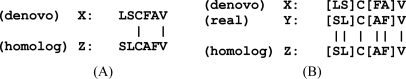Abstract
In proteomics, de novo sequencing is the process of deriving peptide sequences from tandem mass spectra without the assistance of a sequence database. Such analyses have traditionally been performed manually by human experts, and more recently by computer programs that have been developed because of the need for higher throughput. Although powerful, de novo sequencing often can only determine partially correct sequence tags because of imperfect tandem mass spectra. However, these sequence tags can then be searched in a sequence database to identify the exact or a homologous peptide. Homology searches are particularly useful for the study of organisms whose genomes have not been sequenced. This tutorial will present background important to understanding de novo sequencing, suggestions on how to do this manually, plus descriptions of computer algorithms used to automate this process and to subsequently carryout homology-based database searches. This Tutorial is part of the International Proteomics Tutorial Programme (IPTP 1).
Historical Background
Because of the lack of sequence databases, de novo sequencing preceded database search programs by several decades, in which the concept extends back nearly 50 years with the direct evaporation of an unusually volatile peptidolipid, fortuitine, into a mass spectrometer, and subsequent derivation of the sequence from the observed electron impact fragment ions (1). Through the 1960s and 1970s, a number of groups devised various methodologies to produce small volatile peptide derivatives suitable for gas chromatographic separation prior to introduction into a gas chromatography-mass spectrometer, with electron impact ionization and fragmentation (2, 3). This technique was used to completely sequence a small protein of unknown sequence in 1976 (4), and was subsequently used in cases refractory to Edman sequencing, such as blocked N termini (5) or for very hydrophobic proteins (6). Gas chromatography-mass spectrometry produces large numbers of mass spectra, and computer programs reminiscent of those used today for de novo sequencing were written to aid in their interpretation (7). The sensitivity of these methods (requiring 10–100 nmole) could not compete with automated Edman sequencing, and was suitable only to hydrolyzates containing peptides of 2–8 amino acids in length (longer peptide derivatives were not amenable to gas chromatography).
Over the course of subsequent decades, improved methods were developed that allowed for the ionization of higher molecular weight polar molecules without prior derivatization. For example, fast atom bombardment (FAB)1 (8) tended to produce intact singly protonated molecular ions with low intensity fragments; pure peptides at high concentration did exhibit sufficient fragment ion intensity to allow for de novo sequencing (9). A parallel development included work on various combinations of mass analyzers in order to provide mixture analysis via mass selection of precursor ions, fragmentation of the selected ion, and mass analysis of the resulting fragment ions (what is now commonly thought of as tandem MS (MS/MS)). Thus, it was possible to examine multiple peptide ions at once, and the “soft ionization” of FAB with its relatively low fragment ion abundance was ideal. In 1986, Hunt et al. (10) showed how to use a triple quadrupole mass spectrometer with FAB ionization to derive protein sequences from low energy (<200 eV) collision induced dissociation (CID) spectra, and demonstrated this on a known protein, apolipoprotein. Shortly thereafter, tandem mass spectrometry using high energy CID (>2KeV) on a four sector instrument with FAB ionization was used for the first time to sequence a protein of unknown structure (11). A computer program was written to aid in sequencing high energy CID FAB spectra, where the algorithm basically mimicked a manual de novo sequence determination by building up subsequences one amino acid at a time typically starting at the C terminus (12). Sequences were determined for both tryptic and Glu-C peptides, where the overlap between them stitched the individual peptide sequences together into the full length protein sequence. Compared with the triple quadrupole data, sector instruments provided much better precursor selection resolution and product ion resolution, and the fragmentation at high energy was not as affected by the presence and location of basic amino acids. This combination made it possible to unambiguously assign sequences to unknown proteins. In contrast, the fragment ion peak widths in the FAB triple quadrupole spectra were 2–3 Da wide; however, the improved ion transmission made them more sensitive. In the end, the controversy over high energy CID on sectors versus low energy CID on triple quadrupoles was made moot by other developments in instrumentation.
The revolutionary ionization methods of electrospray (13) and matrix assisted laser desorption (14) were not compatible with sector mass analyzers, consequently by the early 1990s nearly all mass spectrometric peptide sequencing was performed using triple quadrupoles with electrospray. Prior to this point, all interpretations of tandem mass spectra of peptides could be considered to be de novo sequencing, either manual or with computer programs. This shifted dramatically with the introduction of database search programs in 1994—Sequest (15) could search uninterpreted spectra and PeptideSearch (16) used a sequence tag algorithm that required a partial manual interpretation of each spectrum. As computers became faster and protein sequence databases became more extensive, this trend away from de novo sequencing accelerated. For a time, intellectual property rights issues inhibited further development of database search programs; however, this somehow has faded away and there are now a plethora of choices (17–23), which has been reviewed by Nesvizhskii (24).
Database search programs have taken much of the wind out of de novo sequencing; however, there remain a few reasons for continuing the tradition (25). There are quite a few unsequenced genomes remaining (i.e. most of them). For example, one of the authors of this manuscript (RJ) managed to show that a cell line was contaminated with a species of mycoplasma whose genome had not yet been sequenced (unpublished results, because no one would report on their contaminated cell lines). Second, given the significant differences between database searching and de novo sequencing approaches, any agreement between them should provide significant validation of the search result. This could be particularly useful for “one-hit wonders” (i.e. proteins identified on the basis of a single peptide identification from a database search). Third, often the majority of high throughput LC-MS/MS tandem mass spectra are not matched in a database search, and it can be prudent to learn why. One can learn about carbamylation, or other sample preparation issues that result in unexpected mass changes. Likewise, autosampler carryover and contamination can lead to the collection of numerous tandem mass spectra that are from a different species other than what was intended. Computer programs that perform de novo sequencing can provide insight into the numbers of high quality unidentified MS/MS spectra for which a plausible peptide sequence can be deduced. In other words, a high quality MS/MS spectrum is one that is sequenceable.
As intimated above, the history of computer programs for de novo sequencing extends back several decades to the days of GCMS of peptide derivatives (7), and during this time there have been a number of algorithms developed. One approach has involved the generation of all amino acid combinations that account for the peptide mass, and then compare predicted fragment ions with observed (26). However, this becomes computationally prohibitive for peptides beyond a length of about eight amino acids. A second approach is what was originally devised for polyamino alcohols (7) and was subsequently used for FAB mass spectra (27), high energy CID (12), and low energy CID (28). This so-called “subsequencing” algorithm tests short sequence segments (beginning at one of the termini) against the observed spectrum, reserving those subsequences that best account for the observed fragment ions, and then extending them by one amino acid and repeating the comparison and extensions until the peptide molecular weight is achieved. A third alternative (29) is more of a computer assisted manual interpretation, where a graphical display shows how various fragment ions in a spectrum connect with others via mass differences matching the common amino acids. The user then chooses a pathway through the ions one amino acid at a time until the calculated mass of the sequence matches the measured peptide mass. A fourth method uses graph theory, and involves the mathematical conversion of the various ion types into a graph containing nodes that represent a single ion type, and then finding pathways through the connecting edges to determine sequence candidates (30). Such an approach was used for data acquired from high energy CID of singly charged peptide ions (31), and was used by one of the authors (RJ) to produce the computer program Lutefisk97 (32), which was later upgraded to Lutefisk1900 (25) sometime around the turn of the millennium. Lutefisk1900 then became a benchmark that allowed others to show the superiority of their own programs. Many de novo sequencing programs (33–43) have been developed since these early programs. Among these programs the better known were Sherenga (39), PEAKS (37) and PepNovo (36), which employ more sensible probabilistic scoring schemes and are generally faster than Lutefisk. Some independent comparisons (44–46) have been made in an attempt to evaluate overall performance on different instruments. Most of these tools are freely available, whereas PEAKS provides both a commercial version and a free web service with the same algorithm.
Despite the tremendous effort researchers have put into de novo sequencing, complete accuracy for every peptide is not possible. The difficulty largely comes from variable data quality; in particular, MS/MS spectra usually do not contain all the fragment ions necessary for deriving a complete peptide sequence, manually or with a computer program. When this happens, the best that one can achieve is a partially correct sequence. These partially correct sequences can still be useful for homology-based database searches, where the sequence candidates, derived from a de novo sequencing program, are searched against sequence databases using programs such as BLAST (47) or FASTA (48). This was first proposed in 1997 (32), where the sequence candidates produced by Lutefisk97 were used as input to a version of FASTA that had been modified to account for the peculiarities and errors common to de novo sequencing (see section “De novo Sequencing Errors and Sequence Tags” for a discussion of common de novo errors). This general concept was subsequently fleshed out by others (49–52). Some researchers have accepted the reality that one cannot count on deriving a complete sequence, especially from low energy CID, and have developed approaches whereby a partial de novo interpretation provides a sequence tag. This tag is then used for a database search. This was the basis for the original PeptideSearch algorithm (16), where the tag was derived by manual inspection with a calculator in hand. There has been much progress since then on automatically generating sequence tags prior to the search, as well as on more accurate search algorithms (22, 53, 54).
Since the 1990s, developments in instrumentation and progress in understanding gas phase chemistry have had important impacts on de novo peptide sequencing. Specifically, the quadrupole/time-of-flight hybrid mass spectrometer was a dramatic improvement (55) over the triple quadrupole, which had been used for shotgun experiments in the 1980s and early 1990s, despite the low duty cycle when scanning the third quadrupole to acquire product ion scans. More recently, the orbitrap has made an impact in that it is possible to rapidly acquire both MS and MS/MS spectra with high mass accuracy and resolution (56). As will be seen, high mass accuracy and resolution are exceedingly important attributes when deriving a peptide sequence. With the large scale commercialization of ion traps (57), notably the LCQ by the Finnigan Corporation, a slight variation on low energy CID became widely available. Although both ion traps and quadrupole collision cells perform low energy CID, fragments that form in an ion trap fall out of resonance and lose kinetic energy such that additional cleavages tend not to be prominent. This is in contrast to a quadrupole collision cell, where a fragment ion that forms near the start of the collision region will likely undergo additional collisions and further fragmentations before exiting. Because y-type ions are more stable than b-type ions, subsequent collisions in a quadrupole cell will reduce the intensity of the longer b-type ions. The upshot is that although both types of collisions produce b/y-type fragments, ion trap MS/MS spectra often contain intense high mass b-type ions, whereas quadrupole MS/MS spectra (e.g. from Q-Tof's) do not (58). This is a good time to point out that the so-called “higher energy C-trap dissociation” or HCD cell (59) that is often sold with the orbitrap does not actually subject precursor ions to high energy collisions, but is more akin to the collision processes seen in quadrupole collision cells. Regardless of whether low energy CID fragments were formed in a trap or a quadrupole (or HCD) collision cell, the mobile proton theory (60, 61) has been an important contribution for understanding this process. Although de novo sequencing has typically been applied to CID spectra, there is no reason why it could not be used with MS/MS spectra obtained using alternative fragmentation methods that also produce contiguous series of fragment ions of the same type. At the moment, the most promising alternatives are electron capture (62) and electron transfer dissociation (63), which are described below in Section “Fragment Ions”.
Basic Concepts
Fragment Ions
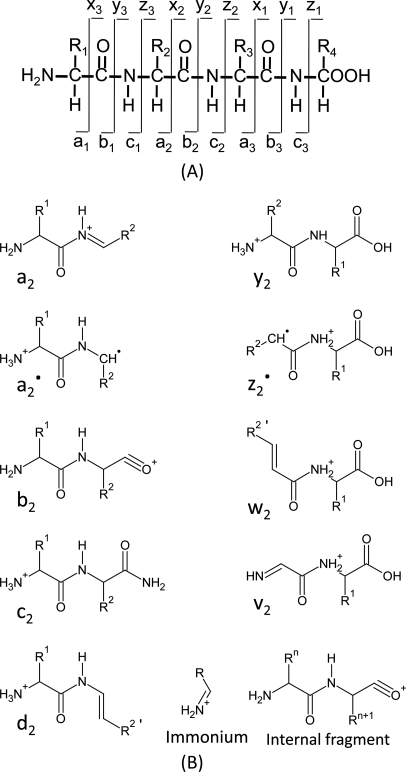
This review only covers de novo sequencing of protonated proteolytic peptides of length less than ∼25 residues, and does not include any discussion of the fragmentation of negatively charged peptides (64), nor fragmentation of intact proteins, as in top-down experiments (65). Three processes will be considered here: vibrational excitation via low energy CID; electronic excitation from high energy CID; and electron transfer dissociation (ETD). Electron capture dissociation spectra exhibit many similarities to ETD spectra; however, instruments that produce ETD data are now much more common and will therefore be the focus here. Fragment ion nomenclature (66, 67) can be a bit confusing with respect to the numbers of additional hydrogen atoms, as well as noting when an ion is a radical or an even electron cation. In this review, we will use the nomenclature in Fig. 1 where the fragment ions are calculated as shown in Table I, and radical cations are distinguished by a superscript dot (e.g. z2·). The ion structures in Fig. 1B are not necessarily the ones thought to be physically present in the mass spectrometer, but they more readily convey how to calculate fragment ion molecular weights.
Fig. 1.
A, A peptide can be fragmented at the peptide backbone, resulting in a, b, c, x, y, and z-ions. B, The structures of different ion types for calculation of the ion molecular weights (see Tables I and II).
Table I. Calculating the masses of positively charged fragment ions. [N] is the mass of the N-terminus (e.g., 1.0078 Da for unmodified peptides and 43.0184 Da for acetylated N-terminus). [C] is the mass of the C-terminus (e.g., 17.0027 Da for unmodified peptides and 16.0187 Da for amidated C-terminus). [M] is the sum of the amino acid residue masses (see Table II) that are contained within the fragment ion. CO is the combined mass of oxygen plus carbon atoms (27.9949 Da) and H is the mass of a proton (1.0078 Da). To calculate the m/z value of a fragment ion, add the mass of the protons to the neutral mass calculated from the table, and divide by the number of protons added.
| Ion Type | Neutral MW of the fragment |
|---|---|
| a | [N] + [M] - CO - H |
| a. | [N] + [M] - CO |
| a-H2O | a – 18.0106 |
| a-NH3 | a – 17.0266 |
| b | [N] + [M] - H |
| b-H2O | b – 18.0106 |
| b-NH3 | b – 17.0266 |
| c | [N] + [M] + NH2 |
| d | a – partial side chain |
| x | [C] + [M] + CO - H |
| y | [C] + [M] + H |
| y-H2O | y – 18.0106 |
| y-NH3 | y – 17.0266 |
| z | [C] + [M] - NH |
| v | y - complete side chain |
| w | z - partial side chain |
At the most simplistic level, low energy CID produces b-type and y-type ions. The concept of a “residue mass” is that this is the mass of an amino acid within a peptide; i.e. it is the mass of an amino acid minus the mass of water, which is lost when amino acids polymerize to form peptides. Table II gives the average and monoisotopic residue masses of the common amino acids, whereas a reverse lookup table from a total monoisotopic mass to the combination of several residues is provided at cs.uwaterloo.ca/∼binma/peaks/masstable.htm. It can be seen from Fig. 1B that a singly-charged b-type ion would be calculated by summing the residue masses and adding the mass of a single hydrogen atom (assuming that the peptide has an unmodified N terminus). Likewise, a singly charged y ion would be calculated by summing the appropriate residue masses, and then adding the mass of water plus a proton. Calculating multiply charged versions involves adding the mass of additional protons and then dividing the sum by the number of charges. The mechanism of formation of b-type ions most likely involves the carbonyl oxygen of the residue N-terminal to the cleavage site (for an extensive review of CID fragmentation see (68)), which explains why one never observes b1 ions in peptides with free N termini. Acylated peptides will produce b1 ions, because there is an N-terminal carbonyl available to induce the cleavage reaction.
Table II. Amino acid three and single letter codes, plus amino acid residue masses and elemental composition. Residue mass is defined as the amino acid minus water (i.e., NH-CHR-CO).
| Residue | 3/1 letter code | Composition | Mono. mass | Avg. mass |
|---|---|---|---|---|
| Alanine | Ala/A | C3N5NO | 71.03712 | 71.08 |
| Arginine | Arg/R | C6H12N4O | 156.10112 | 156.19 |
| Asparagine | Asn/N | C4H6N2O2 | 114.04293 | 114.10 |
| Aspartic acid | Asp/D | C4H5NO3 | 115.02695 | 115.09 |
| Cysteine | Cys/C | C3H5NOS | 103.00919 | 103.14 |
| Cysteine-cma | Cys/C | C5H8N2O2S | 160.03065 | 160.20 |
| Glutamine | Gln/Q | C5H8N2O2 | 128.05858 | 128.13 |
| Glutamic acid | Glu/E | C5H7NO3 | 129.04260 | 129.12 |
| Glycine | Gly/G | C2H3NO | 57.02147 | 57.05 |
| Histidine | His/H | C6H7N3O | 137.05891 | 137.14 |
| Isoleucine | Ile/I | C6H11NO | 113.08407 | 113.16 |
| Leucine | Leu/L | C6H11NO | 113.08407 | 113.16 |
| Lysine | Lys/K | C6H12N2O | 128.09496 | 128.17 |
| Methionine | Met/M | C5H9OS | 131.04049 | 131.19 |
| Methionine-oxb | Met/M | C5H9O2S | 147.03540 | 147.18 |
| Phenylalanine | Phe/F | C9H9NO | 147.06842 | 147.18 |
| Proline | Pro/P | C5H7NO | 97.05277 | 97.12 |
| Serine | Ser/S | C3H5NO2 | 87.03203 | 87.08 |
| Threonine | Thr/T | C4H7NO2 | 101.04768 | 101.10 |
| Tryptophan | Trp/W | C11H10N2O | 186.07932 | 186.21 |
| Tyrosine | Tyr/Y | C9H9NO2 | 163.06333 | 163.18 |
| Valine | Val/V | C5H9NO | 99.06842 | 99.13 |
a Carbamidomethylated cysteine.
b Oxidized methionine.
The concept of a “mobile proton” provides a useful framework for understanding the low energy CID peptide fragmentation process (60). In solution, the sites of peptide protonation are likely to be the N-terminal amino group, the lysine amino group, the histidine imidazole side chain, or the guanido group on arginine. In the gas phase, however, the peptide backbone amides are of comparable basicity to all but the arginine guanido group. Therefore, in the absence of arginine, it takes only a little bit of collisional energy to scramble the site of protonation such that the ionized peptide is actually a population of ions that differ in the site of protonation (e.g. protonation occurring at any of the backbone amides or the side chains). Protonation of the backbone amide is required for the production of b- or y-type fragment ions, and cleavages that require protonation are called “charge promoted” fragmentations. Hence, as long as there is a mobile proton that can be sprinkled across the peptide backbone, one can expect to see a fairly contiguous series of b- and/or y-type ions. When the number of arginine residues match the protons, there are no mobile protons and the CID spectra are atypical and usually difficult to sequence. One can therefore understand why low energy CID of electrospray ionized tryptic peptides has been so successful, because most tryptic peptides will have no more than one arginine at the C terminus, yet be able to take on two protons—one for the arginine side chain and one “mobile” proton to produce the b/y fragment ions. Even for cases where there is a mobile proton, the presence of arginine in the middle of a peptide sequence can have adverse consequences, where b/y-type cleavages near the arginine are of reduced intensity and overall sequence coverage may be sparse.
Low energy CID produces a few additional fragment ion types, and the resulting spectra possess certain characteristics that are useful to note. Under “mobile proton” conditions, the presence of proline in a peptide typically results in intense y-type (and sometimes the corresponding b-type) ions resulting from cleavage on the N-terminal side of proline. Concomitantly, cleavage on the C-terminal side of proline is nonexistent or very much reduced. These effects are because of a combination of increased gas phase basicity of the proline nitrogen, and the unusual ring structure of the proline side chain that inhibits the attack of the carbonyl on the N-terminal side of the proline. Under “mobile proton” conditions, histidine promotes fragmentation at its C-terminal side, resulting in enhanced abundance of the corresponding b/y-type fragments. Sometimes a b/y-type cleavage will occur twice in the same molecule, resulting in a fragment ion that contains neither the peptide's original C- or N terminus. These “internal fragment ions” (Fig. 1B) can be particularly prominent in low energy CID when there is a proline at the N-terminal side of the fragment. The b- and y-type fragment ions can undergo an additional neutral loss of a molecule of water or ammonia, where these are often designated as b-17 or b-18, etc. Under mobile proton conditions (i.e. more protons than arginine residues), these ions are usually less abundant than their corresponding b/y-type ion. There are exceptions, for example, when the N-terminal amino acid is glutamine or carbamidomethylated cysteine, where cyclization of the N-terminal amino acid results in the loss of ammonia to give abundant b-17 ions. Likewise, an N-terminal glutamic acid can cyclize and lose water, and the b-18 ions can be more abundant than the corresponding b fragment ions. In some cases, a b-type fragment ion can lose a molecule of carbon monoxide to form an a-type ion (27.995 Da less than the b-type fragment ion), although these seem to be more prominent for the lower mass fragments (e.g. it is not uncommon to find a2 ions that are of comparable intensity to the b2 ion in low energy CID of tryptic peptides). Single amino acid immonium ions (Fig. 1B) are often seen when MS/MS spectral acquisition includes this low mass region. Certain immonium ions are particularly diagnostic for the presence of their corresponding amino acid—leucine and isoleucine (m/z 86.0970), methionine (m/z 104.0534), histidine (m/z 110.0718), phenylalanine (m/z 120.0813), tyrosine (m/z 136.0762), and tryptophan (m/z 159.0922). For peptide ions undergoing low energy CID that lack a mobile proton, there are some additional fragment ions that become more prominent, such as enhanced cleavage at the C-terminal side of aspartic acid (69). It later became clear that in the absence of a mobile proton, the side chain carboxylic protons from aspartic acid (and to a lesser extent glutamic acid) can provide the necessary proton to catalyze a localized b/y fragmentation (60). Low energy CID of peptide ions lacking a mobile proton also seem to be subject to the formation of a fragment ion that is sometimes called “b+18” (70). This is a rearrangement that occurs where the C-terminal residue is lost, but the C-terminal -OH group, plus a proton, are transferred to the ion. Finally, it should be mentioned that low energy CID of “nonmobile” peptide ions will often give more abundant neutral losses of water and ammonia; for example, one might observe a y-17 ion in the absence of the corresponding y-type fragment ion. Low energy CID spectra of tryptic peptides with a mobile proton are most readily sequenced, as they typically contain contiguous series of b/y-type fragment ions.
The old multisector and the newer Tof-Tof instruments are capable of subjecting peptide ions to much higher collision energy, which results in an initial electronic excitation and produces some different types of fragment ions. In addition to the b/y fragments seen for low energy CID, high energy CID can induce “charge remote” fragmentations (Fig. 1), including the d- and w-type fragment ions that allows for the distinction between leucine and isoleucine (71, 72). In general, high energy CID will produce fragment ions at nearly all peptide bonds regardless of the presence or absence of a mobile proton, which makes it a more robust activation method for de novo sequencing (11). Although CID of FAB-generated singly charged precursors in a multisector instrument is no longer used much for peptide work, CID of MALDI-generated singly charged precursors in a Tof-Tof mass spectrometer will produce very similar data, including the d-, v-, and w-type fragment ions (73, 74). In general, for spectra of singly-charged precursor ions that lack arginine, the b/y-type cleavages are prominent, which suggests that the presence of a mobile proton can still catalyze fragmentation in high energy collisions. The presence of one or more arginine residues in a singly-charged precursor that is subjected to high energy CID results in prominent and informative alternative fragment ions (a/d/w/v-type), which is in contrast to low energy CID under nonmobile proton conditions where spectra are usually difficult to interpret.
Electron capture dissociation is a process whereby an isolated multiply charged peptide ion captures a low energy thermal electron, and the resulting radical cation becomes sufficiently unstable that it fragments to produce predominantly c- and z·-type fragment ions (Fig. 1) (62). In order to produce similar fragmentations in a much cheaper analyzer, the Hunt laboratory developed ETD (63), where anionic molecules are trapped in a linear ion trap (using RF electrical fields) and mixed with multiply charged cationic peptide analyte ions. Given the appropriate anion (one with low electron affinity), an electron is transferred to the peptide cation in an exothermic process that induces the production of the same c- and z·-type fragment ions observed in ECD. It should be noted here that the addition of an electron reduces the charge state of the precursor ion and that this charge reduced cation may or may not fragment any further. The latter are sometimes referred to as “electron transfer no dissociation” (ETnoD), and are particularly prominent when an electron is transferred to a doubly charged precursor (resulting in a net +1 charge). In these cases, peptide bonds may be broken, but there is no dissociation between the neutral fragment and the singly charged fragment because of noncovalent interactions holding the two pieces together. Presumably, the absence of Coulombic repulsion between the neutral and charged fragments allows salt bridges and hydrogen bonds to hold them together. At the moment, low energy CID is sometimes used to shake the pieces apart in a process called “supplemental activation” or SA (75). One of the difficulties with ETD of low charge state precursors, particularly when subjected to SA, is that a hydrogen will to varying extents get transferred from c-type ions to the corresponding z·-type ion to generate what are sometimes referred to as c-1 and z+1 ions (75). The result is that one observes doublets, which can be a problem when deciding how to use most database search engines. For example, should one forget about c-1 and z+1 ions and use a tight fragment ion tolerance, or use a very wide (and less specific) tolerance in case the c-1 and z+1 ions are prominent? A similar problem arises when a secondary electron transfer occurs to an ETD-derived multiply charged fragment ion. For example, a doubly charged fragment can become singly charged either by removal of a proton or addition of an electron, and when both occur one observes singly charged doublets separated by one mass unit. Another unusual ETD fragmentation occurs when a z·-type fragment ion with alkylated cysteine at the C-terminal radical site undergoes homolytic bond cleavage of the beta-carbon-sulfur bond (76). This seems to be a complete conversion that does not occur when the cysteine is located elsewhere in a z·-type fragment ion. In addition to c/z·-type fragment ions, ETD also generates less prominent a·/y-type fragment ions (Fig. 1) (77). One should note that c-type and y-type fragments are even electron species, whereas both a·-type and z·-type ions are radical cations. It has been pointed out that because z·-type ions have an even number of odd valence atoms (e.g. hydrogen and nitrogen) and c-type ions have an odd number of odd valence atoms that it is not possible for the two types of fragment ions to have the same elemental composition (78). If these were the only ions present in ETD spectra, then it should be possible to distinguish one ion type from the other based solely on accurate mass measurements of ETD fragment ions. However, the existence of a·-type fragment ions calls this approach into question. In any case, making this determination of c- versus z-type fragment ions based solely on mass requires high mass accuracy data, which is in itself probably more useful than making this distinction between ion types.
Manual De Novo Sequencing
There are at least two approaches one can take when manually sequencing a peptide from tandem mass spectral data: (1) sequencing from the C terminus and (2) sequencing from an obvious tag. Sequencing from the C terminus may be suitable when the tandem mass spectra includes low m/z fragment ions, and where the peptide was derived from proteolytic cleavage on the C-terminal side of specific amino acids. For example, to begin sequencing a tryptic peptide, one can make an initial assumption that the C terminus of the peptide is either lysine or arginine, and from Tables I and II calculate the corresponding y1 ions as m/z 147.113 or m/z 175.119. If either mass is present, then one attempts to find y2 candidates by subtracting masses of higher m/z ions from the putative y1 to see if there are any mass differences that correspond to an amino acid residue mass. For each y2 candidate the process is repeated in order to determine any candidate y3 ions, and so on. In an ideal case, this process is a matter of finding a pathway through the ions that ends when the observed peptide mass matches what is calculated from the amino acids in the pathway. However, there are countless ways to go wrong, most of which relate to jumping to an ion series other than y-type. The chances of this happening can be greatly reduced when the fragment ions are measured with high mass accuracy. For example, if there are two y2 candidates that differ in mass from the y1 ion by 71.12 and 115.03 Da, then for measurements that are accurate to 0.5 Da these are equally likely (an extension of either alanine or aspartic acid have to be considered). However, if the mass accuracy is within 0.02 Da, only aspartic acid is possible. Another complication is due the absence of a fragment ion. This can occur for several reasons, for example lack of cleavage on the C-terminal side of proline. For most ion trap data, the y1 ions are below the mass cutoff; however, one can sometimes find the corresponding high m/z b-type ion containing all of the residues except the arginine or lysine at the C terminus. These b-type ions are calculated by subtracting 17.002 (one oxygen and one hydrogen) from the peptide molecular weight, and then subtract from this value the residue masses of arginine or lysine. If such a b-type ion is found corresponding to the loss of arginine or lysine, then one can begin there and try to trace out a series of ions toward the lower m/z range. Often it is not possible to get a complete sequence all the way to the N terminus, because it is not uncommon for a CID spectrum to lack fragmentations between the first and second amino acids at the N terminus. Hence, the N terminus of a derived sequence is often not a sequence, but is instead a combined residue mass of two amino acids. One should, however, make sure that this unsequenced mass at the N terminus corresponds to the sum of two amino acid residue masses. For example, an unsequenced N-terminal mass of 150 μ is not possible for an unmodified peptide.
A different approach is to derive a partial sequence from the middle of the peptide from an obvious series of ions differing by amino acid residue masses. For tryptic peptides, one often finds such a short stretch of fairly intense ions at an m/z greater than the precursor ion. In principal, one does not know if these are b-type or y-type ions (and hence, whether the partial sequence goes forward or backwards), but for CID of tryptic peptides in a quadrupole collision cell it is usually safe to guess that this is a partial y-type ion series. For any of these ions, one can subtract their mass from the ‘mass of the peptide plus 2.016 μ (this is the mass of two hydrogens). This calculated mass corresponds to the mass of the lower mass b-type ions (assuming the sequence tag is, in fact, comprised of y-type ions). Identification of such mirror-image ion series should provide some confidence that one is on the right track. Thus, while trying to extend one series, the other series should also be checked out. For data obtained using a quadrupole collision cell, the b-type ion series usually does not extend very far (b2 is prominent, but the series usually does not extend beyond a few residues). At the same time the y-type ion series that defines the C-terminal portion of the peptide is at the low m/z end of the spectrum, which is where spectra typically become more complicated and contain many different types of ions (i.e. immonium, internal, b-type, a-type, and y-type).
One of the difficulties in de novo sequencing is that a contiguous ion series might be identified, but the direction of the sequence may be difficult to establish. In other words, for CID data it may not be clear whether an ion series is y-type or b-type, and for ETD spectra one may not be able to tell if an ions series is c- or z·-type. One way of making this distinction has already been described—trying to link a series to a characteristic ion (or mass difference) that corresponds to an anticipated C-terminal amino acid. If an obvious sequence tag can be linked to a typical tryptic y1, then that would suggest the tag is comprised of y-type ions and the direction of the sequence can therefore be presumed. Alternatively, pairs of ions with characteristic mass differences can indicate the type of fragment ions observed. For example, in low energy CID, b2 ions are often accompanied by intense a2 ion. Thus, if a series of ions are found where the lowest mass one has a satellite peak 27.9949 Da lower, then one could presume that this is a b-type ion series. Obviously, accurate mass measurements will greatly add to the confidence of such presumptions. Likewise, for ETD spectra, ions differing by 16.0182 Da could be z·/y-type pairs, or ions differing by 44.0136 Da could be a·/c-type pairs. Similarly, high energy CID ion series might be distinguished on the basis of a/d-type pairs or y/w-type pairs.
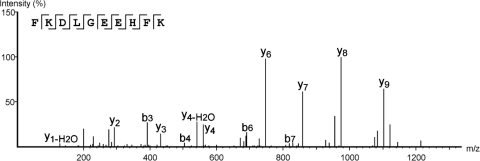
Once a sequence is derived, the final step is to determine whether the majority of the fragment ions (particularly the high intensity ones) can be assigned. First, verify the presence of the sequence-specific fragment ions such as y-type, b-type, and a-type ions, as well as the losses of ammonia or water from these ion types. Check to see if any fragment ions might be multiply charged, which should be verified in high resolution spectra by examining the isotope cluster spacing. Then check to see if any of the remaining ions not accounted for are possibly because of internal fragmentations. Internal fragments are usually short (typically less than five residues), and are calculated by summing the amino acid residue masses together and adding the mass of hydrogen. These ions can also lose water, ammonia, and/or carbon monoxide, although they are usually less intense than the original internal fragment from which there were derived. In particular, check for internal fragments that have proline at the N terminus of the fragment (e.g. the sequence FSTPEDLMNK would very likely have the internal fragments PE, PED, and PEDL). At this point one should have accounted for most of the more abundant ions; in particular, one should be able to account for those at m/z greater than the precursor m/z. There will always be a few ions left over, but these should be few and of minor intensity. For example, Fig. 2 shows a confident de novo sequencing result.
Fig. 2.
A high-quality match between the de novo sequence (inset) and the spectrum.
Automated De Novo Sequencing Algorithms
Manual de novo sequencing is a fun skill to develop if one likes solving puzzles, but with high-throughput generation of MS/MS data, serious work requires automation (Table III lists the current availability of some of the software discussed here). For the purpose of algorithm design, one must formally define an optimization goal for the desired solution. For de novo sequencing, this is achieved via a scoring function that measures the matching quality between a peptide sequence and the MS/MS spectrum. With such a scoring function, the de novo sequencing problem is formulated as:
Table III. The availability of some software reviewed in the article.
De Novo Sequencing
Given a peptide MS/MS spectrum and a mass value M, compute an amino acid sequence P, such that the total residue mass is equal to M, and the matching score between P and the spectrum is maximized.
A simple brute force algorithm for automated de novo sequencing is to enumerate all the possible peptide sequences with the given precursor mass, and report the sequence that achieves the highest score. However, such a naive approach would result in exponential growth of the time complexity (i.e. required computational time) as the length of the peptide increases, and quickly becomes infeasible for peptides longer than ten residues. There are also efforts to use heuristic algorithms such as divide and conquer and pruning to make the exhaustive search faster (38). But these searching algorithms generally do not satisfy the high throughput requirement, and typically require minutes to process one spectrum. Fortunately, more efficient algorithms have been developed. In the rest of this section, the scoring function, and two algorithmic models, spectrum graph and PEAKS, are reviewed.
Scoring Function
The choice of scoring function may greatly influence the accuracy and time complexity of the de novo sequencing algorithm. Superficially, it would seem advantageous to include all knowledge about peptide fragmentation in the scoring function, as this would tend to make the scoring function more accurate. However, inclusion of many additional factors might make finding an optimal sequence computationally intractable, which in turn would make the inclusion of additional fragmentation knowledge ineffective. For example, if internal fragment ions are considered in the scoring function, the problem of finding the optimal sequence was proven to be “NP-hard” (79). NP-hardness is a common technique used in algorithm design to prove the nonexistence of an efficient algorithm to find the optimal solution in a reasonable time period (80). Furthermore, assigning appropriate weighting factors to many additional factors in a scoring function is not trivial. For these practical reasons, efficient scoring functions in today's de novo sequencing algorithms use only a subset of known factors.
Dančík et al. (39) first described a general framework for the de novo sequencing scoring function. Let P = a1a2…an be a peptide sequence of length n with total residue mass M. Fragmentation between amino acids results in two pieces called a prefix (a1 a2…ai) and a suffix (ai+1…an). Suppose the prefix has a total residue mass m (called the prefix mass) and the suffix has a total residue mass M − m. Several fragment ion types (see Section “Fragment Ions”) are expected to form peaks in the spectrum at mass values m + δx and M − m + δy. Here δx = (x = 1,2,…l) and δy = (y = l + 1,…,k) are the mass offsets of the corresponding fragment ion types (see Table I). Thus, by examining the appearance of peaks at these expected mass values, a positive reward or a negative penalty is added toward the score of the sequence P. The precise value of the reward or penalty is computed by the “log-likelihood-ratio” method as follows.
For each fragment ion type, the probability p that the corresponding peak appears in the spectrum can be statistically learned from a large amount of MS/MS spectra with known peptide sequences. Additionally, the background probability q that a peak occurs randomly at a given mass value can also be learned. Thus, the log-likelihood-ratio is defined as log for the event of observing a peak at the expected mass value; and is log for the event of missing a peak at the expected mass value. Because p is normally greater than q, observing a particular fragment ion peak provides a reward to the scoring function, and the missing of the peak causes a penalty.
The score contribution of the fragmentation at prefix mass value m, denoted by f(m), is defined as the total of the log-likelihood-ratio score of all fragment ion types at mass values m + δx and M − m + δy, for x = 1,2,…l and y = l + 1,…,k. The score of a peptide sequence P = a1a2…an, is then defined as sc(P) = Σi = 1n f(mi). Here mi is the total residue mass of the prefix a1a2…ai.
There are extensions of this general scoring framework in the literature. First, such a simple framework assumes independence between different fragment ion types. This is an oversimplification. To account for the correlations between different fragmentation ion types, the PepNovo program (36) used a Bayesian network model to calculate f(m). Additionally, the probability of observing a certain type of fragment ion peak is learned for different mass regions of the spectrum. In the NovoHMM program (34), the dependence between different ion types is also taken into account by a Hidden Markov Model. Second, the relative and absolute intensities of the fragment peaks are ignored in the Dančík framework. The PepNovo program (36) accounted for the intensity information by discretization of the intensity values into high, medium, and low. Liu et al. (81) combined the intensity and the rank of a peak into a significance value, and applied similar statistics on the significance value. Although making the scoring function more accurate, these extensions generally increase the number of parameters to be learned from the MS/MS data, and require a larger number of spectra with known peptide sequences in order to avoid the over-fitting problem.
Another interesting way to define a scoring function without using the above framework is to use computer simulation to predict the MS/MS spectrum from the peptide sequence, and then use the correlation between the predicted and real spectra as a score (38).
Spectrum Graph Model
In mathematics, a graph is an abstract representation of a set of nodes (vertices) and edges that connect pairs of nodes. A significant number of algorithms developed in computer science are graph algorithms, and many practical problems can be formulated as problems on graphs. De novo sequencing is no exception, and the so-called spectrum graph model (30) is widely used for developing de novo sequencing algorithms.
In the spectrum graph model, a node represents a possible interpretation of a peak in the spectrum. In its simplest form, each mass spectral peak generates two nodes, one for the possible y-ion interpretation of that ion, and one for the b-ion. Two nodes in the graph are connected with an edge if they are of the same type (either y or b-ion interpretation), and their corresponding mass values differ by the mass of an amino acid residue. In addition, each node in the graph is assigned a score indicating its significance. The score can be computed by the method discussed in Section “Scoring Function.” Consequently, de novo sequencing is reduced to finding a path in the graph with the maximum total node score.
Although it is entirely possible for a b-type and a y-type ion to have the same mass, some have argued that it is necessary to avoid having a peak be explained as both a y-ion and a b-ion. This can be achieved by finding the optimal “antisymmetric” path in the spectrum graph. An efficient algorithm based on dynamic programming (a popular algorithm design technique in computer science) is available (39, 42). Readers are referred to (30, 39, 42) for more details of the spectrum graph model and its algorithm.
The PEAKS Algorithm
One difficulty encountered with the spectrum graph model is how to handle missing fragment ions, which can result in gaps in the correct path connecting the N and C termini. To avoid this issue, PEAKS used a different algorithm for de novo sequencing (82). Algorithms using the spectrum graph model perform dynamic programming on the nodes of the graph, where each node corresponds to a fragment ion. In contrast, PEAKS' algorithm performs dynamic programming on the mass values regardless of the presence of an observed fragment ion. Consequently, the PEAKS algorithm requires the scoring function f(m) defined in Section “Scoring Function” to be precalculated for every mass value m between 0 and M, even in the absence of any fragment ions (in which case usually f(m) < 0).
The algorithm, as presented here, is a much simplified version of the original PEAKS algorithm (82). For the sake of clarity nominal mass values are assumed. Let m(x) be the mass of an amino acid residue x. Recall that in Section “Scoring Function,” the score of a peptide sequence P = a1a2…an, is defined as sc(P) = Σi = 1n f(mi), where mi is the total residue mass of the prefix a1a2…ai. Note that this score definition can also be used to evaluate an N-terminal partial sequence P of a peptide. Denote BestScore(m) as the best score that an N-terminal partial sequence with total residue mass m can achieve. Because the optimal N-terminal sequence always consists of another optimal N-terminal sequence that is one residue shorter, it can be concluded that BestScore(m) = f(m) + maxresidue x BestScore(m − m(x)). In other words, the fragment ion evidence at mass M, as represented by f(m), is added to the maximum value of BestScore(m − m(x)), where m(x) represents all of the common amino acid residue masses. Thus, the actual algorithm first computes BestScore(m) for every m using the above formula. Then a backtracking procedure is used to repetitively compute the residues of the optimal sequence from C- to N- terminus. In the actual implementation of PEAKS (37), a two-round approach is employed, where a simple score function is used in the first round, which computes 10,000 sequences with the top matching scores. These candidates are further evaluated by a more sophisticated scoring function that takes account other fragment ion types such as immonium ions and the internal cleavage ions.
De Novo Sequencing Errors and Sequence Tags
There are several kinds of sequencing errors. Leucine and isoleucine are isomeric and impossible to distinguish using low energy CID. Low mass accuracy fragment ion measurements cannot distinguish between lysine and glutamine (differ by 0.036 Da) nor between phenylalanine and oxidized methionine (differ by 0.033 Da). For low resolution mass analyzers, precursor ions with higher charges can be difficult to sequence correctly, because of the additional ambiguity resulting from an inability to determine fragment ion charge states. Another problem is that sometimes it can be difficult to determine the directionality of a sequence. In other words, a long contiguous series of ions separated by amino acid residue masses may be identified, but it may not be easy to determine if, for example, it is a y-type or b-type series. If the wrong decision is made, then the derived sequence is backwards.
In order to delineate a sequence, cleavages must occur between every amino acid, but this does not always happen. Cleavage between the two N-terminal amino acids is often nonexistent, since b1 ions are never seen in low energy CID of unmodified peptides and the corresponding y-type ion is often of low abundance and may not be observed either. Poor quality MS/MS spectra may have some fragment ions missing, simply because of poor signal-to-noise. Cleavage on the C-terminal side of proline in low energy CID (or the N-terminal side in ETD data) is usually absent. Nonmobile proton low energy CID spectra are usually of poor quality, with missing fragment ions. In other words, it is not unusual to be missing a ladder peak (e.g. yk). In this case there may be several different amino acid combinations that can explain the gap between yk-1 and yk+1. The other is that there are multiple peaks that can serve as the yk peak, and the software (or person) does not know which one is right. In either case, there may be ambiguity regarding a small segment of a peptide. The software knows the total residue mass, but can easily make incorrect sequence determinations. Sometimes, other evidence such as internal cleavage ions, immonium ions, or neutral loss ions might help make the distinction, but the confidence of the prediction made from these secondary ions is usually low. Hence, it is not unusual for a segment of amino acids in the correct sequence to be substituted by an incorrect segment with the same total residue mass. This is referred to as the mass segment error.
To overcome the problem caused by the mass segment errors, a de novo sequencing program often presents results in one of the following two formats. First, some software such as Lutefisk will replace a segment of low confidence amino acids with their total residue mass, and generate a sequence such as MEG[199.1]CK. By presenting only the high confidence amino acids, the overall accuracy of the software is improved. Other software (e.g. PEAKS) will attempt to predict the entire peptide sequence, but indicate those regions with low confidence. This gives the users more flexibility in that they can decide later whether to keep the low confidence sequence, or convert them to mass segments, as described above. There are also algorithms that are purposely designed for finding high confidence partial sequence tags (83, 84).
Homology Searching with De Novo Sequence Tags
De novo sequencing alone can only derive partial sequence information for individual peptides, but it cannot identify the protein. However, if the protein is in a database, then a tag sequence search can retrieve the protein from the database, as illustrated by Mann and Wilm's early protein identification work with de novo sequence tags (16). To obtain the identity of a protein not present in a database, Taylor and Johnson proposed searching for homologs of de novo sequencing tags by utilizing a modified version of the FASTA homology search tool (32). If a database protein contains a few peptides that are similar to the de novo sequence tags, the protein being studied is likely to be a homolog of the database-derived protein. Although the complete sequence is unknown, one might gain some insight regarding the target protein by identifying a homolog.
Homology (or similarity) searches have long been used in molecular biology. Sequence alignment is the most commonly used model for measuring the similarities between two sequences. In a sequence alignment, extra spaces (often denoted by a ‘-’ symbol) are added to appropriate positions of the two given sequences in order to maximize sequence similarity (see Fig. 3). A pre-defined score matrix such as the BLOSUM matrix (85) specifies the similarity score between any pair of aligned amino acids. A higher BLOSUM score indicates evolutionarily conservative mutations. The sequence alignment score is the sum of the similarity scores. A sequence alignment algorithm such as the Smith-Waterman algorithm (86) is used to construct the optimal alignment with the maximum alignment score (for a more comprehensive review see (87)).
Fig. 3.
An example of a sequence alignment. The dash symbols indicate the inserted spaces, and the vertical bars indicate exact matches.
There are many homology search programs available such as BLAST (88). A few of these homology search programs (BLAST (88), FASTA (48), and Shotgun (89)) have been modified to allow for sequence tag searches (CIDentify (32), MS-BLAST (49), FASTS (51), and MS-Shotgun (50)). MS-Shotgun was later renamed MS-Homology and is included in the Protein Prospector package. Most of these modifications are related to changing the search parameters in accordance with the short length of the query peptide sequences. Although proven to be a powerful approach, a major limitation of the conventional homology search tools for searching de novo sequence tags is that these programs do not take into account the various de novo sequencing errors (Section “De novo Sequencing Errors and Sequence Tags”). These sequencing errors have very different statistical properties compared with evolutionary mutations, and ignoring these differences could significantly affect homology search accuracy. Fig. 4 shows such an example, where in Fig. 4A only evolutionary mutations are taken into account, and the comparison shows very low similarity. In contrast, if de novo sequencing errors and evolutionary mutations are both taken into account, as shown in Fig. 4B, the alignment is much more significant.
Fig. 4.
A, An insignificant match between a de novo sequence tag and a homologous sequence. B, The match becomes significant if the de novo sequencing errors are taken into account. The square brackets indicate the de novo sequence mass segment errors.
The first homology search tool to account for both homology and de novo sequencing errors was CIDentify (32), which was a modification of the FASTA program (90). In this implementation, the best initial match between the query sequence and each protein sequence in the database is rescored to account for sequencing errors. For example, Gly-Gly mismatches to Asn are rescored as perfect matches (likewise for Ala + Gly to Gln), mismatched dipeptides that have identical summed residue masses are rescored as partial matches, and most of the errors described in Section “De novo Sequencing Errors and Sequence Tags” are identified and re-scored. Later on, a few more programs were developed for dealing specifically with these de novo sequencing errors. These include GutenTag (54), OpenSea (52), and SPIDER (53). Among these programs, GutenTag allows de novo sequencing errors but requires the database sequences to be exact. In contrast, OpenSea and SPIDER allow both de novo sequencing errors and inexact database sequences.
OpenSea (52) used a heuristic algorithm to deal with the de novo sequencing errors. If a mismatch between the tag and the database peptide sequence is encountered, the software will examine if a segment of the tag has the same total mass of a segment of the database peptide sequence. If there is, then the software will regard the mismatch as caused by a de novo sequencing error. Otherwise, it regards the mismatch as caused by evolutionary mutations or post-translational modifications. Unlike CIDentify, OpenSea is capable of considering mass segments larger than dipeptides, plus some some additional advantages. This program went a long ways toward solving the de novo sequencing error problem.
Another program, SPIDER (53), uses a more sophisticated model, which additionally deals with the possible overlaps between the de novo sequencing errors and the homology mutations. Also, SPIDER considers the possibility of reconstructing the real peptide sequence by combining both the de novo sequence tag and the homolog. The SPIDER sequencing model takes the de novo sequence tag X and the database sequence Z as input, and tries to compute the real peptide sequence Y. The mismatches between X and Y are explained by the de novo sequencing errors, and the mismatches between Y and Z by the evolutionary mutations. SPIDER's algorithm ensures that the computed sequence Y minimizes the total number of sequencing errors and mutations. With the efficient sequencing algorithm as a subroutine, SPIDER conducts the homology searches by matching the tag X to every peptide Z in the database, and reporting the peptide Z that minimizes the total sequencing errors and mutations. However, because there are millions of database peptides, heuristics are used to speed up the searching. One such heuristics is to call the sequencing algorithm only if there are three or more consecutive letter matches between X and Z. This significantly speeds up the search with only minor effect on the search sensitivity. Such a heuristic (called filtration) has been used in most conventional homology search tools.
Worked Examples
Identifying Peptides Not in a Database
Yin et al. (91) studied a few important sulfur-rich proteins in common beans with the assistance of de novo sequencing. The de novo peptides found by PEAKS were compared with the conceptual translation of the ESTs to identify contigs coding for sulfur-rich proteins, as well as to confirm the N terminus of the mature polypeptide. The de novo peptides covered 48 and 79% of the deduced sequences of the α and β subunits from the major legumin cDNA. In the study of spider hemolymph, Trabalon et al. (92) used PEAKS de novo sequencing to interpret the MS/MS spectra that could not be identified by the Mascot database search software, and reported 64 de novo sequence tags from nine gel bands.
Tag Homology Searches for Studying Related Organisms
Shevchenko et al. (49) developed an implementation of the homology search program Blast to aid in matching de novo sequencing results with homologous database sequences. The method was demonstrated on a couple of organisms (dog and a yeast species) whose genomes, at the time, were not sequenced. More recently, the tag homology search approach has mostly been used in conjunction with the database search approach to identify additional proteins from unsequenced genome. Different combinations of software tools (such as Mascot, PEAKS, PepNovo, MS-BLAST, FASTS, and SPIDER) have been used in the literature. Waridel et al. (93) identified 55 Dunaliella Salina membrane proteins from 32 gel spots using this method, where six proteins were identified only via a tag homology search. Catusse et al. (94) studied the sugarbeet seed proteome, and a total of 759 proteins were identified, including a number of proteins that had not previously been described in seeds. Hatano and Hamada (95) used tag homology searching to identify six of seven detected bands in a gel from the pitcher fluid of the carnivorous plant Nepenthes alata. Tannu and Hemby (96) studied Macaca mulatta proteins and identified 13 human homologs from 30 excised gel spots. A simulation work was published by Habermann et al. (97) to study the effectiveness of the tag homology search approach for cross-species protein identification, where de novo sequencing tags were generated by computer simulation and not experimental mass spectrometry data. They demonstrated that over 80% of the proteins could be positively identified within the mammalian subkingdom.
Identifying Unexpected Chemical Modifications
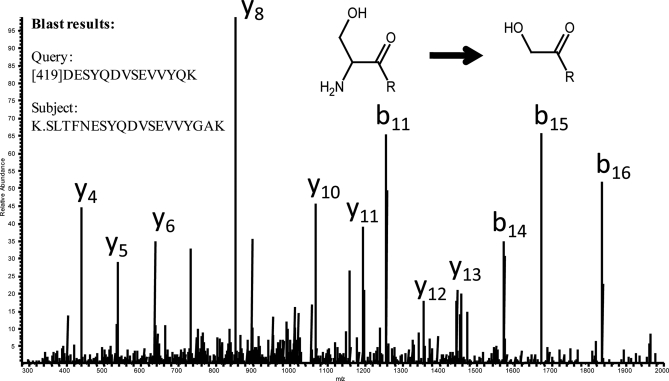
Using LC-MS/MS from an orbitrap/linear trap hybrid mass spectrometer, a data set was acquired from a mouse liver sample that had been subjected to the glycocapture procedure (98). This procedure involves a series of enzymatic and chemical steps. Of particular concern was treatment with sodium periodate, which is supposed to specifically oxidize vicinal diols, but may have additional side reactions. In order to locate unexpected chemical modifications, the MS/MS spectra were subjected to both a database search (X!Tandem) and automated de novo sequencing (PepNovo). Spectra that were not identified in the database search, but were assigned high scoring sequences from PepNovo were subjected to a homology search using BLAST. One example is shown in Fig. 5, which depicts an MS/MS spectrum that did not match to any database sequence. However, PepNovo derived a top scoring sequence of [419.131]DESYQDVSEVVYQK, where the number in brackets indicates a chunk of unsequenced mass on the N-teriminus. The top BLAST result is shown in the figure inset on the left, and matches to a sequence from mouse antithrombin-III. In this match, the query sequence had an Asp that aligned with Asn in the database, which was because of the enzymatic deglycosylation step used in the glycocapture procedure. The query sequence also has the single amino acid Gln whereas the database sequence contains Gly-Ala at this position. As noted already, this is a common error that is made in de novo sequencing, since Gly-Ala has the exact same mass as Gln. Looking toward the N terminus of the database sequence shows the presence of a tryptic cleavage site followed by Ser-Leu-Thr-Phe sequence that precedes NESYQDVSEVVYGAK. However, the database-derived sequence is 29.029 Da greater in mass than the measured mass of this peptide, and the current hypothesis is that this is because of periodate oxidation of the vicinal amine and hydroxyl groups present when serine is at the N terminus of a peptide. The 419 Da at the N terminus is likely to contain the Leu-Thr-Phe, plus an oxidized piece of Ser (see inset of Fig. 5). This demonstration shows how automated de novo sequencing combined with homology searches can lead to a better understanding of side reactions that may be occur during chemical processing of complex samples.
Fig. 5.
Example of using de novo sequencing to identify unexpected chemical modifications. The glycocapture procedure (97) was used to isolate formerly N-glycosylated peptides from mouse liver, which were analyzed using an orbitrap-LTQ hybrid mass spectrometer. MS/MS spectra for which no database-derived sequence could be determined, but that gave high scoring PepNovo-derived sequences, were identified. Shown is one such example, where the PepNovo sequence closely matched mouse antithrombin-III (inset on the left). The difference between the observed mass and that calculated from the database sequence (29.03 Da) suggested that the periodate oxidation step in the glycocapture procedure had modified the N-terminal serine, as shown in the inset in the upper right.
Complete Protein Sequencing
On rare occasions, de novo sequencing has been used to derive complete protein sequences (11, 99). In this method, purified proteins are digested with multiple enzymes to obtain overlapping peptides that are then subjected to tandem mass spectrometry and either manual or computer-assisted de novo sequencing. The peptide sequences are then assembled together according to the sequence overlap. Bandeira et al. has shown how to automate such analyses (100–102). To further improve the protein sequencing accuracy, the CHAMPS algorithm (103) exploited the SPIDER peptide sequencing idea to correct the de novo sequencing errors with a homologous protein sequence. The homologous protein sequence also served as a template for the algorithm to assemble the peptide sequences in the correct order.
Improving Database Searches With De Novo Sequence Tags
Although the most obvious use of de novo sequencing and homology searches is in the study of unsequenced genomes, de novo sequence tags have also been used to improve database searches with known sequence databases. One goal was to improve the database search speed. Matching sequence tags to database sequences can be very efficient when using deliberate index structures. Frank et al. demonstrated that filtration using short sequence tags can be as much as 2000-fold more efficient compared with using only the parent mass as a filter (83). In the InspecT program (22) for identifying modified peptides with unknown PTMs, tag matching is used as a quick way to filter peptide-spectrum match candidates prior to the more time-consuming alignment algorithm that is used for determining the PTM mass. Similar work was presented by Liu et al. (104). A second goal of this approach is to improve the confidence in peptide-spectrum matches. A random match between a long de novo sequence tag and a database peptide is an unlikely event. So, when it happens, the confidence on the correctness of the database peptide is increased. Lutefisk1900 (25) exploits this fact by comparing scores from de novo and database derived sequences, where the assumption is that a correct database sequence out-scores any of the de novo sequences. This property has also been used in the scoring function of the PEAKS DB software (105) to better separate the true and false identifications in the database search.
Current Limitations and Possible Future Developments
Compared with the database search approach, the major limitation of today's de novo sequencing software is the lack of automated statistical validation. Without such an automated validation method, one still needs to inspect the peptide-spectrum matches to decide which de novo sequencing results to take seriously. Thus, although the de novo sequencing software can save time, the evaluation of results still involves a significant amount of manual work. Therefore, a reliable and automated validation method would greatly expand the usability of de novo sequencing in today's proteomics research. There has been some initial research in this direction, but a widely accepted method has not been available. Lutefisk can estimate z-values for de novo sequencing results by generating and scoring sequences from several incorrect peptide MW values (unpublished results). PEAKS computes a confidence score by comparing the top de novo sequencing peptide with the suboptimal peptides (37), and MSNovo (40) estimates a p value by comparing the de novo sequencing score with scores from randomly generated peptides. Additional work along these lines seems warranted. Likewise, evaluation of homologous matches between de novo sequencing results and protein sequence databases from related organisms is not fully automated and manual curation of the results can be time consuming.
Another important limitation of de novo sequencing is in the uncertainty regarding the complete peptide sequence. Database searches might still succeed when fragment ions are missing (if the full peptide is indeed in the database); however, de novo sequencing will not be able to derive a complete sequence or will have uncertainty in a portion of the derived sequence. To confidently sequence a complete peptide sequence, a promising method may be to combine multiple spectra produced by different fragmentation techniques such as CID and ETD. Some preliminary research in this direction have reported improvements in de novo sequencing accuracies (106–108). Also, the use of a MS3 spectra together with the associated MS/MS spectrum (109), or chemical/enzymatic labeling (110) can improve de novo sequence certainty.
Footnotes
* BM has been supported in part by Natural Sciences and Engineering Research Council of Canada (RGPIN 238748). RJ has been supported in part by Federal funds from the National Science foundation (NSF MRI No. 0923536), and the Systems Biology Initiative of the Duchy of Luxembourg.
 This article contains supplemental Presentation.
This article contains supplemental Presentation.
1 The abbreviations used are:
- FAB
- fast atom bombardment
- MS/MS
- tandem MS
- CID
- collision-induced dissociation
- ETD
- electron transfer dissociation.
REFERENCES
- 1. Barber M., Jolles P., Vilkas E., Lederer E. (1965) Determination of amino acid sequences in oligopeptides by mass spectrometry. I. The structure of fortuitine, an acylnonapeptide methyl ester. Biochem. Biophys. Res. Commun. 18, 469–473 [DOI] [PubMed] [Google Scholar]
- 2. Biemann K., Vetter W. (1960) Separation of peptide derivatives by gas chromatography combined with the mass spectrometric determination of the amino acid sequence. Biochem. Biophys. Res. Commun. 3, 578–584 [Google Scholar]
- 3. Biemann K., Gapp G., Seibl J. (1959) Application of mass spectrometry to structure problems. I. Amino acid sequence in peptides. J. Am. Chem. Soc. 81, 2274–2275 [Google Scholar]
- 4. Hudson G., Biemann K. (1976) Mass spectrometric sequencing of proteins. The structure of subunit I of monellin. Biochem. Biophys. Res. Commun. 71, 212–220 [DOI] [PubMed] [Google Scholar]
- 5. Burgus R., Dunn T. F., Desiderio D., Ward D. N., Vale W., Guillemin R. (1970) Characterization of ovine hypothalamic hypophysiotropic TSH-releasing factor. Nature 226, 321–325 [DOI] [PubMed] [Google Scholar]
- 6. Khorana H. G., Gerber G. E., Herlihy W. C., Gray C. P., Anderegg R. J., Nihei K., Biemann K. (1979) Amino acid sequence of bacteriorhodopsin. Proc. Natl. Acad. Sci. U. S. A. 76, 5046–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Biemann K., Cone C., Webster B. R., Arsenault G. P. (1966) Determination of the amino acid sequence in oligopeptides by computer interpretation of their high-resolution mass spectra. J. Am. Chem. Soc. 88, 5598–5606 [DOI] [PubMed] [Google Scholar]
- 8. Barber M., Bordoli R. S., Sedgwick R. D., Tyler A. N. (1981) Fast atom bombardment of solids (F.A.B.): a new ion source for mass spectrometry. J. Chem. Soc., Chem. Commun. 325–327 [Google Scholar]
- 9. Biemann K., Martin S. (1987) Mass spectrometric determination of the amino acid sequence of peptides and proteins. Mass Spectr. Rev. 6, 1–76 [Google Scholar]
- 10. Hunt D. F., Yates J. R., III, Shabanowitz J., Winston S., Hauer C. R. (1986) Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 83, 6233–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson R. S., Biemann K. (1987) The primary structure of thioredoxin from Chromatium vinosum determined by high-performance tandem mass spectrometry. Biochemistry 26, 1209–1214 [DOI] [PubMed] [Google Scholar]
- 12. Johnson R. S., Biemann K. (1989) Computer program (SEQPEP) to aid in the interpretation of high-energy collision tandem mass spectra of peptides. Biol. Mass Spectr. 18, 945–957 [DOI] [PubMed] [Google Scholar]
- 13. Fenn J. B., Mann M., Meng C. K., Wong S. F., Whitehouse C. M. (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 [DOI] [PubMed] [Google Scholar]
- 14. Karas M., Hillenkamp F. (1988) Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60, 2299–2301 [DOI] [PubMed] [Google Scholar]
- 15. Eng J. K., Mccormack A. L., Yates J. R., III (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectr. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 16. Mann M., Wilm M. (1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66, 4390–4399 [DOI] [PubMed] [Google Scholar]
- 17. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 18. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 19. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 20. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox J., Neuhauser N., Michalski A., Scheltema R. A, Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the Maxquant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 22. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 23. Clauser K. R., Baker P., Burlingame A. L. (1999) Role of accurate mass measurement (+/- 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71, 2871–2882 [DOI] [PubMed] [Google Scholar]
- 24. Nesvizhskii A. I. (2007) Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol. Biol. 367, 87–119 [DOI] [PubMed] [Google Scholar]
- 25. Taylor J. A., Johnson R. S. (2001) Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal. Chem. 73, 2594–2604 [DOI] [PubMed] [Google Scholar]
- 26. Sakurai T., Matsuo T., Matsuda H., Katakuse I. (1984) PAAS3: A computer program to determine probable sequence of peptides from mass spectrometric data. Biomed. Mass Spectrom. 11, 396–399 [Google Scholar]
- 27. Ishikawa K., Niwa Y. (1986) Computer-aided peptide sequencing by fast atom bombardment mass spectrometry. Biomed. Environ. Mass Spectrom. 13, 373–380 [DOI] [PubMed] [Google Scholar]
- 28. Yates J. R., III, Griffin P. R., Hood L. E. (1991) in:, Villafranca JJ. (Ed.), vol. 2, Academic Press, San Diego, CA: 1991, pp. 477–485 [Google Scholar]
- 29. Scoble H. A., Biller J. E., Biemann K. (1987) A graphics display-oriented strategy for the amino acid sequencing of peptides by tandem mass spectrometry. Fresenius' Z. Anal. Chem. 327, 239–245 [Google Scholar]
- 30. Bartels C. (1990) Fast algorithm for peptide sequencing by mass spectroscopy. Biomed. Environ. Mass Spectrom. 19, 363–368 [DOI] [PubMed] [Google Scholar]
- 31. Hines W. M., Falick A. M., Burlingame A. L., Gibson B. W. (1992) Pattern-based algorithm for peptide sequencing from tandem high energy collision-induced dissociation mass spectra. J. Am. Soc. Mass Spectr. 3, 326–336 [DOI] [PubMed] [Google Scholar]
- 32. Taylor J. A., Johnson R. S. (1997) Sequence database searches via de novo peptide sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectr. 11, 1067–1075 [DOI] [PubMed] [Google Scholar]
- 33. Grossmann J., Roos F. F., Cieliebak M., Lipták Z., Mathis L. K., Müller M., Gruissem W., Baginsky S. (2005) AUDENS: a tool for automated peptide de novo sequencing. J. Proteome Res. 4, 1768–1774 [DOI] [PubMed] [Google Scholar]
- 34. Fischer B., Roth V., Roos F., Grossmann J., Baginsky S., Widmayer P., Gruissem W., Buhmann J. M. (2005) NovoHMM: a hidden Markov model for de novo peptide sequencing. Anal. Chem. 77, 7265–7273 [DOI] [PubMed] [Google Scholar]
- 35. Pan C., Park B. H., McDonald W. H., Carey P. A, Banfield J. F., VerBerkmoes N. C., Hettich R. L., Samatova N. F. (2010) A high-throughput de novo sequencing approach for shotgun proteomics using high-resolution tandem mass spectrometry. BMC bioinformatics 11, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frank A., Pevzner P. A. (2005) PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 77, 964–973 [DOI] [PubMed] [Google Scholar]
- 37. Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectr. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 38. Zhang Z. (2004) De novo peptide sequencing based on a divide-and-conquer algorithm and peptide tandem spectrum simulation. Anal. Chem.76, 6374–6383 [DOI] [PubMed] [Google Scholar]
- 39. Dancik D., Addona T. A., Clauser K. R., Vath J. E., Pevzner P. A. (1999) De novo peptide sequencing via tandem mass spectrometry. J. Comp. Biol. 6, 327–342 [DOI] [PubMed] [Google Scholar]
- 40. Mo L., Dutta D., Wan Y., Chen T. (2007) MSNovo: a dynamic programming algorithm for de novo peptide sequencing via tandem mass spectrometry. Anal. Chem. 79, 4870–4878 [DOI] [PubMed] [Google Scholar]
- 41. Chi H., Sun R. X., Yang B., Song C. Q., Wang L. H., Liu C., Fu Y., Yuan Z. F., Wang H. P., He S. M., Dong M. Q. (2010) pNovo: de novo peptide sequencing and identification using HCD spectra. J. Proteome Res. 9, 2713–2724 [DOI] [PubMed] [Google Scholar]
- 42. Chen T., Kao M. Y., Tepel M., Rush J., Church G. M. (2001) A dynamic programming approach to de novo peptide sequencing via tandem mass spectrometry. J. Computational Biol. 8, 325–337 [DOI] [PubMed] [Google Scholar]
- 43. DiMaggio P. A., Jr., Floudas C. A. (2007) De novo peptide identification via tandem mass spectrometry and integer linear optimization. Anal. Chem. 79, 1433–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pevtsov S., Fedulova I., Mirzaei H., Buck C., Zhang X. (2006) Performance evaluation of existing de novo sequencing algorithms. J. Proteome Res. 5, 3018–3028 [DOI] [PubMed] [Google Scholar]
- 45. Bringans S., Kendrick T. S., Lui J., Lipscombe R. (2008) A comparative study of the accuracy of several de novo sequencing software packages for datasets derived by matrix-assisted laser desorption/ionisation and electrospray. Rapid Commun. Mass Spectr. 22, 3450–3454 [DOI] [PubMed] [Google Scholar]
- 46. Jung S., Fladerer C., Braendle F., Madlung J., Spring O., Nordheim A. (2010) Identification of a novel Plasmopara halstedii elicitor protein combining de novo peptide sequencing algorithms and RACE-PCR. Proteome Sci. 8, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 48. Pearson W. R., Lipman D. J. (1988) Improved Tools for Biological Sequence Comparison. Proc. Natl. Acad. Sci. U. S. A. 85, 2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shevchenko A., Sunyaev S., Loboda A., Bork P., Ens W., Standing K. G. (2001) Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 73, 1917–1926 [DOI] [PubMed] [Google Scholar]
- 50. Huang L., Jacob R. J., Pegg S. C., Baldwin M. A, Wang C. C., Burlingame A. L., Babbitt P. C. (2001) Functional assignment of the 20 S proteasome from Trypanosoma brucei using mass spectrometry and new bioinformatics approaches. J. Biol. Chem. 276, 28327–28339 [DOI] [PubMed] [Google Scholar]
- 51. Mackey A. J., Haystead T. A., Pearson W. R. (2002) Getting more from less: algorithms for rapid protein identification with multiple short peptide sequences. Mol. Cell. Proteomics 1, 139–147 [DOI] [PubMed] [Google Scholar]
- 52. Searle B. C., Dasari S., Turner M., Reddy A. P., Choi D., Wilmarth P. A., McCormack A. L., David L. L., Nagalla S. R. (2004) High-throughput identification of proteins and unanticipated sequence modifications using a mass-based alignment algorithm for MS/MS de novo sequencing results. Anal. Chem. 76, 2220–2230 [DOI] [PubMed] [Google Scholar]
- 53. Han Y., Ma B., Zhang K. (2005) SPIDER: software for protein identification from sequence tags with de novo sequencing error. J. Bioinformatics Computational Biol. 3, 697–716 [DOI] [PubMed] [Google Scholar]
- 54. Tabb D. L., Saraf A., Yates J. R., 3rd (2003) GutenTag: high-throughput sequence tagging via an empirically derived fragmentation model. Anal. Chem. 75, 6415–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morris H. R., Paxton T., Dell A., Langhorne J., Berg M., Bordoli R. S., Hoyes J., Bateman R. H. (1996) High sensitivity collisionally-activated decomposition tandem mass spectrometry on a novel quadrupole / orthogonal-acceleration time-of-flight mass spectrometer. Rapid Commun. Mass Spectr. 10, 889–996 [DOI] [PubMed] [Google Scholar]
- 56. Makarov A. (2000) Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal. Chem. 72, 1156–1162 [DOI] [PubMed] [Google Scholar]
- 57. March R. E. (1997) An introduction to quadrupole ion trap mass spectrometry. J. Mass Spectr. 32, 351–369 [Google Scholar]
- 58. Lau K. W., Hart S. R., Lynch J. A., Wong S. C., Hubbard S. J., Gaskell S. J. (2009) Observations on the detection of b- and y-type ions in the collisionally activated decomposition spectra of protonated peptides. Rapid Commun. Mass Spectrom. 23, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 59. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 60. Wysocki V. H., Tsaprailis G., Smith L. L., Breci L. A. (2000) Mobile and localized protons: a framework for understanding peptide dissociation. J. Mass Spectr. 35, 1399–1406 [DOI] [PubMed] [Google Scholar]
- 61. Boyd R., Somogyi A. (2010) The mobile proton hypothesis in fragmentation of protonated peptides: a perspective. J. Am. Soc. Mass Spectr. 21, 1275–1278 [DOI] [PubMed] [Google Scholar]
- 62. Zubarev R. A., Kelleher N. L., McLafferty F. W. (1998) Electron capture dissociation of multiply charged protein cations. a nonergodic process. J. Am. Chem. Soc. 120, 3265–3266 [Google Scholar]
- 63. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bowie J. H., Brinkworth C. S., Dua S. (2002) Collision-induced fragmentations of the (M-H)- parent anions of underivatized peptides: an aid to structure determination and some unusual negative ion cleavages. Mass Spectr. Rev. 21, 87–107 [DOI] [PubMed] [Google Scholar]
- 65. Kelleher N. L. (2004) Top-down proteomics. Anal. Chem. 76, 197A–203A [PubMed] [Google Scholar]
- 66. Biemann K. (1990) Appendix 5. Nomenclature for peptide fragment ions (positive ions). Methods Enzymol. 193, 886–887 [DOI] [PubMed] [Google Scholar]
- 67. Roepstorff P., Fohlman J. (1984) Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11, 601. [DOI] [PubMed] [Google Scholar]
- 68. Paizs B., Suhai S. (2005) Fragmentation pathways of protonated peptides. Mass Spectr. Rev. 24, 508–548 [DOI] [PubMed] [Google Scholar]
- 69. Yu W., Vath J. E., Huberty M. C., Martin S. (1993) Identification of the facile gas-phase cleavage of the Asp-Pro and Asp-Xxx peptide bonds in matrix-assisted laser desorption time-of-flight mass spectrometry. Anal. Chem. 65, 3015–3023 [DOI] [PubMed] [Google Scholar]
- 70. Schlosser A., Lehmann W. D. (2000) Five-membered ring formation in unimolecular reactions of peptides: a key structural element controlling low-energy collision-induced dissociation of peptides. J. Mass Spectr. 35, 1382–1390 [DOI] [PubMed] [Google Scholar]
- 71. Johnson R. S., Martin S., Biemann K. (1988) Collision induced dissociation of (m+h)+ ions of peptides. side chain specific sequence ions. Int. J. of Mass Spectrom. Ion Processes 86, 137–154 [Google Scholar]
- 72. Johnson R. S., Martin S., Biemann K., Stults J. T., Watson J. T. (1987) Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 59, 2621–2625 [DOI] [PubMed] [Google Scholar]
- 73. Vestal M. L., Campbell J. M. (2005) Tandem time-of-flight mass spectrometry. Methods Enzymol. 402, 79–108 [DOI] [PubMed] [Google Scholar]
- 74. Satoh T., Sato T., Kubo A., Tamura J. (2011) Tandem time-of-flight mass spectrometer with high precursor ion selectivity employing spiral ion trajectory and improved offset parabolic reflectron. J. Am. Soc. Mass Spectr. 22, 797–803 [DOI] [PubMed] [Google Scholar]
- 75. Swaney D. L., McAlister G. C., Wirtala M., Schwartz J. C., Syka J. E., Coon J. J. (2007) Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 79, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chalkley R. J., Brinkworth C. S., Burlingame A. L. (2006) Side-chain fragmentation of alkylated cysteine residues in electron capture dissociation mass spectrometry. J. Am. Soc. Mass Spectr. 17, 1271–1274 [DOI] [PubMed] [Google Scholar]
- 77. Zubarev R. A., Kruger N. A., Fridriksson E. K., Lewis M. A., Horn D. M., Carpenter B. K., McLafferty F. W. (1999) Electron capture dissociation of gaseous multiply-charged proteins is favored at disulfide bonds and other sites of high hydrogen atom affinity. J. Am. Chem. Soc. 121, 2857–2862 [Google Scholar]
- 78. Hubler S. L., Jue A., Keith J., McAlister G. C., Craciun G., Coon J. J. (2008) Valence parity renders z●-type ions chemically distinct. J. Am. Chem. Soc. 130, 6388–6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xu C., Ma B. (2006) Complexity and scoring function of MS/MS peptide de novo sequencing. Computational systems bioinformatics/Life Sciences Society. Computational Systems Bioinformatics Conference 361–9 [PubMed] [Google Scholar]
- 80. Garey M. R., Johnson D. S. (1979) Computers and Intractability: A Guide to the Theory of NP-Completeness, W. H. Freeman & Co., New York, NY, USA: 1979 [Google Scholar]
- 81. Liu X., Shan B., Xin L., Ma B. (2010) Better score function for peptide identification with ETD MS/MS spectra. BMC Bioinformatics 11 Suppl 1, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ma B., Zhang K., Liang C. (2005) An effective algorithm for peptide sequencing from MS/MS spectra. J. Computer System Sci. 70, 418–430 [Google Scholar]
- 83. Frank A., Tanner S., Bafna V., Pevzner P. (2005) Peptide sequence tags for fast database search in mass-spectrometry. J. Proteome Res. 4, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 84. Jeong K., Kim S., Bandeira N., Pevzner P. A. (2011) Gapped spectral dictionaries and their applications for database searches of tandem mass spectra. Mol. Cell. Proteomics 10, M110.002220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Henikoff S., Henikoff J. G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U. S. A. 89, 10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Smith T. F., Waterman M. S. (1981) Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 [DOI] [PubMed] [Google Scholar]
- 87. Brown D. G., Li M., Ma B. (2004) A tutorial of recent developments in the seeding of local alignment. J. Bioinformatics Computational Biol. 2, 819–842 [DOI] [PubMed] [Google Scholar]
- 88. Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pegg S. C., Babbitt P. C. (1999) Shotgun: getting more from sequence similarity searches. Bioinformatics 15, 729–740 [DOI] [PubMed] [Google Scholar]
- 90. Pearson W. R. (1990) Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183, 63–98 [DOI] [PubMed] [Google Scholar]
- 91. Yin F., Pajak A., Chapman R., Sharpe A., Huang S., Marsolais F. (2011) Analysis of common bean expressed sequence tags identifies sulfur metabolic pathways active in seed and sulfur-rich proteins highly expressed in the absence of phaseolin and major lectins. BMC genoics 12, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Trabalon M., Carapito C., Voinot F., Martrette J. M., Van Dorsselaer A., Gilbert C., Bertile F. (2010) Differences in Brachypelma albopilosa (Theraphosidae) hemolymph proteome between subadult and adult females. J. Exp. Zool. 313, 651–659 [DOI] [PubMed] [Google Scholar]
- 93. Waridel P., Frank A., Thomas H., Surendranath V., Sunyaev S., Pevzner P., Shevchenko A. (2007) Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics 7, 2318–2329 [DOI] [PubMed] [Google Scholar]
- 94. Catusse J., Strub J. M., Job C., Van Dorsselaer A., Job D. (2008) Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc. Natl. Acad. Sci. U. S. A. 105, 10262–10267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hatano N., Hamada T. (2008) Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. J. Proteome Res. 7, 809–816 [DOI] [PubMed] [Google Scholar]
- 96. Tannu N. S., Hemby S. E. (2007) De novo protein sequence analysis of Macaca mulatta. BMC Genomics 8, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Habermann B., Oegema J., Sunyaev S., Shevchenko A. (2004) The power and the limitations of cross-species protein identification by mass spectrometry-driven sequence similarity searches. Mol. Cell. Proteomics 3, 238–249 [DOI] [PubMed] [Google Scholar]
- 98. Zhang H., Li X. J., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 99. Martin-Visscher L. A., van Belkum M. J., Garneau-Tsodikova S., Whittal R. M., Zheng J., McMullen L. M., Vederas J. C. (2008) Isolation and characterization of carnocyclin a, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environmental Microbiol. 74, 4756–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bandeira N., Pham V., Pevzner P., Arnott D., Lill J. R. (2008) Automated de novo protein sequencing of monoclonal antibodies. Nat. Biotechnol. 26, 1336–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bandeira N., Clauser K. R., Pevzner P. A. (2007) Shotgun protein sequencing: assembly of peptide tandem mass spectra from mixtures of modified proteins. Mol. Cell. Proteomics 6, 1123–1134 [DOI] [PubMed] [Google Scholar]
- 102. Bandeira N., Tang H., Bafna V., Pevzner P. (2004) Shotgun protein sequencing by tandem mass spectra assembly. Anal. Chem. 76, 7221–7233 [DOI] [PubMed] [Google Scholar]
- 103. Liu X., Han Y., Yuen D., Ma B. (2009) Automated protein (re)sequencing with MS/MS and a homologous database yields almost full coverage and accuracy. Bioinformatics 25, 2174–2180 [DOI] [PubMed] [Google Scholar]
- 104. Liu C., Yan B., Song Y., Xu Y., Cai L. (2006) Peptide sequence tag-based blind identification of post-translational modifications with point process model. Bioinformatics 22, e307–13 [DOI] [PubMed] [Google Scholar]
- 105. Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G., Ma B. (2011) PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Submitted for publication [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Datta R., Bern M. (2009) Spectrum fusion: using multiple mass spectra for de novo Peptide sequencing. J. Computational Biol. 16, 1169–1182 [DOI] [PubMed] [Google Scholar]
- 107. Bertsch A., Leinenbach A., Pervukhin A., Lubeck M., Hartmer R., Baessmann C., Elnakady Y. A., Müller R., Böcker S., Huber C. G., Kohlbacher O. (2009) De novo peptide sequencing by tandem MS using complementary CID and electron transfer dissociation. Electrophoresis 30, 3736–3747 [DOI] [PubMed] [Google Scholar]
- 108. He L., Ma B. (2010) ADEPTS: advanced peptide de novo sequencing with a pair of tandem mass spectra. J. Bioinformatics Computational Biol. 8, 981–994 [DOI] [PubMed] [Google Scholar]
- 109. Bandeira N., Olsen J. V., Mann M., Pevzner P. A. (2008) Multi-spectra peptide sequencing and its applications to multistage mass spectrometry. Bioinformatics 24, i416–i423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Shevchenko A., Chernushevich I., Ens W., Standing K. G., Thomson B., Wilm M., Mann M. (1997) Rapid “de novo” peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer 11, 1015–1024 [DOI] [PubMed] [Google Scholar]