Abstract
Asthmatic patients are currently classified as either severe or non-severe based primarily on their response to glucocorticoids. However, because this classification is based on a post-hoc assessment of treatment response, it does not inform the rational staging of disease or therapy. Recent studies in other diseases suggest that a classification which includes molecular information could lead to more accurate diagnoses and prediction of treatment response. We therefore measured cytokine values in bronchoalveolar lavage (BAL) samples of the lower respiratory tract obtained from 83 asthma patients, and used bipartite network visualizations with associated quantitative measures to conduct an exploratory analysis of the co-occurrence of cytokines across patients. The analysis helped to identify three clusters of patients which had a complex but understandable interaction with three clusters of cytokines, leading to insights for a state-based classification of asthma patients. Furthermore, while the patient clusters were significantly different based on key pulmonary functions, they appeared to have no significant relationship to the current classification of asthma patients. These results suggest the need to define a molecular-based classification of asthma patients, which could improve the diagnosis and treatment of this disease.
1. Introduction
Asthma is a chronic inflammatory disease of the airways which affects about 300 million individuals worldwide, and results in an estimated 250,000 dying prematurely [1]. The disease is characterized by recurrent airflow obstruction and hyperactivity to nonspecific stimuli [2], which is treated mainly with inhaled glucocorticoid therapy. Although many asthma patients respond well to such therapy, a subset of patients (referred to as “severe”) is unresponsive, and has disproportionately high rates of morbidity and mortality. As a result, medical costs for treating this subset accounts for more than 40% of the total cost of asthma treatment [3].
Unfortunately, relatively little is known about which patients will have poor outcomes to glucocorticoid therapy. For example, although asthma patients are currently classified as severe or non-severe based on their therapeutic response to glucocorticoids [4], this course-grained clinical classification does not explain the varying degrees of lung function compromise, airway hyper-reactivity, gastro-esophageal reflux, and chronic obstructive pulmonary disease (COPD) in patients currently diagnosed with severe asthma. Physicians therefore often use a trial and error process to balance escalating medications with associated side effects in an effort to treat severe asthma patients.
Recent developments in molecular biology and powerful analytical methods such as network analysis provide new opportunities to shift our understanding of diseases from a morphological (based on clinical and histological findings) to a molecular basis [5–6]. For example, gene expression analyses have been shown to improve prediction of treatment response in several diseases such as breast cancer [7–9] and leukemia [10]. Because asthma is a chronic disease associated with innate and T helper lymphocyte-biased inflammation [2], we hypothesized that profiles of airway fluid cytokines that represent major effectors molecules of leukocytic inflammation could provide insights for developing a new molecular-based classification of asthma. Such a classification, based on effector proteins found in lung fluids, could enable more accurate prediction of disease progression and therapeutic response.
We begin by describing our motivation for the current analysis through a brief summary of previous approaches used to analyze asthma patients. Next, we describe how we assembled a dataset of patients and their cytokine profiles, why and how we represented it using networks, and how we analyzed the networks using visualizations and appropriate quantitative measures. We then discuss how the bipartite network analysis revealed complex co-occurrence patterns of cytokine across patients, and how those patterns relate to key attributes of pulmonary function, and known molecular pathways. We conclude by discussing the need to define a molecular-based classification of chronic asthma patients, and the utility of bipartite network analyses to understand complex relationships.
2. Related Work
As stated in the introduction, there is a growing consensus among asthma researchers that the current classification of asthma patients has not been sufficiently predictive to guide treatment. For example, a 2009 World Health Organization panel consisting of 33 asthma researchers from 14 countries concluded that “the use of severity as a single outcome measure has limited value in predicting which treatment will be required and the response to that treatment”. Moreover, they noted that “severity is not a stable feature of asthma but may change with time, whereas the classification by disease severity suggests a static feature” pg. 928 [1].
Despite decades of research in asthma, why has it been so difficult to formulate a classification of asthma patients that can guide effective treatment? We believe this is because majority of the research has either begun with an a priori grouping of patients (using phenotype or molecular information), or has used analytical methods such as hierarchical clustering that assume the existence of disjoint patient clusters [11–14]. For example, Hastie et al., [11] grouped patients based on severity and analyzed how the imposed groups were similar or different based on other phenotype variables. Similarly, Woodruff et al., [12] grouped patients based on high or low expression of IL-13 inducible genes, and compared the imposed groups based on other genes, and lung functions.
To avoid biases based on a priori patient groupings, some researchers have taken a more data-driven approach to identify emergent clusters of patients. For example, Moore et al. [13] used hierarchical clustering to identify five groups of patients based on phenotype information, and then examined which variables were significant between the groups. Similarly, Brasier et al. [14] used hierarchical clustering to identify four groups of patients based on molecular information, but then used the existing severe versus non-severe classification to identify emergent clusters for further analysis. While such data-driven approaches address the limitations of a priori groupings of patients, unsupervised learning methods such as hierarchical clustering and k-means assume the existence of disjoint clusters in the data [15], and therefore could conceal other valid patterns (e.g., uniform distributions or nested clusters) of how patients relate to each other.
Although the above studies have substantially increased our appreciation of the complex multidimensional nature of asthma, to the best of our knowledge none have used data-driven approaches without strong built-in assumptions to analyze how patients are similar or different based on molecular information. Such an approach has the potential to inform the identification of a more clinically useful classification of asthma patients.
3. Method
Our research began with the question: How do cytokines implicated in asthma, co-occur across patients? To address our research question, we made critical decisions regarding data selection, data representation and data analysis as discussed below.
3.1 Data Selection
Our study was based on a secondary analysis of cytokine profiles collected in a consortium-wide study [14]. Levels for 25 cytokine were measured from bronchoalveolar lavage (BAL) samples of the lower respiratory tract obtained from 40 severe, and 43 non-severe asthma patients. The classification of patients was made according to the consensus definition of the American Thoracic Society [4], and the two groups were balanced by age and gender. As shown in Table 1, the dataset included 6 pulmonary function measures determined to be independent by the domain experts. Because 50% of values in 7 cytokines (IL-1b, IL-7, IL-10, IL-12, IL-13, IFN-γ, and GM-CSF) had undetectably low values, they were removed from the dataset, resulting in a total of 18 cytokines (see our earlier publication [14] for details about the data collection and inclusion criteria).
Table 1.
Comparison of six independent pulmonary functions across the three patient clusters identified by the network analysis.
| Pulmonary Function | p value with FDR correction |
|---|---|
| Max FVCpp/MPVLung | 0.006* |
| Max FEV1pp/MPVLung | 0.0375* |
| Baseline FEV1pp | 0.0375* |
| Baseline FEV1/FVC | 0.1944 |
| Max FEV1 Reversal | 0.583 |
| PC20 Methacholine | 0.0375* |
Significant differences between the groups are indicated by asterisks based on a one-way, two-tailed Kruskal-Wallis test with an FDR correction. (FVC=forced vital capacity, FEV1=forced expiratory volume in 1 second, PC20 methacholine=dose of methacholine that produces 20% fall in FEV1, FEV1 albuterol reversal= percent change in FEV1 in response to albuterol inhalation, MPV = maximal postbronchodilator value, pp = percent predicted).
3.2 Data Analysis
Our analysis consisted of two steps: (1) exploratory visual analysis though the use of networks to identify emergent visual patterns of cytokine co-occurrence; and (2) quantitative analysis through the use of methods whose assumptions matched the visual patterns in order to verify them. This two-step method was motivated by our earlier studies [15, 16, 17] using a similar approach which have revealed that co-occurrence relationships can exhibit in different patterns (e.g., nested clusters, disjoint clusters), each prompting the use of quantitative methods that make the appropriate assumptions about the underlying data.
3.2.1 Exploratory Visual Analysis
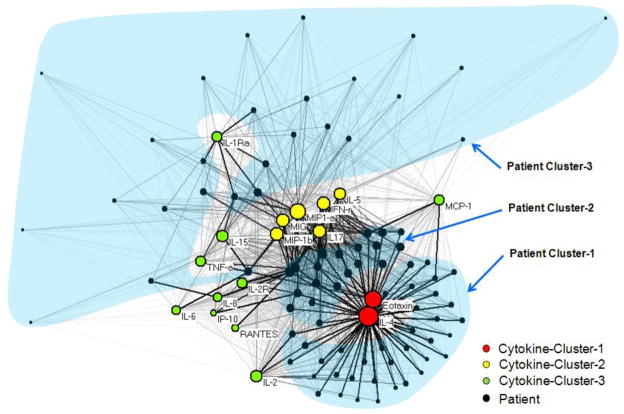
Networks are increasingly being used to analyze a wide range of molecular phenomena such as gene regulation [19], disease-gene associations [20], and disease-protein associations [21]. A network (also referred to as a graph in mathematics) consists of a set of points or nodes, joined in pairs by lines or edges; nodes represent one or more types of entities (e.g., patients or cytokines). Edges between the nodes represent a specific relationship between the entities (e.g., a patient has a particular cytokine expression value). Figure 1 shows a bipartite network (where edges exist only between different types of entities) [22] of patients and cytokines, which was created using Pajek [23] (version 1.23).
Figure 1.
A bipartite network (automatically laid by the Kamada-Kawai algorithm [21]) shows how 18 cytokines (colored nodes) co-occur across 83 patients (black nodes). The thickness of the edges is proportional to the normalized cytokine expression values, and the size of the nodes is proportional to the sum of the edge weights that connect to them. Therefore patients with high total cytokine values have large nodes, and higher cytokine values are represented by thicker edges. For clarity, colors represent cytokine clusters, transparent blue shapes represent patient clusters, and patient IDs are not shown. See Supplementary Figure A, which shows the same network shown here, but with the patient nodes colored by severity to help examine the relationship of the current severe vs. non-severe classification, to the patient clusters.
Node diameter was used to represent the sum of the edge weights connected to it. This enabled a rapid visual inspection to determine for example, which patients have overall high aggregate cytokine values, and how such patients relate to the rest of the network. In addition, using a second network of the same data (see Supplementary Figure A), the node color was used to represent asthma severity (red for severe, and blue for non-severe), which enabled us to analyze how the patterns in the overall network related to the existing classification of asthma.
Edge weights in the network were used to represent the strength of the cytokine values for each patient-cytokine pair. Because the 18 cytokines had different and unknown theoretical ranges, we used the min-max normalization method using the following formula:
where vij is the raw expression value for cytokine i of patient j, v'ij is the corresponding normalized value, and mini and maxi represent the minimum and maximum raw expression values of cytokine i across all patients. This formula performs a linear transformation on the raw data values by converting them to range from 0–1, and therefore preserving the relative distances between the values. The min-max normalization method enables a consistent method to compare the different cytokines values, and is especially useful when outliers are meaningful such as what tends to occur in asthma cytokine expression due to biological diversity [24]. As shown in Figure 1, the edge thicknesses were drawn to be proportional to these normalized cytokine values.
Global patterns in the network were visualized and analyzed using the Kamada-Kawai layout algorithm [25]. The algorithm results in nodes that are connected by high edge weights to be pulled together, and those with low edge weights to be pushed apart. This algorithm is fast but approximate1 and well-suited for small to medium-sized networks consisting of between 50–1000 nodes [26]. As shown, the result is that nodes with a similar pattern of connections (e.g., Eotaxin and IL-4 in the lower right hand side of Figure 1) are placed close to each other.
Network analyses provide two advantages for analyzing complex relationships. (1) They do not require a priori assumptions about the relationship of nodes within the data, such as the hierarchical assumption of hierarchical clustering, or disjoint clusters of k-means. Instead, by using a simple pair-wise representation of nodes and edges, network layouts enable the identification of multiple structures (e.g., hierarchical, disjoint, overlapping, nested) in a single representation [26]. Therefore, while layout algorithms such as Kamada-Kawai depend on the force-directed assumption and its implementation, such algorithms are viewed as less biased for data exploration because they do not impose a particular cluster structure on the data, often leading to the identification of more complex structures in the data [15]. (2) Networks enable the simultaneous visualization of multiple raw values (e.g., patient-cytokine associations, cytokine values, patient attributes), aggregated values (e.g., sum of cytokine values), and emergent global patterns (e.g., clusters) in a uniform visual representation. The overall network representation therefore enables the rapid generation of hypotheses based on complex multivariate relationships, and enables a more informed approach for selecting quantitative methods to verify the patterns in the data.
3.2.2 Quantitative Analysis
The insights derived from the network visualizations were quantitatively analyzed using three methods. (1) Because the network layout suggested the presence of distinct clusters for patients and for cytokines, we used the agglomerative hierarchical clustering method to verify the number of clusters, and to identify the boundaries of the clusters. In addition, we used a heat map to inspect the profiles of specific patients and cytokines. The clustering was done using the Manhattan dissimilarity measure (to handle the weighted edges) with the Ward linkage function [18]. Cluster boundaries were determined based on natural breaks in the patient and cytokine dendrograms. To test whether there were significant breaks in the dendrogram (denoting the existence of disjoint clusters), we compared the variance, skewness, and kurtosis of the dissimilarities in the asthma network, to 1000 permutations of the asthma network. For each network permutation we preserved the number of nodes, and the number of edges connected to each node, in addition to the edge weight distribution of patients when analyzing the cytokine dendrogram, and vice versa. Significant breaks in the asthma patient or cytokine dendrograms would result in a significantly larger variance, skewness, and kurtosis of the dissimilarity measures, compared to the same measures generated from the random networks.
(2) To analyze the relationship between asthma severity and the patient clusters, we used the chi-square test of independence. To analyze the overall significance of 6 independent pulmonary functions, we used the one-way, two-tailed Kruskal-Wallis test (non-parametric ANOVA) to address the skewed values, and the false discovery rate (FDR) procedure to correct for multiple comparisons. (3) To analyze the significance between each pair of clusters for the above patient variables, we used the Dunn’s test procedure.
4. Results
The bipartite network visualization and quantitative analysis revealed distinct patient clusters, and cytokine clusters. For each set of clusters we describe the results of the visual analysis, the cluster analysis, and their significance to clinical attributes and molecular processes.
4.1 Patient Clusters
Exploratory Visual Analysis
As shown in Figure 1, the visual analysis helped to identify three clusters of patients based on their cytokine profiles: (a) Patient-Cluster-1 (shown in the lower right hand corner of Figure 1) had medium to high levels of the Eotaxin and IL-4. However, they had relatively lower values for the rest of the cytokines as shown by their relatively small diameters. (b) Patient-Cluster-2 (shown in the center of the network) had high values of Eotaxin and IL-4, but also high values for another set of six cytokines (IL-5, IFN-γ, MIP1a, MIG, IL-17, MIP-1b) shown in the center of the network. The higher cytokine values result in relatively larger node diameters compared to Cluster-1. (c) Patient-Cluster-3 has overall lower values of many cytokines resulting in them being scattered along the top periphery of the network. The overall lower levels of most cytokines result in relatively smaller node diameters.
Quantitative Analysis
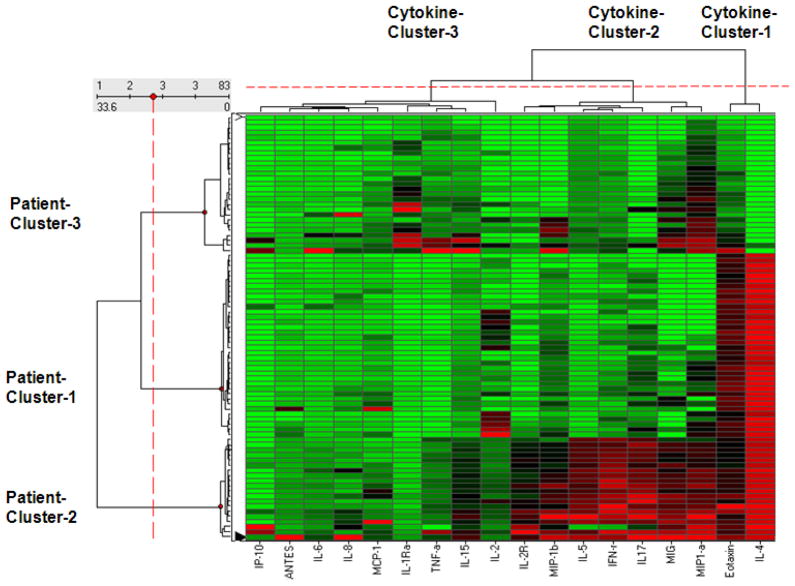
Because the network suggested the existence of distinct patient clusters, we used agglomerative hierarchical clustering to identify the number and boundaries of those clusters. As shown by the patient dendrogram on the vertical axis of Figure 2, the agglomerative hierarchical clustering identified the boundaries of the visual clusters in the network. Furthermore, while Patient-Cluster-1 and Patient-Cluster-2 were intuitively clear from the network, Patient-Cluster-3 was identified as a distinct cluster in the dendrogram because its members have a pattern of similarly low cytokine levels. The clusteredness of the patients in the asthma network was significant as measured by the variance of the dissimilarities (Asthma = 64.95, Random Mean = 20.08, p<.001 two-tailed test), skewness of the distribution of dissimilarities (Asthma = 4.9, Random Mean = 2.81, p<.001 two-tailed test), and kurtosis of the distribution of dissimilarities (Asthma = 30.24, Random Mean = 14.78, p<.001 two-tailed test).
Figure 2.
A heat map where the rows represent patients, the columns represent cytokines, and the colors represent normalized cytokine values (green = 0, red = 1). The rows and columns are ordered based on the results of the agglomerative hierarchical clustering, with dendrograms for the patient and cytokines shown on the vertical and horizontal axes respectively.
Relationship to Clinical Variables
To infer the meaning of the three patient clusters, we analyzed the relationship between each identified cluster to asthma severity, and to pulmonary function.
Asthma Severity
As discussed in the introduction, patients are currently classified as severe or non-severe. Supplementary Figure A shows the same network in Figure 1, but where the patient nodes have been colored based on severity (red for severe, and blue for non-severe). An inspection of the network showed no visual pattern; there appeared to be an even number of both types of severity in each cluster. The chi-square analysis verified this visual result, which showed no significant association in asthma severity between the three patient clusters (χ2(2,N=83)=0.9298, p=0.628). This suggests that a classification of patients based on cytokine profiles does not match the current classification of asthma based on severity.
Pulmonary Function
As shown in Table 1, the Kruskal-Wallis test revealed that 4 out of 6 pulmonary function2 measures were significantly different across the clusters3. The pair-wise inter-cluster analysis revealed that Patient-Cluster-3 had three lung functions (Max FEV1pp/MPVLung, Baseline FEV1pp, and PC20 Methacholine) that were significantly higher than Patient-Cluster-1, and one lung function (Max FVCpp/MPVLung) that was significantly higher than Patient-Cluster-2. In contrast, Patient-Cluster-1 had only one lung function (Max FVCpp/MPVLung) that was significantly higher than Patient-Cluster-2. Patient-Cluster-3 therefore had less baseline airway obstruction (both FEV1 values were significantly higher), less hyper-reactive to methacoline challenge (significantly higher PC20 Methacholine), and preserved pulmonary capacity (significantly higher FVC values) compared to the other two patient clusters.
4.2 Cytokine Clusters
Exploratory Visual Analysis
The bipartite network visualization also revealed three cytokines clusters, which have a complex relationship to the patient clusters. (a) Cytokine-Cluster-1 (in the lower right hand side of the network) consisting of Eotaxin and IL-4 contain cytokines that are pushed together because many patients from Patient-Cluster-1 and -2 have high values of those two cytokines. Their resulting larger diameters suggest that they are over-represented in patients compared to the other cytokines. This observation is also salient by the many red cells (representing high values) in the last two columns (representing Eotaxin and IL-4) of the heat map in Figure 2. (b) Cytokine-Cluster-2 consisting of six cytokines (mentioned earlier) which are pushed together because they have high values of mainly Patient-Cluster-2. Unlike Cytokine-Cluster-1, they have high values for only one patient cluster, and therefore have smaller diameters. (c) Cytokine-Cluster-3 consisting of the remaining cytokines scattered on the left and right hand side of the network have overall lower values across all patients, and therefore have the smallest diameters in the network.
Quantitative Analysis
Similar to the patient clusters, the network suggested the existence of distinct patient clusters. We therefore used agglomerative hierarchical clustering to identify the number and boundaries of the clusters. As shown by the cytokine dendrogram on the horizontal axis of Figure 2, the agglomerative hierarchical clustering identified the boundaries of the visual clusters in the network. While Cytokine-Cluster-1 and Cytokine-Cluster-2 are intuitively clear from the network, Cytokine-Cluster-3 is identified as a distinct cluster in the dendrogram because it has a pattern of similarly weak levels with patients. This observation is salient by the large number of green cells (representing low values) for this cluster in the heat map in Figure 2. The clusteredness of the cytokines in the asthma network was significant as measured by the variance of the dissimilarities (Asthma = 837.62, Random Mean = 46.69, p<.001 two-tailed test), skewness of the distribution of dissimilarities (Asthma = 2.18, Random Mean = 0.49, p<.001 two-tailed test), and kurtosis of the distribution of dissimilarities (Asthma = 7.25, Random Mean = 2.49, p<.001 two-tailed test).
4.3 Discussion
The results suggest that cytokine values can indeed separate patients into distinct clusters. While this result was sufficient on its own for insights to cluster asthma patients, the bipartite network analysis also helped to identify cytokine clusters and their relationship to the patient clusters, which enabled us to infer biological meaning about the patient clusters.
The frequent co-occurrence of Eotaxin and IL-4 (Cytokine-Cluster-1) is congruent with a known sequence of molecular changes in asthma patients who often have a T-helper-2 (TH2) lymphocyte-skewed immune response. This response results in the secretion of IL-4, which in turn induces Eotaxin production by bronchial epithelial cells [27]. The resulting downstream actions include the activation and recruitment of tissue-resident eosinophils, a hallmark of early stage asthma. The presence of Eotaxin and IL-4 in lung fluids therefore appears to represent important sub-stages of a complex molecular pathway in asthma, which explains their frequent co-occurrence in the network.
To understand the biological significance for cytokines in Cytokine-Cluster-2 (IL-5, IFN-γ, MIP1a, MIG, IL-17, and MIP-1b), we entered its members into the Ingenuity Pathway Analysis (IPA) application. The results from IPA suggest that the frequent co-occurrence of these cytokines is regulated by the innate inflammatory nuclear factor-κB pathway (NF-κB). NF-κB is a potent pro-inflammatory transcription factor that activates expression of cytokine networks. Furthermore, persistent NF-κB activation has been linked to uncontrolled/acute exacerbations of asthma [28]. The frequent co-occurrence of this set of cytokines therefore implies the presence of a distinctly different pro-inflammatory state compared to the IL-4 – Eotaxin process.
The above cytokine clusters, along with pulmonary functions of the patients, provide a biological explanation for the patient clusters. The strong relationship of Patient-Cluster-1 to Cytokine-Cluster-1 suggests that patients in this cluster have disease primarily driven by TH2 inflammation. In contrast, Patient-Cluster-2 has a strong relationship to both Cytokine-Clusters-1 and -2. This result implies that patients in Patient-Cluster-2 have a component of activated innate inflammatory pathways. Further evidence for this inference of state-based clusters is provided by differences in pulmonary function across the clusters: Patient-Cluster-3 which has the lowest cytokine values for both of the above cytokine clusters, also has the largest number of significant differences in obstructive airway disease parameters in pulmonary function testing, and lowest airway reactivity response to methacholine compared to Patient-Clusters-1 and -2. This implies that Patient-Cluster-3 represents a subgroup of asthmatics with preserved pulmonary function and greatest response to albuterol without active inflammation. The network analysis of patients and cytokines therefore implies a state-based classification of asthma patients informed by underlying molecular processes. The results also provide evidence for the growing consensus [1] that asthma is a dynamic disease where the same patient could enter different asthmic states based on environmental and other triggers. Future studies that include such information could lead to a better understanding of the relationship between triggers and resulting asthmic states, which could translate into more effective treatment and prevention approaches that are personalized to each patient.
The limitation of our study is that we analyzed only one dataset, and our future research will attempt to replicate the results in a similar dataset. However, the current results suggest that asthma patients can be meaningfully classified using molecular markers such as cytokines.
4.4 Conclusions and Future Research
Cytokines control key processes in asthma including immune activation and T lymphocyte skewing. However, little work has been done to investigate whether and how cytokines could help to classify patients. By using bipartite network visualizations without a priori assumptions of patient classes, combined with appropriate quantitative methods suggested by the patterns in the network, we arrived at a new state-based understanding of asthma.
Our experience suggests that the bipartite network representation was effective because it enabled: (1) the overlaying of multiple raw and aggregated variables in addition to the cluster boundaries, onto the same visualization; (2) the selection of quantitative methods that made the appropriate assumptions about the observed co-occurrence patterns in the data; and (3) the detection of complex relationships between the patient clusters and the cytokine clusters, which were difficult to detect by analyzing just the heat map in Figure 2. These combined features of the bipartite network representation enabled the asthma experts on the team to derive an intuitive understanding of the complex multivariate relationships between molecular and phenotype information, which rapidly led to the proposed state-based classification. The overall approach of using complementary visual and quantitative methods to comprehend complex molecular and phenotype relationships therefore provides an approach that could generalize to other datasets with similar translational goals.
It is important to reiterate that the bipartite network could have revealed co-occurrence patterns without the presence of distinct clusters, prompting us to use other methods to quantify the patterns as we have done in a recent study on cancer patients [15]. Therefore, we believe that bipartite networks provided an important first step to identify the nature of co-occurrence in molecular data, which then guided the use of appropriate quantitative methods to verify those patterns.
In our future research, we plan to extend our understanding of the current results in three ways:
Analyze the significance of the emergent clusters of patients and cytokines by comparison of the bipartite network directly to random networks. This is a non-trivial task as modularity algorithms for bipartite networks [29] (designed to identify and measure the significance of graph partitions or clusters in bipartite networks) currently do not handle edge weights [personal communication Roger Guimerà, Mark Newman].
Explore other complementary visual analytical methods to identify other complex relationships in the data. For example our recent use of three dimensional (3D) immersive visualizations of a renal dataset enabled the identification of a complex relationship of domain importance that was missed in the analysis of a 2D network analysis of the same data [30]. Furthermore, although networks allow multiple variables to be represented using graphical attributes such as color, shape, and size, there are limits on the number of variables that can be simultaneously represented or comprehended, often resulting in the need for multiple networks. We are therefore exploring the use of Circos Ideograms [31, 32] which are explicitly designed to enable a large set of variables to be simultaneously visualized, with the goal of exploring their relationship to the clusters identified through the network analysis, and to each other.
Use the patient clusters and their relationship patient variables to inform the development of classifiers using supervised learning methods. The goal of developing classifiers that are informed by the unsupervised learning methods used in the current study is to enable the resulting classification not only to have predictive power for response to therapy, but also to be meaningful from a domain perspective.
The results of the above multi-method approach, progressing from discovery through visual analytics, verification and validation through quantitative analysis, and prediction through classifiers, could lead in the future to a molecular classification of asthma patients that is based on underlying biological processes and has intuitive domain meaning. Such a classification has a higher probability for successful translation to clinical diagnosis and treatment of this complex disease.
Supplementary Material
Acknowledgments
This work was supported by NIH grants 1U54RR02614 UTMB CTSA (ARB), AI062885 (ARB), NHLBI contract BAA-HL-02–04 (ARB), HL69130 US SARP (WJC), and HL69149 (MC). We thank H. Spratt, M. Sinha, A. Ganesan, and D. Bostick for their suggestions.
Footnotes
The Kamada-Kawai layout algorithm is approximate because it does not guarantee a globally optimal layout. The method is therefore used to explore the data using different starting conditions, and the observed topology verified using appropriate quantitative methods.
FVC and FEV1 are commonly used pulmonary function tests in asthma. Here we used an additional test called maximum postbronchodilatory volume (MPV) to aid us in further characterizing the degree of airflow obstruction.
In contrast, only two (Baseline FEV1pp and MaxFEV1pp/MPVLung) of the six measures were significantly different across the severe and non - severe patients).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
4.6 References
- 1.Bousquet J, Mantzouranis E, Cruz AA, Aït-Khaled N, Baena-Cagnani CE, Bleecker ER, et al. Uniform de nition of asthma severity, control, and exacerbations:Document presented for the World Health Organization Consultation on Severe Asthma. J Allergy Clin Immunol. 2010;126(5):926–38. doi: 10.1016/j.jaci.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 2.Busse WW, Lemanske RF. Asthma. N Engl J Med. 2001;344:350–362. doi: 10.1056/NEJM200102013440507. [DOI] [PubMed] [Google Scholar]
- 3.Godard P, Chanez P, Siraudin L, Nicoloyannis N, Duru G. Costs of asthma are correlated with severity. Eur Respir J. 2000;19:61–67. doi: 10.1183/09031936.02.00232001. [DOI] [PubMed] [Google Scholar]
- 4.American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med. 2000;162:2341–2351. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 5.Coller H, Loh M, Downing J, Caligiuri M, et al. Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 6.Chuang H, Lee E, Liu Y, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Molecular Systems Biology. 2007;3:141. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wulfkuhle JD, Speer R, Pierobon M, Laird J, Espina V, Deng J, Mammano E, Yang SX, Swain SM, Nitti D, et al. Multiplexed Cell Signaling Analysis of Human Breast Cancer Applications for Personalized Therapy. Journal of Proteome Research. 2008;7:1508–1517. doi: 10.1021/pr7008127. [DOI] [PubMed] [Google Scholar]
- 8.van ’t Veer LJ, Dai H, Vijver van de MJ, He YD, Hart AA, Mao M, Peterse HL, Kooy van der K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.Hall P, Ploner A, Bjöhle J, Huang F, Lin CY, Liu E, Miller L, Nordgren H, Pawitan Y, Shaw P, et al. Hormone-replacement therapy influences gene expression profiles and is associated with breast-cancer prognosis: a cohort study. BMC Medicine. 2006;4:16. doi: 10.1186/1741-7015-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cario G, Stanulla M, Fine B, Teuffel O, Neuhoff N, Schrauder A, Flohr T, Schafer B, Bartram C, Welte K, et al. Distinct gene expression profiles determine molecular treatment response in childhood acute lymphoblastic leukemia. Blood. 2005;105:821–826. doi: 10.1182/blood-2004-04-1552. [DOI] [PubMed] [Google Scholar]
- 11.Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, Bleecker ER. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125(5):1028–1036. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, Arron JR, Koth LL, Fahy JV. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180(5):388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, D'Agostino R, Jr, Castro M, Peters SP, Bleecker ER, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181(4):315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brasier AR, Victor S, et al. Molecular phenotyping of severe asthma using pattern recognition of bronchoalveolar lavage derived cytokines. J Allergy Clin Immunol. 2008;121:30–37. doi: 10.1016/j.jaci.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhavnani SK, Bellala G, Ganesan A, Krishna R, et al. The Nested Structure of Cancer Symptoms: Implications for Analyzing Co-occurrence and Managing Symptoms. Methods of Information in Medicine. 2010;49(6):581–591. doi: 10.3414/ME09-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhavnani SK, Carini S, Ross J, Sim I. Network Analysis of Clinical Trials on Depression: Implications for Comparative Effectiveness Research. Proc of AMIA’10. 2010 [PMC free article] [PubMed] [Google Scholar]
- 17.Bhavnani SK, Abraham A, Demeniuk C, Gebrekristos M, Gong A, Nainwal S, Vallabha GK, Richardson RJ. Network Analysis of Toxic Chemicals and Symptoms: Implications for Designing First-Responder Systems; Proc of AMIA’07; 2007. pp. 51–55. [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson RA, Wichern DW. Applied Mutlivariate Statistical Analysis. NJ: Prentice-Hall; 1998. [Google Scholar]
- 19.Albert RK. Complex Networks. 2004. Boolean Modeling of Genetic Regulatory Networks; pp. 459–481. [Google Scholar]
- 20.Goh K, Cusick M, Valle D, Childs B, Vidal M, Barabási A. The human disease network. Proceedings of the National Academy of Sciences. 2007;104:8685. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ideker T, Sharan R. Protein networks in disease. Genome Research. 2008;18:644. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman MEJ. Networks: An Introduction. Oxford University Press; 2010. [Google Scholar]
- 23.Batagelj V, Mrvar A. Pajek – analysis and visualization of large networks. Graph Drawing Software. 2003:77–103. [Google Scholar]
- 24.Brasier AR, Victor S, Ju H, Busse WW, et al. Predicting intermediate phenotypes in asthma using bronchoalveolar lavage-derived cytokines. Clinical and Translational Science. 2010;3(4):147–57. doi: 10.1111/j.1752-8062.2010.00204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamada T, Kawai S. An algorithm for drawing general undirected graphs. Information Processing Letters. 1989;31(1):7–15. [Google Scholar]
- 26.Nooy W, Mrvar A, Batagelj V. Exploratory Social Network Analysis with Pajek. Cambridge University Press; 2005. [Google Scholar]
- 27.Fujisawa T, Kato Y, et al. Chemokine production by the BEAS-2B human bronchial epithelial cells: Differential regulation of eotaxin, IL-8, and RANTES by TH2- and TH1-derived cytokines. J Allergy Clin Immunol. 2001;105(1):126–133. doi: 10.1016/s0091-6749(00)90187-8. [DOI] [PubMed] [Google Scholar]
- 28.Gagliardo R, Chanez P, Mathieu M, et al. Persistent Activation of Nuclear Factor– B Signaling Pathway in Severe Uncontrolled Asthma. Am J Respir Crit Care Med. 2003;168(10):1190–1198. doi: 10.1164/rccm.200205-479OC. [DOI] [PubMed] [Google Scholar]
- 29.Guimera R, Sales-Pardo M, Amaral LAN. Module identification in bipartite and directed networks. Phys Rev E. 2007;76:1–8. doi: 10.1103/PhysRevE.76.036102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhavnani SK, Arunkumaar G, Hall T, Maslowski E, et al. Discovering Hidden Relationships between Renal Diseases and Regulated Genes through 3D Network Visualizations. BMC Research Notes. 2010;3:296. doi: 10.1186/1756-0500-3-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krzywinski M, Schein J, Birol I, Connors J, et al. Circos: an Information Aesthetic for Comparative Genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhavnani SK, Pillai R, Calhoun WJ, Brasier AR. How Circos Ideograms Complement Networks: A Case Study in Asthma. Proc of AMIA Summit on Translational Bioinformatics. 2011 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.