar responses to A431 cells. HCT116 cells show increased autocrine production of the EGFR-stimulating ligand transforming growth factor-α and carry a mutant allele (G13D) of the KRAS protoncogene (52, 53). Because KRAS mutations block clinical and cellular responses to EGFR inhibitors, such as cetiximab, we hypothesized that an EGFR inhibition signature would be minimal or absent in the HCT116 cells.
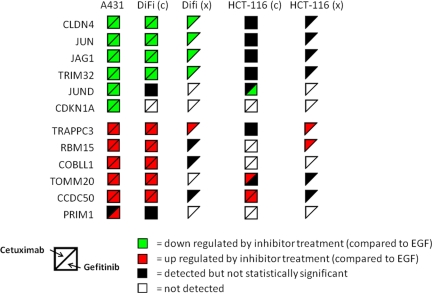
Both cell lines were treated with EGF and inhibitors as described above, and the 12 EGFR inhibition signature proteins were analyzed. Peptides for 11 of the 12 proteins were successfully monitored in DiFi cells, whereas peptides for only eight of the proteins were detected in HCT116 cells (Fig. 4). The COBLL1, PRIM1, and RBM15 peptides monitored in HCT116 cells produced no detectable signals, and CDKN1A peptide signals were absent in both cell lines. Most of the peptide signals in DiFi cells monitored showed statistically significant differences between EGF only and inhibitor-treated cells (Fig. 4, DiFi(c)) consistent with the trends noted in A431 cell experiments (Fig. 4, A431). Only JUND and PRIM1 failed to show significant changes. HCT116 cells were largely resistant to changes in EGFR inhibition signature proteins, with significant differences detected only for CCDC50, JUND, and TOMM20. CCDC50 peptide signals were significantly up-regulated in both inhibitor treated conditions when compared with EGF only samples—in agreement with A431 and DiFi cell MRM data—whereas JUND and TOMM20 peptides showed substantial differences for only gefitinib or cetuximab treatments, respectively (Fig. 4, HCT116(c)). Plots of MRM data for DiFi and HCT116 cells are presented in supplemental Figs. S4 and S5, and supplemental Tables S8 and S9 list the peptide peak areas, normalized peak areas, averages, and CVs for the EGFR inhibition signature peptides monitored in the two cell lines.
Fig. 4.
Summary of expression changes for EGFR inhibition signature proteins in cell lines and mouse xenograft models. Symbol colors indicate decreased expression (green); increased expression (red); detected, but no significant change (black) compared with EGF treatment in cells (c) or compared with vehicle control in xenografts (x); and not detected (white). Only cetuximab treatment was used in xenograft experiments.
To confirm the expression of the EGFR inhibition signature in the DiFi and HCT116 cells, we required at least half of the detectable signature proteins to show expression changes consistent with changes measured in the A431 model. The MRM data for DiFi cells demonstrate expression changes consistent with those in A431 cell—11 of the 12 signature proteins were detected, and nine showed significant expression differences. None of the proteins showed changes opposite those observed in A431 cells. This result for DiFi cells is consistent with their overexpression of EGFR and responsiveness to EGFR inhibitors. In contrast, the HCT116 cells display expression changes sensitive to both inhibitors in only one of the eight detectable signature proteins (CCDC50) and responded only to a single inhibitor for two others (JUND and TOMM20). By our criteria, the HCT116 cells do not display an EGFR inhibition signature, a result that is consistent with the effect of the heterozygous G13D KRAS mutation, which blocks responses to EGFR inhibitors (53).
Immunoblot Confirmation of EGFR Inhibition Signature
Further confirmation of protein changes in response to EGFR perturbation was done by immunoblotting with antibodies to p-EGFR, JAG1, JUN, and CLDN4 in the A431, DiFi, and HCT116 cell lines across all treatments (supplemental Fig. S6). In all cell lines, an increase was observed in Tyr(P)-998 and Tyr(P)-1173 signals in EGF-treated cells relative to proliferating. In A431 and DiFi cells treated with gefitinib, EGFR Tyr(P)-998 and Tyr(P)-1173 both were decreased to basal (proliferating) levels, whereas a less profound, yet still detectable decrease was observed in HCT116 cells. Cetuximab treatment (at 10 μg/ml) in all three cell lines partially inhibited EGFR autophosphorylation. For EGFR, Tyr(P)-1173 in DiFi cells and Tyr(P)-998 in HCT116 cells, no change in phosphotyrosine immunoblot signal was detected between EGF only and cetuximab-treated cells (supplemental Fig. S6). Protein expression changes in all three cell lines measured by immunoblot were consistent with MRM data. In A431 and DiFi cell lines, JAG1, JUN, and CLDN4 were increased in EGF treated compared with proliferating cells. In both A431 and DiFi cells, gefitinib decreased JAG1, JUN, and CLDN4 expression, whereas cetuximab produced partial reversal of EGF stimulation for these proteins. In HCT116 cells, EGF elevated JAG1, but the inhibitors did reverse this effect.
Confirmation of EGFR Inhibition Signature Protein Changes in Mouse Xenograft Tumors
To further confirm protein changes in the EGFR inhibition signature in tissue specimens, we analyzed mouse xenograft tumors derived from DiFi and HCT116 cell lines. These xenograft samples come from our previous study, in which we used novel optical imaging probes to study therapeutic responses to cetuximab in vivo (29). DiFi and HCT116 xenografts were grown in athymic nude mice. Tumor-bearing mice were then treated with 40 mg/kg cetuximab or saline vehicle intraperitoneally every 3 days for 1 week (three total injections). Image analysis demonstrated a significant decrease in EGF probe uptake in DiFi xenografts and increased apoptosis in cetuximab-treated mice, whereas HCT116 xenografts showed neither effect. Gefitinib was not employed in this study.
We analyzed FFPE sections from the xenograft tumors by MRM to measure the 12 EGFR inhibition signature proteins. Analyses of xenograft tumors from vehicle-treated mice were used for reference to assess the effects of cetuximab treatment. MRM analyses detected 8 of the 12 EGFR inhibition signature proteins in each xenograft type (see Fig. 4, DiFi(x) and HCT116(x)). DiFi-derived tumors from cetuximab-treated mice showed decreased expression of CLDN4, JAG1, JUN, and TRIM32 and an increase in TRAPPC3 when compared with tumors from vehicle-treated mice. These differences are similar to those between EGF-treated and only to cetuximab/EGF-treated DiFi cells, although the xenograft analyses yielded no statistically significant differences for CCDC50, COBLL1, or RBM15. MRM data for these experiments are provided in supplemental Fig. S7 and Table S10.
HCT116 xenografts from cetuximab-treated mice showed a significant increase in TRAPPC3 and RBM15 compared with vehicle-treated tumors, but the other proteins in the EGFR inhibition signature were either unchanged or were not detected (Fig. 4, HCT116(x)). Of note are the differences between the EGFR inhibition signatures for the treated HCT116 cells and the HCT116 mouse tumors. In HCT116 cells, cetuximab increased CCDC50 and TOMM20, but not in the HCT116 xenografts. The lack of consistent inhibitor-induced protein expression differences in the HCT116 xenograft samples is consistent with the lack of response to cetuximab in vivo (29). MRM data for these experiments are provided in supplemental Fig. S8 and in Table S11. An important aspect of this study is the analysis of the EGFR inhibition signature in archival FFPE tissue specimens. Although a few of the measured peptides yielded CVs ∼40%, most yielded CVs <30%.
Confirmation of the EGFR inhibition signature was assessed by the criteria described above—changes concordant with the A431 model in at least 50% of the measurable signature proteins. The signature was confirmed in DiFi xenografts (five concordant changes in eight detectable signature proteins) but not in HCT116 xenografts (two concordant changes in eight detectable signature proteins).
Use of the EGFR Inhibition Signature to Assess Therapeutic Response to Cetuximab in Ménétrier's Disease
A potential use of the EGFR inhibition signature is to assess a response to drug treatment. To test the applicability of the EGFR inhibition signature to assess the clinical response to EGFR inhibition, we analyzed serial biopsies from a single Ménétrier's disease patient treated with cetuximab, as described in our recent study (28). Ménétrier's disease is a rare hypertrophic gastropathy characterized at the molecular level by high expression of transforming growth factor-α and constitutively activated EGFR signaling (54, 55). Symptoms of Ménétrier's disease are highly responsive to cetuximab therapy, which rapidly and dramatically reverses most effects of the disease (28, 56).
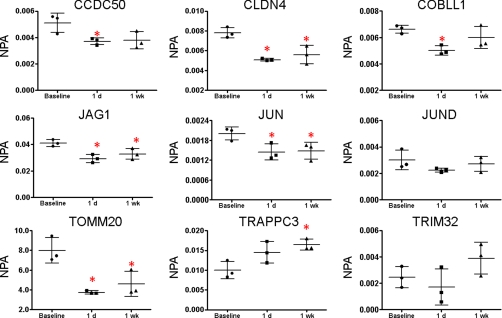
One patient from our recent study (28) showed improved symptoms and a histologically normal stomach after the initial 1-month trial. Baseline (no treatment) specimens as well as serial biopsies from 1 day and 1 week after the initial cetuximab treatment were analyzed in triplicate (three separate analyses of pooled biopsies from several sites in the stomach) from this patient. MRM analyses detected peptides from 9 of the 12 EGFR inhibition signature proteins (Fig. 5). (The MRM data are presented in expanded form in supplemental Table S12.) Because our cell model for EGFR inhibition reflected an acute response to EGF and inhibitors, we focused on protein expression changes after 1 day and 1 week of cetuximab treatment (biopsies were also collected at 1 and 4 months of continued treatment). CLDN4, JAG1, and JUN all were significantly decreased relative to baseline after 1 day. JUND and TRIM32 appeared decreased, although the effect was not significant. These changes all are consistent with the effects of cetuximab treatment in the A431 and DiFi cell models. In contrast, CCD50, COBBL1, and TOMM20 all were decreased by 1 day of cetuximab treatment, in contrast to the increases in these proteins produced by cetuximab in the A431 and DiFi models. At 1 week of cetuximab treatment, CLDN4, JAG1, and JUN were still decreased relative to pretreatment, consistent with responses to cetuximab treatment in A431 cells, DiFi cells, and the DiFi xenograft model. Expression of JAG1, JUN, and CLDN4 expression was also measured by immunoblotting (supplemental Fig. S9). Consistent with MRM data, JAG1 and CLDN4 were significantly decreased from the baseline sample at 1 day and 1 week post-treatment. JUN expression measured by immunoblotting did not change with cetuximab treatment.
Fig. 5.
MRM analyses of EGFR inhibition signature proteins in tissue biopsies from a Ménétrier's disease patient treated with cetuximab. Gastric tissue biopsies were collected from three separate locations prior to treatment (baseline control) and at 1 day and 1 week after treatment with cetuximab. The NPA, which is the total MRM transition peak area for the target peptide divided by the peak area for the β-actin-labeled reference peptide. The data points are representative of three separate biopsies taken from a single patient with mean and standard deviation shown. *, significant difference between baseline and time post initial treatment as determined by Student's two-tailed t test.
By the criteria described above, these results do not confirm the EGFR inhibition signature. Three of the signature proteins (CCD50, COBBL1, and TOMM20) showed changes opposite those in the A431 model. Nevertheless, the three proteins (CLDN4, JUN, and JAG1) showing changes concordant with the A431 model were most consistently responsive to EGFR inhibition in all of our experiments.
DISCUSSION
Direct analysis of phosphorylated receptor proteins and their downstream effectors is the most commonly employed method to assess signaling networks. This approach is complicated by the transient nature of protein post-translational modifications, their low abundance relative to unmodified proteins and potential artifacts caused by uncontrolled preanalytical variables. We asked whether analysis of changes in protein expression could indicate responses of a signaling network. The rationale for this concept is 2-fold. First, protein expression is easier to measure and is less labile than phosphorylation or other post-translational modifications. Second, signaling pathways ultimately drive gene and protein expression changes, which are most directly linked to phenotypes. A protein expression-based analysis could be routinely applied to clinical specimens, including archival FFPE tissues, where direct measurements of phosphorylated intermediates may be neither practical nor valid. Here we tested the hypothesis that manipulation of the EGFR signaling axis with clinically used drugs generates characteristic protein expression changes. Our results confirm the hypothesis and demonstrate a prototypical approach to derive and test protein expression signatures for drug action on signaling networks.
The A431 cell model provided a reproducible and extensively studied (57–61) system to study EGFR-driven signaling events. Four treatment conditions were selected for comparisons: normal proliferating cells, cells stimulated with EGF, and cells preincubated with either gefitinib or cetuximab and then treated with EGF in the presence of the inhibitors. Our initial immunoblot analyses (Fig. 1) confirmed EGFR activation and inhibition. All treated A431 cells were serum-starved overnight to synchronize the cells and to limit potential interference by alternate EGFR ligands, which could complicate results. Serum starving the cells nevertheless introduced an inherent experimental difference between the proliferating (nontreated) cells harvested in the presence of 10% fetal bovine serum and the EGF-treated A431 cells incubated in serum-free medium. Shotgun and MRM analyses (data not shown) verified elevation of two proteins (HMGCR and HMCS1) and a decrease of one protein (A2MG) reported to be induced by serum starvation. These proteins were expressed at similar levels in all EGF-treated A431 cells (even in the presence of the inhibitors) compared with proliferating cells. This example illustrates the potential for experimental artifacts and underscores the importance of understanding the underlying biology of the discovery model.
Shotgun proteomic analyses of treated and nontreated A431 cells yielded highly reproducible proteomic inventories—CVs for numbers of protein identifications were less than 5% across three separate cultures for each treatment condition (Table I). The combined inventories comprised nearly 4,300 proteins identified at a protein level FDR of 4.7%. Quasi-likelihood analyses generated three pairwise comparisons (EGF versus proliferating, EGF versus gefitinib, and EGF versus cetuximab) based on shotgun spectral count data. These data sets were then filtered on the basis of spectral counts (≥11), fold-change (≥2), and p value (≤0.20) to generate the three protein groups representing EGF-simulated proteins, cetuximab-sensitive proteins and gefitinib-sensitive proteins (Fig. 2). A Venn diagram comparison of these proteins generated the EGFR inhibition signature proteins.
Three important points should be emphasized about the comparison process. First, the application of a minimum spectral count threshold (here ≥11 across all analysis runs in each data set comparison) is required to control protein level FDR. Without the threshold, protein FDR exceeded 30%, whereas with the threshold, protein FDR was ∼3%. Second, an important consideration in data set comparisons is the stringency of the cut-off criteria applied. We employed a relatively loose p value threshold (≤0.2) to maximize detection of differences in protein group comparisons. We reasoned that subsequent MRM analyses would identify false positive differences and confirm true positives that might not have been detected in the spectral count comparisons at higher stringency. We employed a 2-fold expression cut-off for the same comparisons. We experimented with different p value and fold change thresholds (supplemental Fig. S2) and ultimately settled on criteria that yielded an EGFR inhibition signature compatible in number of proteins with our capacity to perform subsequent MRM studies. Third, the interplay between p value and fold change threshold variation also affected the number and distribution of proteins in the overlap categories in Venn diagram analysis. For example, at thresholds of p ≤ 0.2 and a 2-fold difference, the central overlap (EGFR inhibition signature) in the Venn diagram contained 13 proteins, but at p ≤ 0.1, the central overlap contained only four proteins (JUN, JUND, CDKN1A, and RBM15) (supplemental Fig. S2, bottom row middle Venn diagram).
Despite the relatively loose criteria used to derive the EGFR expression signature, 11 of the 13 proteins were verified by MRM with expression changes similar to those observed from shotgun data (Fig. 4). One protein (GFPT2) proved difficult to monitor selectively by MRM and was not further studied, and another (PRIM1) showed significant reversal of EGF-induced change by gefitinib, but not by cetuximab. These MRM analyses targeted a single peptide for each of the proteins, but a parallel set of analyses targeting a second unique peptide for 10 of the EGFR inhibition signature proteins also verified the results of the spectral count comparisons (supplemental Fig. S3).
Although our shotgun analyses represented an empirical approach to discover a candidate signature, the component proteins have interesting and biologically plausible relations to EGFR signaling. A core group of proteins (e.g. JAG1, JUN, and CLDN4) displayed the most robust responses to EGFR inhibition. JAG1 (Jagged-1) is a Notch1 receptor ligand with reported links to EGFR expression (62, 63); moreover, up-regulation of JAG1 expression and Notch1 signaling has been noted in human colon adenocarcinomas and intestinal tumors (64–66) and associated with poor prognosis in breast cancer (67). An increase in JUN detection in EGF-treated samples is consistent with literature reports that indicate an increase in mRNA levels for the transcription factor upon EGF stimulation (68–70). Overexpression of JUN has also been linked with increased invasiveness and hormone resistance in breast cancer cells (71). Increased expression of the tight junction protein CLDN4 upon EGF treatment (72) is in agreement with our data. JUN and CLDN4 also are linked to EGF via the transcription factor Sp1 (73, 74). EGFR manipulation dramatically affected CDKN1A, a TP53-regulated cyclin-dependent kinase inhibitor (75), but only in A431 cells, which lack a functional TP53 protein. This protein may have utility to assess EGFR inhibition only in TP53 mutant tumors.
Once a candidate EGFR expression signature was established, we asked whether this set of protein expression changes can consistently represent the output of the EGFR signaling network in cells and tissues that differ in responsiveness to inhibitors. The EGFR inhibition signature in the DiFi cells was nearly identical to that in the A431 model, which is consistent with the high EGFR expression and responsiveness to inhibitors for both lines. In contrast, the HCT116 cells displayed only one consistent change (increase in CCDC50 by both inhibitors) observed in the A431 cells. This result is consistent with the impact of the KRAS mutant status of these cells, which blocks response to EGFR inhibition (53).
Analyses of DiFi- and HCT116-derived mouse xenograft models extended confirmation of the EGFR inhibition signature to tissue specimens. Although fewer targeted EGFR inhibition signature proteins were detected overall (8 of 12 for each xenograft type), there was again a clear difference in response of the signature proteins. Five of eight detected signature proteins displayed the expected expression changes in DiFi xenografts, whereas only two did in HCT116 xenografts (Fig. 5). An important aspect of the xenograft studies is that they demonstrate the potential to analyze protein expression signatures in FFPE specimens. Although we have previously demonstrated that shotgun proteome inventories of frozen and FFPE tissues are equivalent (27), the yield of proteins may be decreased. Indeed, comparison of the cell line and xenograft data also suggests a “loss of signal,” in that fewer of the signature proteins were quantifiable in the xenografts.
We evaluated the EGFR inhibition signature in Ménétrier's disease because of the dramatic response of this syndrome to cetuximab therapy (28). Our results indicate that the three most consistent elements of the signature (JUN, JAG1, and CLDN4) respond to cetuximab treatment in Ménétrier's disease. However, CCDC50, COBBL1, and TOMM20 expression changes were opposite to those in the A431 model. This discrepancy could reflect that fact that Ménétrier disease is a hyperproliferative disorder, but not a cancer, and thus may differ in other ways from the biology of the A431 discovery model and the DiFi and HCT116 models, which are all cancer cell lines. This point underscores the importance of matching the discovery model as closely as possible to the clinical application of an expression signature.
One potential complication in comparing EGFR-mediated responses in cell models and tissues is that endogenous levels of EGF and exposure cannot be controlled in tissues. Moreover, signaling effects driven in vivo by other cell types such as stroma may influence the expression of candidate signature proteins. These factors could contribute to a lack of complete agreement between cell and in vivo models. However, the most EGF- and EGFR inhibitor-responsive protein changes observed (JUN, JAG1, and CLDN4) were consistent across all of the model systems studied. Thus, proteins whose expression was significantly elevated by EGF and reversed by both inhibitors proved to be the most consistent indicators of EGFR inhibition in the other models.
Our main objective in this work was to determine whether protein expression signatures can represent the effects of drugs on a signaling network rather than to develop and refine a clinically useful EGFR inhibition signature. This latter objective would entail a more elaborate process that includes 1) multiple discovery systems to generate a more broadly based candidate signature, 2) integration of quantitative measurements with a valid statistical model to establish score thresholds and account for variability, and 3) systematic validation in a carefully selected, valid patient cohort. Nevertheless, some of the proteins identified in this study would appear likely to contribute to a clinically useful EGFR inhibition protein profile. Our results establish proof of concept for this approach by demonstrating that protein expression signatures detect activation and inhibition of dynamic signaling networks. Clinically useful response signatures developed through this approach could have broad impact in the field of cancer therapeutics.
Footnotes
* This work was supported by National Institutes of Health Grants U24CA126479 and U24CA159988 (to D. C. L.) and P50CA095103 and P50CA128323 (to R. J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- EGFR
- epidermal growth factor receptor
- FA
- formic acid
- MRM
- multiple reaction monitoring
- FFPE
- formalin-fixed, paraffin-embedded
- TFE
- 2,2,2-trifluoroethanol
- FBS
- fetal bovine serum
- MOPS
- 4-morpholinepropanesulfonic acid
- MES
- 4-morpholineethanesulfonic acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- NPA
- normalized peak area
- CV
- coefficients of variation.
REFERENCES
- 1. Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. (2005) Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250 [DOI] [PubMed] [Google Scholar]
- 2. Hinsby A. M., Olsen J. V., Mann M. (2004) Tyrosine phosphoproteomics of fibroblast growth factor signaling. J. Biol. Chem. 279, 46438–46447 [DOI] [PubMed] [Google Scholar]
- 3. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rexer B. N., Ham A. J., Rinehart C., Hill S., de Matos Granja-Ingram N., González-Angulo A. M., Mills G. B., Dave B., Chang J. C., Liebler D. C., Arteaga C. L. (2011) Phosphoproteomic mass spectrometry profiling links Src family kinases to escape from HER2 tyrosine kinase inhibition. Oncogene 30, 4163–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villén J., Elias J. E., Gygi S. P. (2007) Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 6, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 6. Espina V., Edmiston K. H., Heiby M., Pierobon M., Sciro M., Merritt B., Banks S., Deng J., VanMeter A. J., Geho D. H., Pastore L., Sennesh J., Petricoin E. F., 3rd, Liotta L. A. (2008) A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol. Cell. Proteomics 7, 1998–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Espina V., Mueller C., Edmiston K., Sciro M., Petricoin E. F., Liotta L. A. (2009) Tissue is alive: New technologies are needed to address the problems of protein biomarker pre-analytical variability. Proteomics Clin. Appl. 3, 874–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Silvestri A., Colombatti A., Calvert V. S., Deng J., Mammano E., Belluco C., De Marchi F., Nitti D., Liotta L. A., Petricoin E. F., Pierobon M. (2010) Protein pathway biomarker analysis of human cancer reveals requirement for upfront cellular-enrichment processing. Lab. Invest. 90, 787–796 [DOI] [PubMed] [Google Scholar]
- 9. Ferguson K. M. (2008) Structure-based view of epidermal growth factor receptor regulation. Annu. Rev. Biophys. 37, 353–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hynes N. E., MacDonald G. (2009) ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 11. Hynes N. E., Lane H. A. (2005) ERBB receptors and cancer: The complexity of targeted inhibitors. Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 12. Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., Varmus H. (2005) Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oxnard G. R., Arcila M. E., Sima C. S., Riely G. J., Chmielecki J., Kris M. G., Pao W., Ladanyi M., Miller V. A. (2011) Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clin. Cancer Res. 17, 1616–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esteva F. J., Yu D., Hung M. C., Hortobagyi G. N. (2010) Molecular predictors of response to trastuzumab and lapatinib in breast cancer. Nat. Rev. Clin. Oncol. 7, 98–107 [DOI] [PubMed] [Google Scholar]
- 15. Benvenuti S., Sartore-Bianchi A., Di Nicolantonio F., Zanon C., Moroni M., Veronese S., Siena S., Bardelli A. (2007) Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 67, 2643–2648 [DOI] [PubMed] [Google Scholar]
- 16. Rebucci M., Peixoto P., Dewitte A., Wattez N., De Nuncques M. A., Rezvoy N., Vautravers-Dewas C., Buisine M. P., Guerin E., Peyrat J. P., Lartigau E., Lansiaux A. (2011) Mechanisms underlying resistance to cetuximab in the HNSCC cell line: Role of AKT inhibition in bypassing this resistance. Int. J. Oncol. 38, 189–200 [PubMed] [Google Scholar]
- 17. Jhawer M., Goel S., Wilson A. J., Montagna C., Ling Y. H., Byun D. S., Nasser S., Arango D., Shin J., Klampfer L., Augenlicht L. H., Perez-Soler R., Mariadason J. M. (2008) PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 68, 1953–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hennessy B. T., Lu Y., Gonzalez-Angulo A. M., Carey M. S., Myhre S., Ju Z., Davies M. A., Liu W., Coombes K., Meric-Bernstam F., Bedrosian I., McGahren M., Agarwal R., Zhang F., Overgaard J., Alsner J., Neve R. M., Kuo W. L., Gray J. W., Borresen-Dale A. L., Mills G. B. (2010) A technical assessment of the utility of reverse phase protein arrays for the study of the functional proteome in non-microdissected human breast cancers. Clin. Proteomics 6, 129–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pernas F. G., Allen C. T., Winters M. E., Yan B., Friedman J., Dabir B., Saigal K., Mundinger G. S., Xu X., Morris J. C., Calvo K. R., Van Waes C., Chen Z. (2009) Proteomic signatures of epidermal growth factor receptor and survival signal pathways correspond to gefitinib sensitivity in head and neck cancer. Clin. Cancer Res. 15, 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pierobon M., Calvert V., Belluco C., Garaci E., Deng J., Lise M., Nitti D., Mammano E., Marchi F. D., Liotta L., Petricoin E. (2009) Multiplexed cell signaling analysis of metastatic and nonmetastatic colorectal cancer reveals COX2-EGFR signaling activation as a potential prognostic pathway biomarker. Clin. Colorectal Cancer 8, 110–117 [DOI] [PubMed] [Google Scholar]
- 21. Tsavachidou-Fenner D., Tannir N., Tamboli P., Liu W., Petillo D., Teh B., Mills G. B., Jonasch E. (2010) Gene and protein expression markers of response to combined antiangiogenic and epidermal growth factor targeted therapy in renal cell carcinoma. Ann. Oncol. 21, 1599–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thelemann A., Petti F., Griffin G., Iwata K., Hunt T., Settinari T., Fenyo D., Gibson N., Haley J. D. (2005) Phosphotyrosine signaling networks in epidermal growth factor receptor overexpressing squamous carcinoma cells. Mol. Cell. Proteomics 4, 356–376 [DOI] [PubMed] [Google Scholar]
- 23. Pandey A., Podtelejnikov A. V., Blagoev B., Bustelo X. R., Mann M., Lodish H. F. (2000) Analysis of receptor signaling pathways by mass spectrometry: Identification of Vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc. Natl. Acad. Sci. U.S.A. 97, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo A., Villén J., Kornhauser J., Lee K. A., Stokes M. P., Rikova K., Possemato A., Nardone J., Innocenti G., Wetzel R., Wang Y., MacNeill J., Mitchell J., Gygi S. P., Rush J., Polakiewicz R. D., Comb M. J. (2008) Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. U.S.A. 105, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peréz-Soler R., Saltz L. (2005) Cutaneous adverse effects with HER1/EGFR-targeted agents: Is there a silver lining? J. Clin. Oncol. 23, 5235–5246 [DOI] [PubMed] [Google Scholar]
- 26. Slebos R. J., Brock J. W., Winters N. F., Stuart S. R., Martinez M. A., Li M., Chambers M. C., Zimmerman L. J., Ham A. J., Tabb D. L., Liebler D. C. (2008) Evaluation of strong cation exchange versus isoelectric focusing of peptides for multidimensional liquid chromatography-tandem mass spectrometry. J. Proteome Res. 7, 5286–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sprung R. W., Jr., Brock J. W., Tanksley J. P., Li M., Washington M. K., Slebos R. J., Liebler D. C. (2009) Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol. Cell. Proteomics 8, 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fiske W. H., Tanksley J., Nam K. T., Goldenring J. R., Slebos R. J., Liebler D. C., Abtahi A. M., La Fleur B., Ayers G. D., Lind C. D., Washington M. K., Coffey R. J. (2009) Efficacy of cetuximab in the treatment of Menetrier's disease. Sci. Translational Med. 8, 8ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manning H. C., Merchant N. B., Foutch A. C., Virostko J. M., Wyatt S. K., Shah C., McKinley E. T., Xie J., Mutic N. J., Washington M. K., LaFleur B., Tantawy M. N., Peterson T. E., Ansari M. S., Baldwin R. M., Rothenberg M. L., Bornhop D. J., Gore J. C., Coffey R. J. (2008) Molecular imaging of therapeutic response to epidermal growth factor receptor blockade in colorectal cancer. Clin. Cancer Res. 14, 7413–7422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gross M. E., Zorbas M. A., Danels Y. J., Garcia R., Gallick G. E., Olive M., Brattain M. G., Boman B. M., Yeoman L. C. (1991) Cellular growth response to epidermal growth factor in colon carcinoma cells with an amplified epidermal growth factor receptor derived from a familial adenomatous polyposis patient. Cancer Res. 51, 1452–1459 [PubMed] [Google Scholar]
- 31. Licklider L. J., Thoreen C. C., Peng J., Gygi S. P. (2002) Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal. Chem. 74, 3076–3083 [DOI] [PubMed] [Google Scholar]
- 32. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang H., Liu Q., Zimmerman L. J., Ham A. J., Slebos R. J., Rahman J., Kikuchi T., Massion P. P., Carbone D. P., Billheimer D., Liebler D. C. (2011) Methods for peptide and protein quantitation by liquid chromatography-multiple reaction monitoring mass spectrometry. Mol. Cell. Proteomics 10, M110.006593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma Z. Q., Tabb D. L., Burden J., Chambers M. C., Cox M. B., Cantrell M. J., Ham A. J., Litton M. D., Oreto M. R., Schultz W. C., Sobecki S. M., Tsui T. Y., Wernke G. R., Liebler D. C. (2011) Supporting Tool Suite for Production Proteomics. Bioinformatics 27, 3214–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: Highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang B., Chambers M. C., Tabb D. L. (2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qian W. J., Liu T., Monroe M. E., Strittmatter E. F., Jacobs J. M., Kangas L. J., Petritis K., Camp D. G., 2nd, Smith R. D. (2005) Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: The human proteome. J. Proteome Res. 4, 53–62 [DOI] [PubMed] [Google Scholar]
- 38. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 39. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 40. Li M., Gray W., Zhang H., Chung C. H., Billheimer D., Yarbrough W. G., Liebler D. C., Shyr Y., Slebos R. J. (2010) Comparative shotgun proteomics using spectral count data and quasi-likelihood modeling. J. Proteome Res. 9, 4295–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Breslow N. (1990) Further studies in the variability of pock counts. Stat. Med. 9, 615–626 [DOI] [PubMed] [Google Scholar]
- 42. Pawluk-Kolc M., Zieba-Palus J., Parczewski A. (2006) Application of false discovery rate procedure to pairwise comparisons of refractive index of glass fragments. Forensic Sci. Int. 160, 53–58 [DOI] [PubMed] [Google Scholar]
- 43. Masui H., Castro L., Mendelsohn J. (1993) Consumption of EGF by A431 cells: Evidence for receptor recycling. J. Cell Biol. 120, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liao H. J., Carpenter G. (2007) Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol. Biol. Cell 18, 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liao H. J., Carpenter G. (2009) Cetuximab/C225-induced intracellular trafficking of epidermal growth factor receptor. Cancer Res. 69, 6179–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nishimura Y., Bereczky B., Ono M. (2007) The EGFR inhibitor gefitinib suppresses ligand-stimulated endocytosis of EGFR via the early/late endocytic pathway in non-small cell lung cancer cell lines. Histochem. Cell Biol. 127, 541–553 [DOI] [PubMed] [Google Scholar]
- 47. Chinkers M., McKanna J. A., Cohen S. (1979) Rapid induction of morphological changes in human carcinoma cells A-431 by epidermal growth factors. J. Cell Biol. 83, 260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chinkers M., McKanna J. A., Cohen S. (1981) Rapid rounding of human epidermoid carcinoma cells A-431 induced by epidermal growth factor. J. Cell Biol. 88, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu P. H., Kuo T. C., Chang K. C., Chang C. H., Chu C. Y. (2011) Gefitinib-induced epidermal growth factor receptor-independent keratinocyte apoptosis is mediated by the JNK activation pathway. Br. J. Dermatol. 164, 38–46 [DOI] [PubMed] [Google Scholar]
- 50. Mitsos A., Melas I. N., Siminelakis P., Chairakaki A. D., Saez-Rodriguez J., Alexopoulos L. G. (2009) Identifying drug effects via pathway alterations using an integer linear programming optimization formulation on phosphoproteomic data. PLoS Comput. Biol. 5, e1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Olive M., Untawale S., Coffey R. J., Siciliano M. J., Wildrick D. M., Fritsche H., Pathak S., Cherry L. M., Blick M., Lointier P., et al. (1993) Characterization of the DiFi rectal carcinoma cell line derived from a familial adenomatous polyposis patient. In Vitro Cell. Dev. Biol. 29A, 239–248 [DOI] [PubMed] [Google Scholar]
- 52. Awwad R. A., Sergina N., Yang H., Ziober B., Willson J. K., Zborowska E., Humphrey L. E., Fan R., Ko T. C., Brattain M. G., Howell G. M. (2003) The role of transforming growth factor α in determining growth factor independence. Cancer Res. 63, 4731–4738 [PubMed] [Google Scholar]
- 53. van Houdt W. J., Hoogwater F. J., de Bruijn M. T., Emmink B. L., Nijkamp M. W., Raats D. A., van der Groep P., van Diest P., Borel Rinkes I. H., Kranenburg O. (2010) Oncogenic KRAS desensitizes colorectal tumor cells to epidermal growth factor receptor inhibition and activation. Neoplasia 12, 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Palmer W. E., Bloch S. M., Chew F. S. (1992) Menetrier disease. Am. J. Roentgenol. 158, 62. [DOI] [PubMed] [Google Scholar]
- 55. Rich A., Toro T. Z., Tanksley J., Fiske W. H., Lind C. D., Ayers G. D., Piessevaux H., Washington M. K., Coffey R. J. (2010) Distinguishing Menetrier's disease from its mimics. Gut 59, 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Burdick J. S., Chung E., Tanner G., Sun M., Paciga J. E., Cheng J. Q., Washington K., Goldenring J. R., Coffey R. J. (2000) Treatment of Menetrier's disease with a monoclonal antibody against the epidermal growth factor receptor. N. Engl. J. Med. 343, 1697–1701 [DOI] [PubMed] [Google Scholar]
- 57. Wahl M., Carpenter G. (1988) Regulation of epidermal growth factor-stimulated formation of inositol phosphates in A-431 cells by calcium and protein kinase C. J. Biol. Chem. 263, 7581–7590 [PubMed] [Google Scholar]
- 58. Xu Y., Guo D. F., Davidson M., Inagami T., Carpenter G. (1997) Interaction of the adaptor protein Shc and the adhesion molecule cadherin. J. Biol. Chem. 272, 13463–13466 [DOI] [PubMed] [Google Scholar]
- 59. Bianco R., Shin I., Ritter C. A., Yakes F. M., Basso A., Rosen N., Tsurutani J., Dennis P. A., Mills G. B., Arteaga C. L. (2003) Loss of PTEN//MMAC1//TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene 22, 2812–2822 [DOI] [PubMed] [Google Scholar]
- 60. Mariner D. J., Davis M. A., Reynolds A. B. (2004) EGFR signaling to p120-catenin through phosphorylation at Y228. J. Cell Sci. 117, 1339–1350 [DOI] [PubMed] [Google Scholar]
- 61. Kim S. H., Song Y. C., Kim S. H., Jo H., Song Y. S. (2009) Effect of epidermal growth factor receptor inhibitor alone and in combination with cisplatin on growth of vulvar cancer cells. Ann. N.Y. Acad. Sci. 1171, 642–648 [DOI] [PubMed] [Google Scholar]
- 62. Choi K., Ahn Y. H., Gibbons D. L., Tran H. T., Creighton C. J., Girard L., Minna J. D., Qin F. X., Kurie J. M. (2009) Distinct biological roles for the notch ligands Jagged-1 and Jagged-2. J. Biol. Chem. 284, 17766–17774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Purow B. W., Sundaresan T. K., Burdick M. J., Kefas B. A., Comeau L. D., Hawkinson M. P., Su Q., Kotliarov Y., Lee J., Zhang W., Fine H. A. (2008) Notch-1 regulates transcription of the epidermal growth factor receptor through p53. Carcinogenesis 29, 918–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gao J., Liu J., Fan D., Xu H., Xiong Y., Wang Y., Xu W., Wang Y., Cheng Y., Zheng G. (2010) Up-regulated expression of Notch1 and Jagged1 in human colon adenocarcinoma. Pathol. Biol. 59, 298–302 [DOI] [PubMed] [Google Scholar]
- 65. Reedijk M., Odorcic S., Zhang H., Chetty R., Tennert C., Dickson B. C., Lockwood G., Gallinger S., Egan S. E. (2008) Activation of Notch signaling in human colon adenocarcinoma. Int. J. Oncol. 33, 1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guilmeau S., Flandez M., Mariadason J. M., Augenlicht L. H. (2010) Heterogeneity of Jagged1 expression in human and mouse intestinal tumors: Implications for targeting Notch signaling. Oncogene 29, 992–1002 [DOI] [PubMed] [Google Scholar]
- 67. Reedijk M., Odorcic S., Chang L., Zhang H., Miller N., McCready D. R., Lockwood G., Egan S. E. (2005) High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 65, 8530–8537 [DOI] [PubMed] [Google Scholar]
- 68. Ben-Ari E. T., Bernstein L. R., Colburn N. H. (1992) Differential c-jun expression in response to tumor promoters in JB6 cells sensitive or resistant to neoplastic transformation. Mol. Carcinog. 5, 62–74 [DOI] [PubMed] [Google Scholar]
- 69. Kayahara M., Wang X., Tournier C. (2005) Selective regulation of c-jun gene expression by mitogen-activated protein kinases via the 12-O-Tetradecanoylphorbol-13-acetate-responsive element and myocyte enhancer factor 2 binding sites. Mol. Cell. Biol. 25, 3784–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bartel D. P., Sheng M., Lau L. F., Greenberg M. E. (1989) Growth factors and membrane depolarization activate distinct programs of early response gene expression: Dissociation of Fos and Jun induction. Genes Dev. 3, 304–313 [DOI] [PubMed] [Google Scholar]
- 71. Smith L. M., Wise S. C., Hendricks D. T., Sabichi A. L., Bos T., Reddy P., Brown P. H., Birrer M. J. (1999) c-Jun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype. Oncogene 18, 6063–6070 [DOI] [PubMed] [Google Scholar]
- 72. Singh A. B., Harris R. C. (2004) Epidermal growth factor receptor activation differentially regulates claudin expression and enhances transepithelial resistance in Madin-Darby canine kidney cells. J. Biol. Chem. 279, 3543–3552 [DOI] [PubMed] [Google Scholar]
- 73. Chen B. K., Chang W. C. (2000) Functional interaction between c-Jun and promoter factor Sp1 in epidermal growth factor-induced gene expression of human 12(S)-lipoxygenase. Proc. Natl. Acad. Sci. U.S.A. 97, 10406–10411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ikari A., Atomi K., Takiguchi A., Yamazaki Y., Miwa M., Sugatani J. (2009) Epidermal growth factor increases claudin-4 expression mediated by Sp1 elevation in MDCK cells. Biochem. Biophys. Res. Commun. 384, 306–310 [DOI] [PubMed] [Google Scholar]
- 75. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]