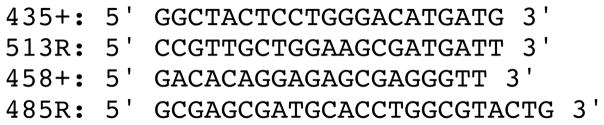Abstract
A short (259 nucleotide) conserved intronic sequence (CIS) is surprisingly informative for delineating deep phylogenetic relationships in cone snails. Conus species previously have been assigned to clades based on the evidence from mitochondrial 12S and 16S rRNA gene sequences (1129 bp). Despite their length, these genes lack the phylogenetic information necessary to resolve the relationships among the clades. Here we show that the relationships can be inferred from just 46 sites in the very short CIS sequence (a portion of “intron 9” of the γ-glutamyl carboxylase gene). This is counterintuitive because in short sequences sampling error (noise) often drowns out phylogenetic signal. The intron 9 CIS is rich in synapomorphies that define the divergence patterns among eight clades of worm- and fish-hunting Conus, and it contains almost no homoplasy. Parsimony, maximum-likelihood and Bayesian analyses of the combined sequences (mt rRNA + CIS) confirm most of the relationships among 23 Conus sequences. This phylogeny implies that fish-hunting behavior evolved at least twice during the history of Conus -once among New World species and independently in the Indo-Pacific clades.
Keywords: episodic evolution, conserved intron, Conus, evolution
INTRODUCTION
A recurring problem in phylogeny is the resolution rapid radiations that occurred relatively far in the past. Our laboratories have been engaged in a long-term study of venomous marine snails belonging to the superfamily Conoidea, a group of probably more than 12,000 species. We focus especially on one cone snail lineage of around 700 species (Röckel et al., 1995) that seems to have radiated very quickly.
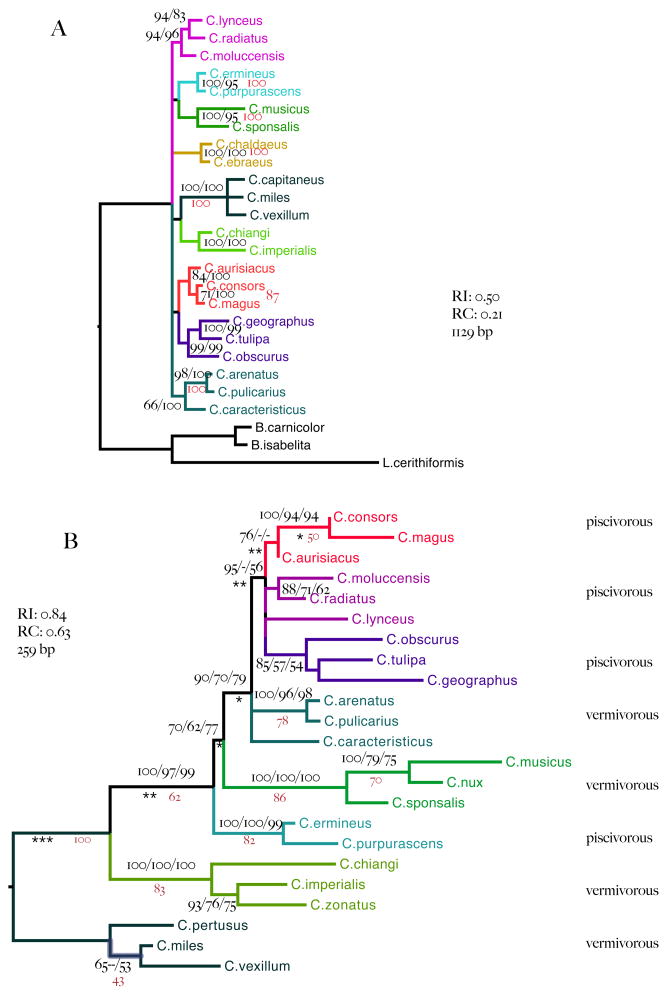
The present framework for understanding Conus evolution is based on both the fossil record and molecular phylogenetics. The fossil record suggests two major bursts of speciation among cone snails (Kohn, 1990, Duda and Kohn, 2005), an initial one in the Eocene and a later radiation in the Miocene (with an apparent contraction in Conus biodiversity in the Oligocene). Understanding the tempo of change through integration of the fossil record with the molecular phylogeny has so far depended on a single gene (Calmodulin, Duda and Kohn, 2005) that follows a molecular clock but that has left unresolved the evolutionary history describing the order of divergence among the major clades. Neither does our molecular phylogenetic analysis of 12SrRNA and 16SrRNA sequences provide sufficient resolution to delineate the history of divergence among these clades (Figure 2A). This failure to resolve the deep evolutionary relationships is not an isolated problem in phylogenetics because saturation can obscure what little molecular change may have occurred during a short burst of speciation. Often neither the fossil record nor molecular data provide sufficient data to detect the order of divergence events among such clades.
Figure 2.
A. Tree inferred from 12S and 16S mt rRNA sequences. Branches are labeled with Bayesian support values (right; posterior probabilities expressed as percentages) and maximum likelihood bootstrap proportions (left). B. Tree inferred from intron 9 CIS sequences alone. Branches are labeled with posterior probabilities (left) maximum likelihood bootstrap proportions (middle) and maximum parsimony bootstrap proportions (right). Missing values indicate that the corresponding support value is less than 50%. Maximum likelihood and Bayesian estimated branch lengths depart significantly from strict molecular clock assumptions. Local clock rates better describe sequence evolution at branches marked with asterisks (departures from a strict clock rate significant at p<0.5 *, p <0.01 ** and p<0.0001 ***).
Common wisdom suggests using longer sequences to increase the probability of including informative sites. In contrast, we present here an approach to resolving these problematic divergence events using a very short but highly informative sequence that we designate as intron 9. This 259 base pair sequence lies between coding sequence intervals that are homologous to exons 9 and 10 of the human γ-glutamyl carboxylase, a posttranslational modification enzyme expressed in Conus venom ducts. Recent parsimony-based analysis of this short conserved region defined four different fish-hunting cone snail (Imperial et al., 2007a).
We extend these earlier results by adding a sample of worm-hunting Conus species. We show that the relationships of these clades to each other and to the fish-hunting clades can be resolved by the pattern of substitutions in the conserved intronic sequence even though the alignment is much shorter than those of more conventional markers such as the 12S and 16S rRNA genes (>1129 bp.). The successful extension of the CIS analysis to our larger group of Conus species suggests that tests of tempo and mode of evolution of Conus and their relatives may be well within reach.
METHODS
We analyzed the three clades of fish-hunting species (Table 1) for which the conserved sequence in intron 9 had been previously analyzed (Imperial et al., 2007b). To these we added presumptive vermivorous Conus species. These are classically defined as in Table 1.
Table 1.
Conus clades analyzed in this study with tests of the global molecular clock against local clocks.
| Clade Name | Species | Global vs Local Clock | Global vs Local Clock | Global vs Local Clock | Global vs Local Clock | Global vs Local Clock |
|---|---|---|---|---|---|---|
| Pionoconus Mörch, 1852 |
C. consors C. magus* C. aurisiacus |
−2ln(L1/L2)= 23.4 p=0.0094 |
− 2ln(L1/L2 ) =27.8 p=0.0332 |
−2ln(L1/L2)= 39.6 p=0.0118 |
−2ln(L1/L2 )= 48.5 p=0.0095 |
− 2ln(L1/L2 )= 86.1 p<0.0001 |
| Phasmoconus Mörch, 1852 |
C. radiatus* C. lynceus C. molucconsis |
|||||
| Gastridium Modeer, 1793 |
C. geographus* C. tulipa C. obscurus |
|||||
| Puncticulus Swainson, 1840 |
C. arenatus* C. pulicarius C. caracteristicus |
|||||
| Harmoniconus DaMotta, 1991 |
C. musicus* C. sponsalis C. nux |
|||||
| Chelyconus Mörch, 1852 |
C. ermineus* C. purpurascens |
|||||
| Stephanoconus Mörch, 1852 |
C. imperialis C. chiangi C. zonatus |
|||||
| Rhizoconus Mörch, 1852 |
C. miles* C. vexillum C. pertusus |
Type species of designated clade name.
High molecular weight DNA was isolated from body tissue of the snails. Genomic DNA was prepared from 20 mg of frozen tissue from each species using the Gentra PUREGENE DNA Isolation Kit (Gentra Systems, Minneapolis, MN) according to the manufacturer’s standard protocol.
The complete intron 9 sequences were amplified from genomic DNA by polymerase chain reaction using primers corresponding to conserved amino acid residues in exons 9 and 10 (Figure 1). PCR was carried out using Phusion™ high fidelity DNA polymerase (New England Biolabs, MA, USA). 10ng of genomic DNA was used in 50μl reactions with 1X Phusion GC Buffer, 200 μM dNTPs, 0.5μM each forward and reverse primer, 3% DMSO, and 1 unit of Phusion polymerase. PCR was carried out in two steps. The initial amplification was carried out using primers 435+ and 513R (Figure 1). The product was diluted 20 fold and subsequently amplified using 458+ and 485R. The first PCR reaction was done in a PTC 100 thermocycler (MJ Research, MA, USA) using the following conditions: an initial denaturation step of 98°C for 2 min, followed by 34 cycles of 98°C for 30 sec, 52°C for 30 sec, 72°C for 30 sec, and a final extension of 72°C for 5 min. The second PCR reaction was under the following conditions: 98°C for 2 min, followed by 40 cycles of 98°C for 30 sec, 52°C for 30 sec, 72°C for 30 sec, and a final extension at 72°C of 5 min.
Figure 1.
Nucleotide sequences of primers used for PCR amplification.
Amplified DNA was purified by agarose gel electrophoresis and isolated using the Qiagen gel extraction kit (Qiagen, CA, USA). Purified amplicons were cloned into pGEM-T Easy vector (Promega, WI, USA). The nucleotide sequence of the insert was determined using oligonucleotide primers corresponding to T7 and SP6 promoter sequences at the Health Sciences Center Core Sequencing Facility, University of Utah. Sequences from 4–5 clones were compared to assess conservation and to rule out the presence of multiple alleles.
All complete sequences were assembled using SeqMan (DNASTAR, Inc., WI, USA). DNA sequences of intron 9 from the following species were determined and deposited in GenBank under accession numbers FJ461229-FJ461251: C. arenatus, C. pulicarius, C. caracteristicus C. musicus, C. sponsalis, C. chaldeus, C. nux, C. miles, C. vexillum, C. pertusus, C. chiangi., C. radiatus, C. moluccensis, C. lynceus, C. purpurascens, C. ermineus, C. magus, C. consors, C. aurisacus, C. geographus, C. obscurus, C. zonatus and C. tulipa.
For phylogenetic analyses, sequences were aligned using Clustal X (Larkin et al., 2007), Muscle and Tcoffee (Notredame, et al. 2000) or Rcoffee (Moretti, et al. 2008) as appropriate. We compared the fit of these alternative alignments using the color-coded scoring system of T-Coffee. The best alignments were concatenated using MacClade (Maddison and Maddison, 2005).
For all unpartitioned analyses, maximum likelihood parameters describing sequence evolution were optimized with a GTR+I+G model that includes six possible substitution types and allows unequal base frequencies (GTR, Tavaré, 1986), invariant sites (I), and across-site rate heterogeneity (G). Trees were inferred using maximum likelihood methods (PhyML, Guindon and Gascuel, 2003) and for partitioned maximum-likelihood inference of the concatenated sequences, the GTR+G model parameters were estimated independently for each gene (RaxML, Stamatakis, et al. 2008).
Trees were also inferred using maximum parsimony (PAUP*, Swofford 2002). To infer the pattern of synapomorphic changes and to calculate homoplasy indices we used MacClade 4.0 (Maddison and Maddison, 2005). Bootstrap proportions were calculated from the consensus of maximum parsimony trees and maximum-likelihood trees inferred from 1000 bootstrap replicates of the data for each method.
Bayesian analyses (Mr. Bayes, Huelsenbeck et al., 2001; Ronquist and Huelsenbeck, 2003) comprised 1,000,000 generations with the first 25% of the sampled generations discarded as burn-in trees. Two MCMCMC runs (Metropolis-Coupled Markov Chain Monte Carlo) of four chains each were used to thoroughly explore tree space. Convergence of the likelihoods was determined by comparing the average standard error of the difference (ASED) in split frequencies between the two runs and by comparing plots of the log-likelihood scores. Optimality was also judged adequate when the potential scale reduction factor (PSRF) for the total tree length and for each model parameter reached 1.00.
Trees were rooted on the basis of a previous, larger-scale analysis of Conus species (available from PSC). To determine if the sequences we tested showed the same pattern of rapid change as the fossil record, we tested strict molecular clock assumptions against a local clock using maximum likelihood methods (Yoder and Yang, 2000, HyPhy 2.0, Kosakovsky et al., 2005), against a Bayes-estimated relaxed, uncorrelated log-normal clock with a Yule speciation process, and against an exponential clock describing episodic evolution (Beast 1.5.3, Drummond, et al. 2006).
We assessed the sources and strength of the phylogenetic signal along each of the sequences by examining Buneman trees and Neighbor Networks using Splits Tree 4.11.3 (Huson and Bryant, 2006) built from 1000 bootstrap replicates of the sequences.
RESULTS AND DISCUSSION
We find that 12S and 16S rRNA sequences define relationships within eight Conus clades (Figure 2A) and that these are consistent with earlier classification schemes (Table 1). However, as in previous analyses of these sequences and of calmoudulin (Duda and Kohn, 2005), the standard markers completely fail to reveal the order of divergence among the Conus clades, despite their length.
Utility of short intron sequences in other analyses
Several previous phylogenetic studies have used relatively conserved short intron sequences to infer similar problematic phylogenies that cover the same time frame as that of the Conus species. Prychitko and Moore (2000, 2003) analyzed the γ-fibrinogen intron 7 sequences of 28 species of birds (18 families in 9 orders). During the estimated 55–90 Myr history of this group, indels have accumulated and most nodes in the most-parsimonious trees have strong bootstrap support (~90%). This intron sequence also yielded a well-resolved gene tree based on base changes, indels and clusters of conserved bases for 7 to 8 million years of woodpecker evolution. As in the case of the γ-carboxylase intron 9, the length of the γ-fibrinogen intron 7 varies in size (619 bp to 1012 bp) and related species carry identical changes.
Pecon-Slattery (2004) analyzed the intron sequences in three single-copy Y-chromosome genes, ZFY (zinc protein on Y, 999 bp), SMCY (select mouse cDNA on Y, 1395 bp), and UBEIY (ubiquitin activating enzyme E1 on Y, 1210 bp) in 36 species of Felidae and identified eight monophyletic lineages. The authors also identified diagnostic sites, including SINE retroelements that define each of the lineages.
Characterization of the conserved region of intron 9
We previously determined genomic sequences encoding γ-glutamyl carboxylase from Conus textile and have designated as “intron 9” the DNA sequences between coding sequence intervals that are homologous to exons 9 and 10 of the human γ-glutamyl carboxylase. Nucleotide sequences in exons 9 and 10 are highly conserved among different phyla (Supplementary Figure 2A). In C. textile the intron is 1112 bp in length although the total length of the intron varies between 850 and 1179 bp in other species that we analyzed. The region between positions 440 and 674 of C. textile intron 9 (Supplementary Figure 1C, shown in italics) is conserved among the different Conus species studied here. In all subsequent figures, position 440 of the C. textile sequence is designated position #1, and all of the sequences are aligned accordingly.
Phylogenetic analysis with intron 9
Figure 2B shows the relationships of the eight worm-hunting and fish-hunting clades inferred from the conserved region of intron 9 (the CIS). Shared-derived deletions (Supplementary Figure 2) were observed between nucleotides 122 and 132 in C. radiatus, C. lynceus and C. moluccensis (the Phasmoconus clade). Members of the Pionoconus clade share a similar deletion at sites 76 to 85.
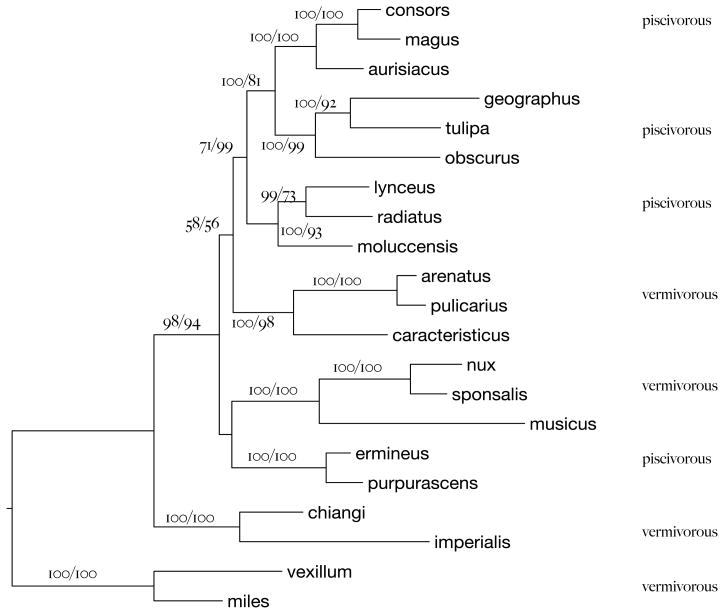
Nucleotide substitutions at 46 sites (Table 2) in the CIS are informative for the relationships within and among the eight clades. These are shown as an annotated alignment of all 259 positions of the CIS in Supplementary Figure 2. Collectively, changes at 28 sites in the CIS resolve most of the deeper relationships among the eight named clades (Figure 2B, Table 2, starred sites). The intron9 fragment adds sufficient phylogenetic signal to the mitochondrial rRNA sequences to resolve most among- and within-clade relationships with strong statistical support (Figure 3).
Table 2.
Nucleotide states at 46 intron9 CIS sites that are informative for the relationships among the Conus species. Twenty eight starred sites are responsible for defining the eight species-group clades. These represent 69% of the phylogenetically informative sites (those that are not invariant or singletons).
| Clade | Species | |||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pionoconus | Conus | consors | A | G | A | T | G | T | A | A | C | T | G | G | A | T | G | C | G | G | A | T | A | T | A | G | C | A | T | G | T | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | A |
| Conus | magus | A | G | A | T | G | T | A | A | C | T | G | G | A | T | G | C | G | G | A | T | A | T | A | G | C | A | T | G | T | G | T | T | C | T | T | T | A | G | A | T | C | G | T | G | G | A | |
| Conus | aurisacus | A | G | A | T | G | T | A | A | T | T | G | G | A | T | A | C | G | G | A | T | A | T | A | G | C | A | T | G | T | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | A | |
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Gastridium | Conus | geographus | A | G | A | T | G | T | A | A | T | T | G | G | A | T | A | C | G | G | A | T | A | T | A | G | C | A | T | G | C | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | C |
| Conus | tulipa | A | G | A | T | G | T | A | A | T | T | G | G | A | T | A | C | G | G | A | T | A | T | A | G | C | A | T | G | C | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | C | |
| Conus | obscurus | A | G | A | T | G | T | A | A | T | T | G | G | A | T | A | C | G | G | A | T | A | T | A | G | C | A | T | G | C | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | C | |
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Phasmoconus | Conus | lynceus | A | G | A | T | G | T | A | A | T | T | G | G | A | T | A | C | G | G | A | T | A | T | A | G | - | - | - | - | - | G | T | T | C | T | T | T | A | G | A | T | C | T | T | C | G | A |
| Conus | radiatus | A | G | A | T | G | T | A | A | T | T | G | G | A | T | A | C | G | A | A | T | A | T | A | G | - | - | - | - | - | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | A | |
| Conus | moluccensis | A | G | A | T | G | T | A | A | T | T | G | G | A | G | A | C | G | A | A | T | A | T | A | G | - | - | - | - | - | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | A | |
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Puncticulus | Conus | arenatus | A | G | A | T | C | G | A | A | T | T | G | G | A | A | A | C | G | G | - | T | A | T | A | G | - | A | G | G | T | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | A |
| Conus | pulicarius | A | G | A | T | C | G | A | A | T | T | G | G | A | A | A | C | G | G | - | T | A | T | A | G | - | A | G | G | T | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | A | |
| Conus | caracterisicus | A | G | A | T | C | T | A | A | T | T | G | G | A | T | A | C | G | G | - | T | A | T | A | G | - | A | T | G | T | G | T | T | C | T | T | T | A | G | A | T | C | T | T | G | G | A | |
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Harmoniconus | Conus | musicus | G | G | G | T | C | T | A | A | T | T | T | G | A | T | A | C | A | G | A | T | A | T | A | G | - | A | T | G | T | G | T | T | C | C | T | T | A | A | A | T | C | A | T | A | A | A |
| Conus | nux | G | G | G | T | C | T | A | A | T | T | T | G | A | T | A | C | A | G | A | T | A | T | A | G | - | A | T | G | T | G | T | C | C | C | T | T | A | A | A | T | C | A | T | A | A | A | |
| Conus | sponsalis | A | G | A | T | C | T | A | A | T | T | T | G | A | T | A | C | A | G | A | T | A | T | G | G | - | A | T | G | T | G | T | C | C | C | T | T | A | A | A | T | C | A | T | A | G | A | |
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Chelyconus | Conus | erminius | A | G | A | T | C | T | A | A | T | T | G | G | G | T | A | C | A | G | A | T | A | T | A | G | - | A | T | G | T | G | T | T | T | T | T | T | A | G | A | T | C | T | T | G | G | A |
| Conus | purpurascens | A | G | A | T | C | T | A | A | T | T | G | G | G | N | A | C | A | G | A | T | A | T | A | G | - | A | T | G | T | G | T | T | T | T | T | T | A | G | A | T | C | T | T | G | G | A | |
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Stephanoconus | Conus | chiangi | A | G | A | A | C | T | A | G | T | T | G | G | A | T | A | C | A | G | G | T | T | T | C | G | A | G | T | G | T | G | T | T | C | T | A | T | G | G | G | C | A | T | A | G | G | A |
| Conus | imperialis | A | G | A | A | C | T | A | G | T | T | G | T | A | T | T | C | A | G | G | T | T | T | C | G | A | G | T | G | T | G | T | T | C | T | A | T | G | G | G | C | A | T | A | G | G | A | |
| Conus | zonatus | A | G | C | A | C | T | A | G | T | T | G | T | A | T | T | C | A | G | G | T | T | T | C | G | - | G | T | G | T | G | T | T | C | A | A | T | G | G | G | C | A | T | A | G | G | A | |
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Rhizoconus | Conus | pertusus | A | G | A | C | C | T | G | A | T | A | G | G | A | T | A | A | A | G | A | C | T | T | A | A | - | G | T | T | T | A | C | T | C | T | T | C | G | G | G | T | A | T | T | G | G | A |
| Conus | miles | A | A | A | C | C | T | G | A | T | A | G | G | A | T | A | A | A | G | A | C | T | C | A | A | - | G | T | T | T | A | C | T | C | T | T | C | G | G | G | T | A | T | T | G | G | A | |
| Conus | vexillum | A | A | A | C | C | T | G | A | T | A | G | G | A | T | A | A | A | G | A | C | T | C | A | A | - | G | T | T | T | A | C | T | C | T | T | T | G | G | G | T | A | T | T | G | G | A | |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||||||||||||||||
Figure 3.
Tree inferred from the concatenated sequence of 12S and 16S rRNA and intron 9. Branches are labeled with Bayesian support values (left) and maximum likelihood bootstrap values (right). The addition of intron 9 resolves the branching order among most of the species-group clades. The relationships of Harmoniconus and Chelyconus to each other and the group of four clades to which they are most closely related differ slightly but non-significantly from the arrangement favored by intron 9 alone.
Source of the strong signal in the CIS
Most of the intron 9 sequence is conserved (74% invariant sites and singletons). The small proportion of synapormophies (10%, Table 2, starred sites), however, is concentrated on the branches that describe the deeper evolutionary relationships among the eight clades (Figure 2B). This suggests that the substitutions occur only intermittently. Indeed maximum likelihood analysis rejects a strict molecular clock, favoring instead local-clock rates (0.0001 ≤ p ≤ 0.0332 Table 1) on the branches that join the major clades deep within the tree (Figure 2B, starred branches). Similarly Bayesian analysis favored a relaxed log-normal clock (−2lnLΔ=20.4). And yet the evolution of intron 9 is less consistent with a truly episodic pattern of divergence described with an exponential clock model that might be expected during a burst of speciation (difference from a strict clock: −2lnLΔ=11.2). Application of a relaxed local clock shows promise for resolving the tempo and mode of evolution of a larger sample of Conus species because, unlike earlier studies (e.g. Duda and Kohn, 2005), these data that also resolve the relationships among the major clades of Conus species. Hence detailing the timing and order of divergence over the period from the early Cenozoic to the present should be quite tractable.
Intron 9 CIS has a higher substitution rate than the mitochondrial 12S and 16S genes also analyzed here (45% and 74% invariant sites respectively). On the other hand, the informative CIS substitutions occur on short branches deep in the tree, with very little conflicting signal on the branches that describe relationships among the clades (Lento plot, Supplementary Figure 3A). The rRNAs show strong signal with little conflict at the sites that describe the relationships within each of the clades, but little consistent signal for deeper relationships, and overall they are more saturated than the intron 9 CIS (Supplementary Figure 3B). As a result, the CIS shows higher levels of homology (rescaled consistency index=0.63, retention index= 0.84) than the12S and 16S rRNAs (rescaled consistency index=0.22, retention index= 0.49). These appear to be the reasons why the short nuclear CIS proves so informative and allows us to resolve deep branches with confidence.
To further assess the quality of the phylogenetic signal, we present results from bootstrapped Buneman trees and from Neighbor Net goodness-of-fit tests (Huson and Bryant, 2006). To resolve clades, the conservative algorithm for Buneman trees requires strong signal without conflict. We find that, despite this restriction, the Buneman tree for intron9 is resolved at the very nodes that define the early divergence events that the 12S and 16S rRNA genes fail to resolve (Figure 2B, red bootstrap values). Similarly, a well resolved neighbor network fits the intron 9 data with an astonishing 96.6% confidence level. Using 12SrRNA and 16SrRNA, both the Buneman tree and the neighbor net have fewer resolved clades than does the intron 9 CIS data; the early divergence events among the eight clades are completely unresolved (data and results available from PSC).
An application to fish-hunter behavior
Figures 2B and 3 show how substitutions in the CIS (alone, and together with those in the mt rRNA genes) define the relationships among eight Conus clades, thus extending the results of Imperial, et al. (2007). A substitution at position 27 (C->G) unites three of the four fish hunters (Pionoconus, Phasmoconus, Gastridium) with strong support (Figure 2B). Two other CIS substitutions (63:A->G and 150:A->C) unite the worm-hunting Puncticulus clade with these three fish-hunters with strong posterior probability support in the combined analysis.
Seven substitutions in the CIS unite these four clades with the worm-hunting Harmoniconus and the fish-hunting Chelyconus. A substitution in the CIS (84:A->G) implies that the worm-hunting Harmoniconus is sister to the group-of-four, and we consider this resolution the most plausible of the three possibilities. It is modestly supported by Bayes, maximum-likelihood and maximum parsimony analyses of the CIS alone (Figure 2B). Analysis of the combined data set weakly supports the alternative arrangement in which Harmoniconus and Chelyconus are sisters, forming a clade that is itself sister to the group-of-four. However, this arrangement does not have a significantly higher likelihood than the alternative arrangement implied by the CIS data alone (Figure 2B, Shimodaira-Hasegawa, −lnL = 0.45, p ≤ 0.3). Additional sequence information will be needed to securely resolve this part of the tree, but both of the seemingly most plausible resolutions imply that fish-hunting habits were twice derived from worm-hunting habits.
The Chelyconus clade consists of New-World fish-hunting species, while the Pioconus-Gastridium-Phasmoconus clade consists of Indo-Pacific fish hunters. Their last common ancestor appears to have been a worm hunter. Thus all stages in the transition from vermivory to piscivory must have occurred independently in the ancestors of Chelyconus, on the one hand, and the Indo-Pacific fish hunters, on the other hand. Consistent with this hypothesis, there are substantial molecular differences between the peptide toxins used in fish capture by Chelyconus and by the three clades of Indo-Pacific fish hunters (for a review, see Terlau and Olivera, 2004).
Future research
The 259 base-pair CIS provides a surprising number of substitutions that are informative for the deepest splits in the tree, which are not resolved at all by the much longer 12S/16S data set. How general this will be across the whole superfamily Conoidea, which includes more than 12,000 species remains to be assessed, Presumably the answer will depend on how broadly the CIS has been used in similar functional roles - whether only in the cone-snail lineage or across the whole superfamily Conoidea. A related question is whether conserved intronic sequences in other genes will turn out to evolve in the manner seen here. If so, then a much more detailed account of evolutionary change during the Cenozoic will be within reach. Our preliminary analysis of more distantly related Conus species and of members of the closely related Turridae suggests that this pattern of conserved sites with just a few very key synapomorphic changes is widespread.
A deliberate search for other conserved intronic intervals could be highly productive if, like intron 9, they yield more signal per base pair than the much longer genes that often fail to resolve relationships of interest. Resolution of evolutionary history of a broad assortment of other problematic groups that diverged during the Cenozoic, when biogeographic and climatic events seem to have resulted in bursts of evolutionary change (e.g. modern birds and mammals), may be possible with further research into the function and the evolutionary history of this and similar introns.
Supplementary Material
Acknowledgments
This work was supported by a program project grant (GM48677) from the National Institute of General Medical Sciences. We are grateful to Terry Merritt and Tuong Huynh for help in preparing the manuscript. Three reviewers provided very helpful remarks that greatly improved the clarity of the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- Bandyopadhyay PK, Garrett JE, Shetty RP, Keate T, Walker CS, Olivera BM. g-Glutamyl carboxylation: an extracellular post-translational modification that antedates the divergence of molluscs, arthropods and chordates. Proc Natl Acad Sci USA. 2002;99:1264–1269. doi: 10.1073/pnas.022637099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daMotta AJ. A Systematic Classification of the Gastropod Family Conidae at the generic level. Rome: La Conchiglia; 1991. [Google Scholar]
- Duda TF, Jr, Kohn AJ, Palumbi SR. Origins of diverse feeding ecologies within Conus, a genus of venomous marine gastropods. Biol J Linnean Soc. 2001;73:391–409. [Google Scholar]
- Duda TF, Jr, Kohn AJ. Species-level phylogeography and evolutionary history of the hyperdiverse marine gastropod genus Conus. Mol Phyl Evol. 2005;34:257–272. doi: 10.1016/j.ympev.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu DJD, Watkins M, Dia-Monje V, Cartier GE, Cruz LJ, Olivera BM. Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon. 2001;39:1899–1916. doi: 10.1016/s0041-0101(01)00175-1. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Heralde FM, 3rd, Imperial J, Bandyopadhyay PK, Olivera BM, Concepcion GP, Santos AD. A rapidly diverging superfamily of peptide toxins in venomous Gemmula species. Toxicon. 2008;51:890–897. doi: 10.1016/j.toxicon.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of Phylogenetic Networks in Evolutionary Studies. Molecular Biology and Evolution. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. software available from www.splitstree.org. [DOI] [PubMed]
- Imperial JS, Kantor Y, Watkins M, Heralde FM, 3rd, Stevenson B, Chen P, Hansson K, Stenflo J, Ownby JP, Bouchet P, Olivera BM. Venomous auger snail Hastula (Impages) hectica (Linnaeus, 1758): molecular phylogeny, foregut anatomy and comparative toxinology. J Exp Zoolog B Mol Dev Evol. 2007a;308:744–756. doi: 10.1002/jez.b.21195. [DOI] [PubMed] [Google Scholar]
- Imperial JS, Silverton N, Olivera BM, Bandyopadhyay P, Sporning A, Ferber M, Terlau H. Using chemistry to reconstruct evolution: On the origins of fish-hunting in venomous cone snails. Proceedings of the American Philosophical Society. 2007b;151:185–200. [Google Scholar]
- Iredale . Mem Queensland Museum. 1930. p. 10. [Google Scholar]
- Kohn AJ. Tempo and mode of evolution in Conidae. Malacologia. 1990;32:57–67. [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy:hypothesis testing uising phylogenies. Bioinformatics. 2005;21(5):676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Maddison D, Maddison W. MacClade 4.08. Sinauer Assoc; Sunderland, MA: 2005. [Google Scholar]
- Morch . Alphonso d’Aguirra et Gadea Comes de Yoldi. 1852. Catalogus conchyliorum quae reliquit D. [Google Scholar]
- Moretti S, Wilm A, Higgins DG, Xenarios I, Notredame C. R-Coffee a web server for accurately aligning noncoding RNA sequences. Nucleic Acids Res. 2008 Jul 1;:36. doi: 10.1093/nar/gkn278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for gast and accurate multiple seqeunce alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Pecon-Slattery J, Pearks Wilkerson AJ, Murphy WJ, O’Brien SJ. Phylogenetic assessment of introns and SINEs within the Y chromosome using the cat family felidae as a species tree. Mol Biol Evol. 2004;21:2299–2309. doi: 10.1093/molbev/msh241. [DOI] [PubMed] [Google Scholar]
- Prychitko TM, Moore WS. Comparative evolution of the mitochondrial cytochrome b gene and nuclear beta-fibrinogen intron 7 in woodpeckers. Mol Biol Evol. 2000;17:1101–1111. doi: 10.1093/oxfordjournals.molbev.a026391. [DOI] [PubMed] [Google Scholar]
- Prychitko TM, Moore WS. Alignment and phylogenetic analysis of beta-fibrinogen intron 7 sequences among avian orders reveal conserved regions within the intron. Mol Biol Evol. 2003;20:762–771. doi: 10.1093/molbev/msg080. [DOI] [PubMed] [Google Scholar]
- Röckel D, Korn W, Kohn AJ. Manual of the Living Conidae. Wiesbaden, Germany: Verlag Christa Hemmen; 1995. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Stamatakis P, Hoover J, Rougemont A Rapid Bootstrap Algorithm for the RAxML Web-Servers. Systematic Biology. 2008;75(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Swainson W. A treatise on malacology; or the natural classification of shells and shellfish. London: Longman, Orme, Brown, Green; 1840. [Google Scholar]
- Swofford DL. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates; Sunderland, Massachusetts: 2002. PAUP*. [Google Scholar]
- Tavaré S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. American Mathematical Society: Lectures on Mathematics in the Life Sciences. 1986;17:57–86. [Google Scholar]
- Terlau H, Olivera BM. Conus Venoms: A Rich Source of Novel Ion Channel-Targeted Peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Yoder A, Yang Z. Estimation of Primate Speciation Dates Using Local Molecular Clocks. Mol Biol Evol. 2000;17(7):1081–1090. doi: 10.1093/oxfordjournals.molbev.a026389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.