Abstract
Recent findings suggest that Ring finger protein 146 (RNF146), also called Iduna, have neuroprotective property due to its inhibition of Parthanatos via binding with Poly(ADP-ribose) (PAR). The Parthanatos is a PAR dependent cell death that has been implicated in many human diseases. RNF146/Iduna acts as a PARsylation-directed E3 ubquitin ligase to mediate tankyrase-dependent degradation of axin, thereby positively regulates Wnt signaling. RNF146/Iduna can also facilitate DNA repair and protect against cell death induced by DNA damaging agents or γ-irradiation. It can translocate to the nucleus after cellular injury and promote the ubiquitination and degradation of various nuclear proteins involved in DNA damage repair. The PARsylation-directed ubquitination mediated by RNF146/Iduna is analogous to the phosphorylation-directed ubquitination catalyzed by Skp1-Cul1-F-box (SCF) E3 ubiquitin complex. RNF146/Iduna has been found to be implicated in neurodegenerative disease and cancer development. Therefore modulation of the PAR-binding and PARsylation dependent E3 ligase activity of RNF146/Iduna could have therapeutic significance for diseases, in which PAR and PAR-binding proteins play key pathophysiologic roles.
Key words: E3 ubquitin ligase, ring finger protein 146, PAR-binding proteins, PARsylation, Parthanatos inhibitors, ubiquitination
Ring finger proteins contain ring fingers, which are considered to be the functional module for E3 ubiquitin ligase activity.1,2 Ring finger protein 146 (RNF146), a novel PARsylation-directed ring finger E3 ubiquitin ligase, is located at 6q22.1-q22.33 of human chromosome.3 RNF146 encodes a protein of 359 amino acids with a predicted molecular weight of 39.8 kDa.3 The molecular structure of RNF146 contains one N-terminus C3HC4 ring finger domain (35–77 aa) as well as one WWE domain (91–167 aa).3 The poly(ADP-ribose) (PAR) binding motif (144–167 aa) at the tail of WWE domain of RNF146 is involved in various important functions. The detailed molecular structure of RNF146 is illustrated in Figure 1. This PAR-binding protein was recently demonstrated to protect against Parthanatos and function as PARsylation-directed E3 ubquitin ligase to ubiquitinate PARsylated substrates.3–5 Here, we provide a concise summary of these novel functions of RNF146 and discuss the potential pathophysiological and therapeutic significance of novel functions of RNF146 for human diseases.
Figure 1.
Molecular structure of RNF146. The RNF146 is a novel ring finger E3 ubiquitin ligase with 359 amino acids. The molecular structure of RNF146 contains one N-terminus C3HC4 ring finger domain (35–77 aa, 7 cysteine and 1 histidine residues involved in ring finger formation is highlighted in red) as well as one WWE domain (91–167 aa, the highly conserved 2 tryptophan and 1 glutamic acid, which WWE domain is named after, are highlighted in red). The PAR binding motif (144–167 aa) is at the C-terminus tail of WWE domain of RNF146 (highlighted in blue at the end of WWE sequence).
RNF146, a PAR-Binding Dependent Neuroprotective Inhibitor of Parthanatos
Parthanatos is a special form of cell death dependent on poly(ADP-ribose) polymerase-1 (PARP-1) activation which has been recently identified and defined via combination of names of PARP-1 and Thanatos, the Greek personification of death and mortality.6,7 In Parthanatos, stimulus induces rapid activation of PARP-1 in nucleus which functions to synthesize PAR polymer.6,7 The synthesized PAR polymer can be used for post-translational modifications of protein residues.8 However the accumulated free PAR polymer synthesized in nucleus can trans-locate to cytoplasm where it binds with apoptosis inducing factor (AIF) and promotes AIF release from mitochondria outer membrane.6,7,9 The AIF then translocates back to nucleus where it induces degradation of DNA and executes the death sentence for cells.6,7 The Parthanatos has been implicated in the pathogenesis of many human diseases such as inflammation, diabetes, neurodegenerative disease, heart attack, stroke and ischemia reperfusion injury.6,7
Recently, RNF146 has been identified to be the first novel endogenous inhibitor of Parthanatos to rescue neurons from glutamate excitotoxicity in brain.4 Furthermore the neuroprotective function of RNF146 was dependent on its binding with PAR in cytosol.4 The authors named RNF146 the Iduna, a mythological Norwegian goddess who guards a tree full of golden apples used to restore health to sick and injured gods.4 They showed that the expression level of RNF146 in mouse cortical neuronal cultures increases significantly after NMDA challenge.4 They further showed that RNF146 protects neurons against NMDA induced toxicity in a PAR binding dependent manner.4 The protection of RNF146 has been verified in in vitro and in vivo models. Previous studies demonstrated that binding of PAR with AIF was the key event to induce AIF release from the cytosolic side of the mitochondrial outer membrane and functions to trigger Parthanatos.10 Thus RNF146 functions as a PAR binding protein to preemptively bind with PAR in cytoplasm, therefore RNF146 prevents interaction of PAR with AIF in mitochondria and abrogates AIF release and subsequent induction of Parthanatos.4 One limitation of these findings was that a synthesized peptide based on PAR binding sequence of RNF146 (144–167 aa) was used to confirm the binding activity of this sequence with PAR in vitro.4 It isn't clear whether this short synthesized peptide is enough to protect cells against Parthanatos similar to full length RNF146. Nevertheless, these new findings have potential therapeutic applications targeting human disorders where Parthanatos is a key player. The possible neuroprotective mechanism of RNF146 against Parthanatos is illustrated in Figure 2.
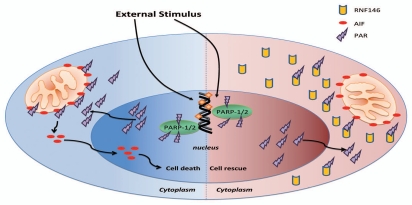
Figure 2.
Potential molecular mechanism for RNF146 induced neuroprotection against Parthanatos. Extensive extra-cellular stimulus can activate PARP-1/2 in nucleus. The activated PARP-1/2 can lead to production of large sum of PAR in nucleus. These PAR can translocated to cytoplasm where can bind with AIF in outer membrane of mitochondria (left). The binding of PAR with AIF can trigger the release of AIF from mitochondria (left). The AIF then translocates to nucleus and induces Parthanatos of cells (left). However in the presence of RNF146, the PAR will bind to RNF146 preemptively, hereby prevent the binding of PAR with AIF in mitochondria (right). The abrogation of binding between PAR and AIF by RNF146 will inhibit Parthanatos and contribute to cell rescue (right).
RNF146 has recently been shown to be a PARsylation dependent E3 ligase.3,5 Therefore it is possible that RNF146 promotes ubiquitination and degradation of AIF after PAR binding with AIF. However, Andrabi et al. showed that overexpression of RNF146 without RING domain still can partially block glutamate induced excitotoxicity.4 This protective effect by RNF146 without RING domain is weaker compared with the protective effect by wild type RNF146. Furthermore the protective effect by RNF146 without RING domain is likely to be dependent on the overexpressed level of RNF146 without RING domain that is far above endogenous level of RNF146 in cells.4 Further research to determine if the total AIF protein levels can be increased by RNF146 RNAi and decreased by overexpression of RNF146 will be useful.
RNF146, a PAR-Binding Dependent PARsylation-Directed E3 Ubquitin Ligase that Positively Regulates Wnt Signaling
The Wnt signaling pathway is important to normal development and cancer.11,12 In the absence of Wnt ligand, the β-catenin binds with degradation complex including GSK-3β, CKI, APC and axin.11,12 In degradation complex, GSK-3β and CKI phosphorylate β-catenin.11,12 After being phosphorylated, the β-catenin can be recognized by β-Trcp and finally contributes to phosphorylation-directed ubiquitination of β-catenin by Skp1-Cul1-F-box (SCF) E3 ubiquitin complex and gets degraded.11–13 However in the presence of Wnt ligand, the binding of Wnt ligand to its receptor can disrupt the degradation complex and spares β-catenin from phosphorylation.11,12 The spared β-catenin then enters nucleus and activates Wnt-targeted genes.11,12 As axin is the concentration-limiting factor for β-catenin degradation complex, tight regulation of the concentration of axin is important to Wnt signaling.14
Previous studies demonstrated that inhibition of tankyrase activity can stabilize axin and downregulate Wnt signaling.15 It was proposed that tankyrase could lead to PARsylation of axin and promote ubiquitination and degradation of axin in cells.15,16 Recently, RNF146 was confirmed to be the novel PARsylation-directed E3 ubiqitin ligase to induce ubiquitination of PAR modified axin and positively regulate Wnt signaling.3,17 It was found that tankyrases catalyzed transferring of PAR to residue of axin (PARsylation).3,15,17 After PARsylation of axin, RNF146 binds with PARsylated axin via the PAR binding motif of RNF146.3 RNF146 then functions as a PARsylation-directed E3 ligase to recruit E2 ubiquitin-conjugating enzyme and contributes to ubiquitination of PARsylated axin.3 This leads to downregulation of axin level, dissociation of degradation complex for β-catenin, increased level of β-catenin and upregulation of Wnt signaling in cells.3,17 Thus RNF146 positively regulates Wnt signaling via its novel PARsylation-directed E3 ubiquitination and degradation of axin.3 The mechanisms for positive regulation of Wnt signaling via tankyrase-dependent PARsylation and ubiquitination of axin by RNF146 are summarized in Figure 3. RNF146 was also shown to promote PARsylation-directed ubiquitination and degradation of tankyrases.3,17 However the tankyrase can PARsylate RNF146, which subsequently ubiquitinates PARsylated RNF146, leading to RNF146 degradation.17
Figure 3.
Molecular mechanism for RNF146 induced positive regulation of Wnt signaling. In the absence of Wnt ligand, the β-catenin binds with degradation complex including GSK-3β, CK1, APC and axin. In the degradation complex, GSK-3β and CK1 phosphorylate β-catenin sequentially. After phosphorylation, the β-catenin can be degradated via phosphorylation-directed ubiquitination (A). However tankyrase (PARP-5) can PARsylate axin. After PARsylation of axin, RNF146 can bind with PARsylated axin via PAR-binding motif of RNF146. Then RNF146 function as PARsylation-directed E3 ubiquitin ligase and contribute to degradation of axin. This leads to downregulation of axin level, dissociation of degradation complex for β-catenin and increased β-catenin level. The β-catenin can enter nucleus and activate Wnt targeted genes (B).
So far proteins BLZF1 and CASC3 have also been shown to be substrates targeted by tankyrase and RNF146 for degradation.3 BLZF1 is required for normal Golgi structure and for protein transport from the endoplasmic reticulum (ER) through the Golgi apparatus to the cell surface.18 BLZF1 is upregulated during retinoid-induced maturation of NB4 promyelocytic leukemia.19 It has been suggested that BLZF1 might act as a transcription factor, or a coregulator, involved in either cell growth control and/or maturation.19 On the other hand, CASC3 protein plays a role in the stress response by participating in cytoplasmic stress granule assembly and by favoring cell recovery following stress.20,21 CASC3 has been implicated in breast cancer development and metastasis, as well as gastric cancers.22 Furthermore recent findings by Kang et al. demonstrated that RNF146 is a PARsylation dependent E3-ligase and it plays a role in DNA damage and repair.5 Therefore physiological functions of RNF146 concerning its PARsylation-directed ubiquitination of targeted PARsylated substrates might be involved in complicated cellular processes.
The identification of RNF146 as a PARsylation-directed E3 ligase establishes a novel molecular paradigm that links tankyrase-dependent PARsylation to ubiquitination and degradation of proteins. The mechanism of PARsylation-directed ubiquitination of degradation substrates is similar to that of phosphorylation-directed ubiquitination of substrates by SCF E3 ubiquitin complex. In SCF induced ubiquitination, the C-terminal of scaffolding protein Cullin1 binds with ROC1 (small ring finger protein), while the N-terminal of Cullin1 binds with SKP1 which further recruits F-box proteins (FBP) as the recognizing protein to bind with phosphorylated protein substrates.23,24 Then ROC1 recruits E2 and contributes to ubiquitination of phosphorylated substrates.23,24 However for RNF146 induced PARsylation-directed ubiquitination of axin, RNF146 directly recognizes PARsylated substrates via its PAR-binding motif in WWE domain. The RNF146 can further recruit E2 ubiquitin-conjugating enzyme via its ring finger domains, and contribute to ubiquitination of PARsylated substrates. Therefore the PARsylation-directed ubiquitination of substrates emerges as another post-translational modification markers directed ubiquitination of protein substrates, besides the phosphorylationdirected ubiquitination of substrates. The catalytic mechanisms of phosphorylation-directed ubiquitination of substrates and PARsylation-directed ubiquitination of substrates are summarized in Figure 4.
Figure 4.
Comparison between PARsylation-directed ubiquitination by E3 ubiquitin ligase RNF146 and phosphorylation-directed ubiquitination by E3 ubiquitin ligase SCF complex. In phosphorylation-directed ubiquitination of substrates, the Cullin1 bind with ROC1 at its C-terminal, while at its N-terminus it bind with SKP1 which further recruit FBP to recognize phosphorylated protein substrates. The ROC1 can recruit E2 ubiquitin-conjugating enzyme and contribute to ubiquitination of phosphorylated substrates. In PARsylation-directed ubiquitination of axin, RNF146 directly recognize PARsylated substrates via its PAR-binding motif in WWE domain and recruit E2 via its ring finger domains, finally contributing to ubiquitination of PARsylated substrates.
RNF146, a PAR-Binding Dependent PARsylation-Directed E3 Ubquitin Ligase in DNA Damage and Repair
The role of PARP-1 catalyzed PARsylation, which is vital to DNA repair, has been extensively studied for decades and detailed mechanism of PARsylation related DNA repair has been summarized in several well-written review papers.25–27 In brief, after detection of DNA damage by PARP-1, the PARP-1 can be activated.25–27 The activated PARP-1 synthesizes PAR at the DNA damage site and transfers PAR to amino acid residues of various nuclear proteins including PARP-1 itself, PARP-2, XRCC-1, Aurora B, DNA pol β, Topo I, II, DNA pol α, H1, H2B, TRF2, Cenpa and Bub3.25–27 PARsylation of histones H1, H2B, TRF2, Cenpa and Bub3 can change the chromatin structure and increase chromatin accessibility at the actual site of DNA damage thus promoting DNA repair.25–27 PARsylation of PARP-1 itself, PARP-2, XRCC-1, Aurora B, DNA pol β, Topo I, II and DNA pol α is related to repair complex assembly at the damage site and finally completion of DNA damage repair.25–27
Recent findings by Kang et al. demonstrated that RNF146 is a PARsylation dependent E3-ligase and it played a role in DNA damage and repair.5 In their report, they verified that RNF146 has PARsylation dependent E3 ligase activity.5 Furthermore they showed that RNF146 can translocate to nuclear after cellular injury and can bind to a number of nuclear proteins that are either PARsylated or bind with PAR.5 These RNF146 interacting nuclear proteins include PARP-1, PARP-2, nucleolin, DNA ligase III, KU70, KU86, XRCC1 and histones.5 These nuclear proteins are all essential to PARsylation dependent DNA repair.25–27 They also demonstrated that PAR binding is required for RNF146 ubiquitination and degradation of these nuclear proteins.5 Dependent on the PAR binding and ubiquitin E3 ligase activity of RNF146, RNF146 facilitates DNA repair and protects against cell death induced by the DNA damaging agent and promotes cell survival after γ-irradiation.5
Mortusewicz et al. reported that the PAR degrading enzyme PAR glycohydrolase (PARG) is recruited to DNA damage sites through PAR- and PCNA-dependent mechanisms.28 Previous studies have shown that PARG is required for efficient repair of single- and double-strand breaks and oxidized bases of DNA strands.29–31 It is known that exorbitant activation of PARP-1 can deplete cellular ATP and lead to cell death.25 Therefore to strictly control PAR signals in cells seems to be an indispensable process during PARsylation directed DNA repair. The PAR signals need to be eliminated in time after DNA repair has been completed. Therefore it can be hypothesized that the RNF146 might function as another modulation approach to control PAR signals in nuclear during the PARsylation directed DNA repair process, besides the PARG modulating pathway. It was demonstrated that after DNA damage, both PARG and RNF146 will translocate from cytosol to nuclear to play essential roles in the process of DNA repair.5,28 However it is still unclear how the two modulation pathways cooperate with each other in DNA repair and whether functions of one modulation pathways in DNA repair can be substituted by another. Future researches are needed to clarify these.
The Roles of RNF146 in Cancer Development and Neurological Disorders
Two recent findings suggest a potential link between RNF146 and cancer development.32,33 A genome-wide association study (GWAS) identified a risk locus at chromosome 6q22.33 for breast cancer.32 Candidate genes in the 6q22.33 region include ECHDC1 and RNF146.32 The subsequent study on Jewish Ashkenazi breast cancer high risk women demonstrated that an intronic sequence variation was detected in 4/105 genotyped patients in the third intron of ECHDC1 gene.33 No other sequence variations in the genomic regions of RNF146 and ECHDC1 genes were found in any of studied participants.33 However recent findings by Campa et al. showed no association of one single-nucleotide polymorphisms (SNP) of RNF146 (rs2180341) with breast cancer risk based on studies of breast cancer samples from population not involving Ashkenazi Jews.34 The finding that RNF146 can positively regulate Wnt signaling via PARsylation dependent degradation of axin provides new supporting evidences for the potential roles of RNF146 in mammary cancer development.3,17 An association between Wnt signaling and breast cancer has been established by previous studies in references 35–38. Furthermore BLZF1 and CASC3, the two new identified RNF146 targeted substrates was reported to be implicated in cancer development and cell proliferation.19–22 On the other hand, the breast cancer associated gene-1 and -2 (BRCA1/2) deficient are implicated in genetic instability and even breast cancer formation.39,40 The PARP-1 inhibitors can kill BRCA1/2 deficient cells with extremely high efficiency (summarized in a review paper).41 The RNF146 can promote PARsylation-directed ubiquitination and downregulate levels of tankyrases as well as PARP-1 in cells.3,5,17 Therefore RNF146 might be implicated in breast cancer with BRCA1/2 deficiency via downregulation of PARP-1. Taken together, a positive link between RNF146 and cancer development seemed to be biologically plausible. Further studies are required to verify the potential roles of RNF146 in cancer development, especially for mammary tumors.
On the other hand, investigators have found that RNF146 is upregulated early in brains of Alzheimer disease (AD) patients, compared with aged controls.42 They used subtractive hybridization of transcripts from human frontal cortex, which degenerates in the late stage of AD, against transcripts of the inferior temporal cortex, which is influenced in the early stage of AD. Their findings suggest that RNF146 might function early in the progression of AD.42 However it is not clear whether the early increased expression of RNF146 in AD brain contributes to neurodegeneration of AD or just a protective response to neurons injury in early AD. This will be a research area worth exploring in the future.
The Potential Novel Functions of Other PAR-Binding Proteins
In a unbiased proteomic screen for PAR-binding proteins, hundreds of PAR-binding proteins have been identified.43 These PAR-binding proteins are found to be involved in DNA damage signaling and DNA repair, modification of chromatin structure, regulation of transcription, replication, RNA metabolism, RNA splicing, protein synthesis, cell death and cell cycle and mitosis.43 It is possible that some of these PAR binding proteins might also have similar functions as RNF146 and might also act as inhibitors of Parthanatos or function as PARsylation-directed E3 ligases.
Cullin1 was found to have PAR binding activity.43 Furthermore overexpression of Cullin1 could promote cell proliferation.44 The increased Cullin1 has been associated with cancer development.45,46 Therefore it was suggested that Cullin1 might function as an inhibitor of Parthanatos. Overexpression of Cullin1 might also be neuroprotective as RNF146. Furthermore Cullin1 is the key protein in SCF phosphorylation-directed E3 ubiquitin ligase complex. Therefore Cullin1 might also be involved in PARsylation-directed ubiquitination of PARsylated substrates. It is possible that after Cullin1 recruits ROC1 and E2 to its C-terminus, the Cullin1 might bind with PARsylated substrates via its PAR-binding motif and functions as the PARsylation-directed E3 ubiquitin ligase. Another possible model is that the PAR-binding motif of Cullin1 might cooperate with FBP to recognize phosphorylated plus PARsylated substrates and promotes ubiquitination of these substrates. These hypotheses need future verifications. The potential mechanisms involving PARsylation-directed ubiquitination of substrates by Cullin1 are summarized in Figure 5.
Figure 5.
Possible mechanisms of Cullin1 involvement in PARsylation-directed ubiquitination of substrates. (A) The PAR-binding motif of Cullin1 might cooperate with F-box protein at the N-terminus of Cullin1 to recognize phosphorylated plus PARsylated substrates and promote ubiquitination of these substrates. In this model, the PARsylation-directed ubiquitination cooperate with phosphorylation-directed ubiquitination in SCF complex mediated ubiquitination and degradation of phosphorylated plus PARsylated substrates. (B) The Cullin1 might recruit ROC1 to introduce E2 at its C-terminus, while Cullin1 bind with PARsylated substrates via its PAR-binding motif and function as PARsylation-directed E3 ubiquitin ligase. The predicted PAR-binding motif of Cullin1 (from 460–481 aa) is shown at the bottom, based on consensus sequence for PAR-binding motif. Key amino acids are highlighted. The dipeptide KR is shown in white highlighted with a black background, while proximal residues that fit the consensus are indicated in black but highlighted with a gray background. Amino acids not important are normally shown in black on a white background.
Proteins similar to RNF146 with WWE domain as well as E3 ubiquitin ligase domains such as ring finger domains or HECT domains should have functions of PARsylation-directed E3 ubiquitin ligases. Domain searching identifies proteins DTX1, DTX2 (ring finger protein 58), DTX4 (ring finger protein 155), TRIP12 (probable E3 ubiquitin-protein ligase TRIP12) and HUWE1 (E3 ubiquitin-protein ligase HUWE1) can be potential candidates for PARsylation-directed E3 ubiquitin ligases.3 Based on the consensus sequence proposed for PAR-binding motifs,43 these proteins also have predicted PAR-binding motifs at the end of their WWE domains (Table 1). Some proteins have more than one PAR-binding motif at the end of their WWE domains (Table 1). The HUWE1 has been confirmed to be a E3 ubiquitin-protein ligase with WWE domain and HECT domains.47 The DTX family proteins have also been verified to have E3 ubiquitin ligase activity with WWE and ring finger domains.48 The TRIP12 is a probable E3 ubiquitin ligase with WWE and HECT domains. Therefore it is highly possible that these proteins can function as PARsylation-directed E3 ubiquitin ligases similar to RNF146. The molecular structures of DTX1, DTX2, DTX4, TRIP12 and HUWE1 with respective domains are illustrated in Figure 6.
Table 1.
The putative PAR-binding motif in RNF146, DTX-1, DTX2, DTX4, TRIP12 and HUWE1
 |
The putative PAR-binding motif of these proteins is acquired based on the consensus sequence for PAR-binding motif: [HKR]1− X2− X3− [AIQVY]4− [KR]5− [KR]6− [AILV]7− [FILPV]8.43 More than one predicted PAR-binding motif according to the consensus sequence can be discerned in some proteins.
Figure 6.
Molecular structures of DTX1, DTX2, DTX4, TRIP12 and HUWE1. The molecular structure of DTX1, DTX2, DTX4, TRIP12 and HUWE1 containing WWE domains, ring finger domains, HECT domain as well as other domains or motif is illustrated in different color.
Future Perspectives for Potential Applications of PAR-Related Strategies
The free PAR can induce AIF release from mitochondria and mediate Parthanatos of cells. However RNF146 can bind with PAR in cytosol and can abrogate PAR induced Parthanatos.4 Therefore, in principle, any agents that functions along the pathway of PAR induced Parthanatos may have therapeutic potential. Agents that can disrupt nucleus translocation of AIF, interfere with the binding between PAR and AIF or directly bind with PAR as well as agents that can promote degradation of PAR or inhibit PAR production (PARP-1 inhibitors) might function as cellular protective agents against Parthanatos. Therefore screening for nature or synthetic compounds which could regulate PAR induced Parthanatos might present a promising therapeutic approach in the future. On the contrary, PAR mimics with longer lifespan or PARP-1 activators might become effective therapeutic agents for cancer therapy as they can activate Parthanatos and promote cell death. On the other hand, as binding of PAR with RNF146 is requested to activate RNF146 PARsylation dependent E3 ligase activity, small-molecule PAR mimics that can activate RNF146 E3 ligase activity might be utilized as Wnt signaling activators for bone anabolism. Small-molecule PAR mimics that can bind with RNF146 but function to abrogate RNF146 E3 ligase activity might be applied to block Wnt-dependent tumor growth.
However some caveats need to be considered in the therapeutic application of PAR-related strategies. First, PAR-binding proteins have been implicated in cancer, thus this potential link needs to be fully investigated before they can be considered as therapeutic targets for other diseases. Second, the PAR-binding proteins have complicated and essential functions in cells, especially for repair of DNA, synthesis of DNA, DNA directed RNA synthesis, therefore the PAR-related strategies should also be proven to be safe from serious disturbance of these intracellular functions, before it can be practically applied. Despite these caveats, recent evidences have provided cautious optimism that PAR-binding proteins may be potential therapeutic targets for many human diseases and further studies to address these are warranted.
Acknowledgments
We thank National Medical Research Council, Singapore Millennium Foundation and Duke-NUS Graduate Medical School for their support.
References
- 1.Fang S, Lorick KL, Jensen JP, Weissman AM. RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin Cancer Biol. 2003;13:5–14. doi: 10.1016/S1044-579X(02)00095-0. [DOI] [PubMed] [Google Scholar]
- 2.Ardley HC. Ring finger ubiquitin protein ligases and their implication to the pathogenesis of human diseases. Curr Pharm Des. 2009;15:3697–3715. doi: 10.2174/138161209789271807. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 4.Andrabi SA, Kang HC, Haince JF, Lee YI, Zhang J, Chi Z, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17:692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagné JP, et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci USA. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci. 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang LB, Zhang LY, Shan CH. A new form of cell death: Parthanatos. Yi Chuan. 2010;32:881–885. doi: 10.3724/sp.j.1005.2010.01187. [DOI] [PubMed] [Google Scholar]
- 8.Yung TM, Sato S, Satoh MS. Poly(ADP-ribosyl)ation as a DNA damage-induced post-translational modification regulating poly(ADP-ribose) polymerase-1-topoisomerase I interaction. J Biol Chem. 2004;279:39686–39696. doi: 10.1074/jbc.M402729200. [DOI] [PubMed] [Google Scholar]
- 9.Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann NY Acad Sci. 2008;1147:233–241. doi: 10.1196/annals.1427.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4:20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 12.Pecina-Slaus N. Wnt signal transduction pathway and apoptosis: a review. Cancer Cell Int. 2010;10:22. doi: 10.1186/1475-2867-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latres E, Chiaur DS, Pagano M. The human F box protein beta-Trcp associates with the Cul1/Skp1 complex and regulates the stability of beta-catenin. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- 14.Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1:10. doi: 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 16.Fearon ER. PARsing the phrase “all in for Axin”-Wnt pathway targets in cancer. Cancer Cell. 2009;16:366–368. doi: 10.1016/j.ccr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, et al. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS ONE. 2011;6:22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Short B, Preisinger C, Körner R, Kopajtich R, Byron O, Barr FA. GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155:877–884. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duprez E, Tong JH, Dérré J, Shen SJ, Berger R, Chen Z, et al. JEM-1, a novel gene encoding a leucine-zipper nuclear factor upregulated during retinoid-induced maturation of NB4 promyelocytic leukaemia. Oncogene. 1997;14:1563–1570. doi: 10.1038/sj.onc.1200995. [DOI] [PubMed] [Google Scholar]
- 20.Baguet A, Degot S, Cougot N, Bertrand E, Chenard MP, Wendling C, et al. The exon-junction-complexcomponent metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci. 2007;120:2774–2784. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- 21.Noble CG, Song H. MLN51 stimulates the RNAhelicase activity of eIF4AIII. PLoS ONE. 2007;2:303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Degot S, Régnier CH, Wendling C, Chenard MP, Rio MC, Tomasetto C. Metastatic Lymph Node 51, a novel nucleo-cytoplasmic protein overexpressed in breast cancer. Oncogene. 2002;21:4422–4434. doi: 10.1038/sj.onc.1205611. [DOI] [PubMed] [Google Scholar]
- 23.Jackson PK, Eldridge AG. The SCF ubiquitin ligase: an extended look. Mol Cell. 2002;9:923–925. doi: 10.1016/S1097-2765(02)00538-5. [DOI] [PubMed] [Google Scholar]
- 24.Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/S0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 25.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. doi: 10.1016/S0301-472X(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 26.Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst) 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Vidakovic M, Poznanovic G, Bode J. DNA break repair: refined rules of an already complicated game. Biochem Cell Biol. 2005;83:365–373. doi: 10.1139/o05-044. [DOI] [PubMed] [Google Scholar]
- 28.Mortusewicz O, Fouquerel E, Ame JC, Leonhardt H, Schreiber V. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 2011;39:5045–5056. doi: 10.1093/nar/gkr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher AEO, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates sin-gle-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erdélyi K, Bai P, Kovács I, Szabó E, Mocsár G, Kakuk A, et al. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. FASEB J. 2009;23:3553–3563. doi: 10.1096/fj.09-133264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, et al. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gold B, Kirchhof T, Stefanov S, Lautenberger J, Viale A, Garber J, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menachem TD, Laitman Y, Kaufman B, Friedman E. The RNF146 and ECHDC1 genes as candidates for inherited breast and ovarian cancer in Jewish Ashkenazi women. Fam Cancer. 2009;8:399–402. doi: 10.1007/s10689-009-9255-7. [DOI] [PubMed] [Google Scholar]
- 34.Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103:1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nusse R, van Ooyen A, Cox D, Fung YK, Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984;307:131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 36.Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 37.Mohinta S, Wu H, Chaurasia P, Watabe K. Wnt pathway and breast cancer. Front Biosci. 2007;12:4020–4033. doi: 10.2741/2368. [DOI] [PubMed] [Google Scholar]
- 38.Veeck J, Bektas N, Hartmann A, Kristiansen G, Heindrichs U, Knüchel R, et al. Wnt signalling in human breast cancer: expression of the putative Wnt inhibitor Dickkopf-3 (DKK3) is frequently suppressed by promoter hypermethylation in mammary tumours. Breast Cancer Res. 2008;10:82. doi: 10.1186/bcr2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng CX. Tumorigenesis as a consequence of genetic instability in Brca1 mutant mice. Mutat Res. 2001;477:183–189. doi: 10.1016/S0027-5107(01)00119-1. [DOI] [PubMed] [Google Scholar]
- 41.De Soto JA, Deng CX. PARP-1 inhibitors: are they the long-sought genetically specific drugs for BRCA1/2associated breast cancers? Int J Med Sci. 2006;3:117–123. doi: 10.7150/ijms.3.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Rotz RC, Kins S, Hipfel R, von der Kammer H, Nitsch RM. The novel cytosolic RING finger protein dactylidin is upregulated in brains of patients with Alzheimer's disease. Eur J Neurosci. 2005;21:1289–1298. doi: 10.1111/j.1460-9568.2005.03977.x. [DOI] [PubMed] [Google Scholar]
- 43.Gagné JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen G, Li G. Increased Cul1 expression promotes melanoma cell proliferation through regulating p27 expression. Int J Oncol. 2010;37:1339–1344. doi: 10.3892/ijo_00000786. [DOI] [PubMed] [Google Scholar]
- 45.Chen G, Cheng Y, Martinka M, Li G. Cul1 expression is increased in early stages of human melanoma. Pigment Cell Melanoma Res. 2010;23:572–574. doi: 10.1111/j.1755-148X.2010.00725.x. [DOI] [PubMed] [Google Scholar]
- 46.Bai J, Zhou Y, Chen G, Zeng J, Ding J, Tan Y, et al. Overexpression of Cullin1 is associated with poor prognosis of patients with gastric cancer. Hum Pathol. 2011;42:375–383. doi: 10.1016/j.humpath.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Parsons JL, Tait PS, Finch D, Dianova II, Edelmann MJ, Khoronenkova SV, et al. Ubiquitin ligase ARFBP1/Mule modulates base excision repair. EMBO J. 2009;28:3207–3215. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeyama K, Aguiar RC, Gu L, He C, Freeman GJ, Kutok JL, et al. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J Biol Chem. 2003;278:21930–21937. doi: 10.1074/jbc.M301157200. [DOI] [PubMed] [Google Scholar]