Abstract
We have established a model system of hormone action, in an Sf9 cell transfection system, using defined enhancer motifs and natural core promoters of metamorphosis-associated genes. The DR1 enhancer, that is an established DNA binding site for the ecdysone receptor/ultraspiracle heterodimer, was necessary for transcriptional activation by 20-OH ecdysone. For this activated transcription, a natural sequence closely 5′ to the TATA box is necessary. Cotreatment with juvenile hormone III strongly suppressed the steroid activation of transcription. However, in the absence of the sequence located closely 5′ to the TATA box, cotreatment with juvenile hormone instead increased transcription over that occurring due to 20-hydroxy-ecdysone alone. This sensitivity to activation by cotreatment with juvenile hormone could be transferred to a related, but otherwise unresponsive, hexamerin core promoter simply by transferring to the unresponsive promoter the five base transcription start site (ACAGT) from the responsive hexamerin gene. These are the first reports that the direction of JH action on 20-OH ecdysone-activated transcription can be reversed by removal of a sequence at the core promoter, and that modulatory action of juvenile hormone can be transferred to a different gene by transferring the transcription start site motif.
Keywords: Ecdysone, Juvenile Hormone, Core Promoter, Metamorphosis, Enhancer, Transcription, Ultraspiracle, Ecdysone Receptor, RXR
1. Introduction
Physiological processes are coordinated by steroid and terpenoid regulators in both vertebrates and invertebrates. Vertebrate fetal brain development, gonadal differentiation and mammogenesis are all regulated by steroid hormones (Cooke et al. 1998; Hayes 1998; Shyamala 1999). Carboxylated terpenoids, i.e., retinoids, are also proximal regulators of differentiation and formation of tissues and limbs (Smith et al. 1998; Maden et al. 2001). In comparison, in the invertebrates, ecdysteroid hormones (e.g., 20 OH ecdysone) time the molting-based growth of larval insects and the cellular commitment to particular morphogenetic programs (Nakagawa and Henrich 2009). The methyl ester of a carboxylated terpenoid, methyl epoxyfarnesoate (= insect ‘juvenile hormone,’ JH), is a primary regulator of the particular morphogenetic program to which the cells become committed (Truman and Riddiford 2007). Its central importance to cellular commitment is readily evident by the fatal derangement and deformation of organs and appendages that occur under conditions of juvenile hormone pathway disregulation (Liu et al. 2009; Jones et al. 2010a; Riddiford et al. 2010). Invertebrate development may also be regulated by additional juvenile hormone-related compounds (e.g., methyl farnesoate) (Wang and LeBlanc 2009; Jones et al. 2010b).
The regulation of insect morphogenetic programs by ecdysteroids and JH provides evidence at the physiological level of the principle of integration of hormonal signaling pathways in the control of cellular differentiation. While exposure of cells to 20-OH ecdysone activates a particular program of differentiation, the simultaneous exposure to both 20-OH ecdysone and juvenile hormone suppresses expression of that program and activates another (Riddiford 1996). In the classic model, exposure of larval cells to 20-OH ecdysone stimulates differentiation of cells toward a pupal phenotype, except that the presence of juvenile hormone during the larval exposure to ecdysteroid promotes a continued larval phenotype and larval to larval molt (Truman and Riddiford 2002). In vertebrate models systems, the observations of crosstalk between hormonal signaling pathways at the physiological level have become now understood at the molecular level of resolution, with the revelation that there are a number of different mechanisms by which this crosstalk is achieved. For example, it may occur at the level of competition between different nuclear receptors for a common binding site (Vasudevan et al. 2001a), or at the level of one hormone receptor (e.g. thyroid hormone receptor) squelching activity of another (estrogen receptor) by competition for coactivators (Vasudevan et al. 2001b), or by different receptors competing for binding to the common RXR heterodimer partner (Hunter et al. 1996; Kakizaki et al. 2002), or by direct competition of the two ligands for the same receptor (e.g., oxytocin and progesterone, Grazzini et al. 1998), or by the two hormones, each binding to the respective partner of the heterodimer and modifying what would have otherwise been the outcome of single hormone binding (Calleja et al. 2006).
While the integration of steroid and juvenile hormone signaling pathways at the physiological level has been established for decades (Willis 1974; Riddiford 1996), the underlying molecular basis for the signaling integration is only just now beginning to be discerned at the level of participating hormone receptors (Konopova and Jindra 2007; Bitra and Palli 2009; Li et al. 2011; Zhang et al. 2011). Even less is understood about how information contained in DNA promoter sequences integrates with multiple hormone signaling during insect development (Nakagawa and Henrich 2009).
A major limitation for development of a molecular paradigm for integration of larval ecdysteroid and JH signaling pathways has been that very few core promoters have been isolated, that are responsive to both ecdysteroid signaling and JH signaling through identified nuclear receptor binding sites (Berger et al. 1992; 2005; Kethidi et al. 2004). We have sought to resolve this gap by use of several related genes encoding hexamerin proteins from a lepidopteran (Trichoplusia ni) that become very highly expressed at the onset of the metamorphic larval instar (Jones et al. 1987, 1996). These genes appear to be induced in vivo by 20-hydroxyecdysone (Manohar et al. 2010), and the expression of several can be suppressed by exogenous juvenile hormone (Jones et al. 1993). We have isolated the core promoters of these genes, as candidates for promoters that would be competent to recruit a transcription apparatus that is responsive to JH signaling. These core promoters, and defined hormone response elements, provide a powerful and straightforward system for elucidating a molecular paradigm for integration of ecdysteroid and JH signaling.
2. Materials and Methods
2.1 Cell culture, transfections and hormone treatment
Spodoptera frugiperda (Noctuidae) cell line, Sf9, was maintained and transfected, in triplicate in each replication, as described previously (Jones et al., 1998). Test promoter constructs were prepared in pGL2 vector (firefly luciferase reporter). Normalized activity was calculated as the amount of firefly luciferase activity divided by the activity reported by a cotransfected plasmid constitutively expressing Renilla luciferase, as per manufacturer protocol (Promega). Between 24 and 48 h after the transfection, the cells were treated with 1 μM 20-OH ecdysone (“20E), or with 100 μM juvenile hormone III (“JH III”, a naturally occurring form of JH in Noctuidae; Steinera et al., 1999), or with both hormones (both obtained from Sigma-Aldrich), or with just EtOH carrier only (final EtOH concentration 1–2%). At the indicated hours after hormone treatment, the cells were harvested and the reported activity measured.
2.2 Reporter Promoter Constructs
The sequences and characteristics of the promoters of the AJHSP1, BJHSP1 and arylphorin (ARYL) genes of the lepidopteran Trichoplusia ni (Noctuidae) were previously described (Jones et al., 1996). Promoter regions (containing the TATA box, transcription start site (= position +1), and 5′ untranslated region up to the first base of the translation start codon, of AJHSP1 (−66 to + 10), BJHSP1 (−109 to +24) and arylphorin (−67 to +28) were cloned into the Kpn I/Bgl II sites of pGL2 vector. For each of these “core” promoters, an Nhe I site was then placed immediately 5′ to the Kpn I site, and direct repeat sequences with a single intervening base (“DR1”) were cloned into the Nhe I site by the following method (DR1 was used because it has been reported as a transcriptionally active EcR/USP heterodimer binding site, e.g., Vogtlii et al., 1998; Fang et al., 2005). Complementary oligonucleotides encoding the DR1 motif were synthesized, with each oligonucleotide possessing at its 5′ end a four base overhang of an Nhe I restriction site (CTAG). Upon annealing, the double stranded oligonucleotides would then have a CTAG overhang at each 5′ end. The annealed oligonucleotides were then ligated into concatamers, fractionated by native PAGE and the gel fractions corresponding to concatamer forms recovered and ligated into the Nhe I site. The specific sequence for the DR1 oligonucleotide was (upper strand) : 5′-CAAGGTCAAAGGTCAG-3′ (repeat motif underlined; CTAG on 5′ ends not shown).
Point mutations to the BJHSP1 promoter made from the TATA box to the translation start codon were done with the Quick Change Site Directed Mutagenesis kit (Stratagene). Truncation of AJSHP1 and BJHSP1 to 9 bases 5′ to the start of the TATA box and extending to the first base of the translation start codon, was accomplished by chemical synthesis of the complimentary DNA strands, with the appropriate overhangs for cloning into NheI, HindIII cut pGL2, and ligating the annealed oligos into that vector in those sites. Next, complimentary oligos containing two tandem copies of the DR1 motif were prepared with flanking, overhanging sequences for a 5′ KpnI site and 3′ NheI site (Upper strand: CCTAGAGGTCAAAGGTCACCTAGGTCAAAGGTCAG), for cloning into KpnI/NheI cut pGL2 containing the truncated AJHSP1 or BJHSP1 core promoters. The same procedure of core promoter preparation with a 5′-placed tandem DR1 was used to prepare constructs in which the AJHSP1 was mutated to possess the BJHSP1 initiator motif (ACAGT), and the BJHSP1 was made to possess the AJHSP1 initiator motif (ACTGG). All constructs were confirmed by sequencing.
3. Results
3.1 Potential Regulatory Motifs in Core Hexamerin Promoters
Previous studies have established the transcription start point and corresponding TATA boxes of the three JH-sensitive hexamerin promoters in T. ni (Jones et al. 1996). The BJHSP1, BJHSP2 and ARYL genes possess exactly the same motif that contains the transcription start point: ACAGT (the italicized first A is the start point). The AJHSP1 gene instead has the sequence ACTGG. Sequence alignment of the core promoters shows a number of motifs that are conserved in by BJHSP1, BJHSP2 and ARYL that are not conserved by AJHSP1 (Fig. 1). Our further experiments here focused on how the differences in core promoter sequence between BJHSP1 and AJHSP1 might contribute to modulation of core promoter transcriptional activity by the hormones 20E and JH III.
Figure 1.
Alignment of the regions of the four hexamerin genes of Trichoplusia ni, from the TATA box (bold, underlined) to the first base (A) of the ATG translation start codon. The transcription start point (italicized A) and the four subsequent bases are underlined. A region of similarity of three genes immediately 5′ to the TATA box is in italics, whereas a region of similarly on the 3′ side is shown in bold only. A motif common to the three genes that is between the transcription and translation start points is shown in underline only. Shown above (AJHSP1) and below (BJHSP1) the specific wild type bases are the mutations to those bases that were prepared in certain experiments.
3.2 20E-induction of the DR1-enhanced core promoter
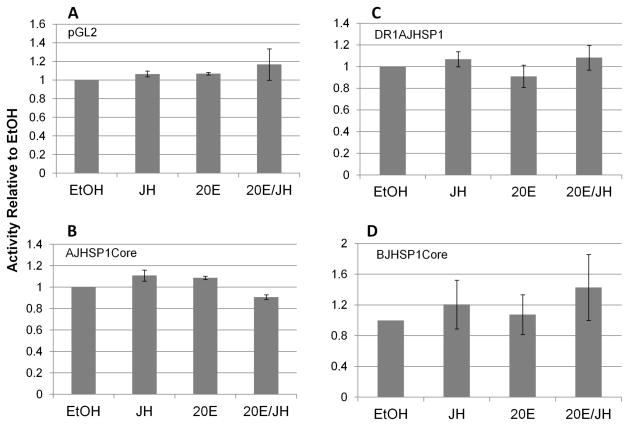
The pGL2-firefly luciferase reporter vector into which the core promoters were cloned does not itself transcriptionally respond to treatment of the transfected cells with 20E, JH III, or 20E/JH III (Fig. 2A). Similarly, the core promoters of AJHSP1 and BJHSP1 do not themselves transcriptionally respond to treatment of the SF9 cells with any of these hormone regimens (Fig. 2B, Fig. 2D). Hence, particular motifs in the core promoters may be necessary (see below), but are not sufficient, for transcriptional response to any of these hormone regimens.
Figure 2.
Transcriptional activities of the pGL2 basic vector (“pGL2”, luciferase reporter), AJHSP1 core promoter (“AJHSP1core”), BJHSP1 core promoter (“BJHSP1core”) and AJHSP1core with DR1 enhancer (two tandem copies) placed immediately 5′ to the AJHSP1core (“DR1AJHSP1”). Shown are means and standard error of transcriptional activities, 24–48 h after hormone treatment, relative to the ethanol carrier control (“EtOH, which is shown as value of 1.0). Experimental treatments were 1 μM 20-hydroxy ecdysone (“20E”), 100 μM juvenile hormone III (“JH III”), or a combined treatment of the two hormones at these concentrations (“20E/JH”). For each of the four constructs in panels A–D, none of the three hormone regimens exerted a significant effect on transcription different than the EtOH control (ANOVA, p > 0.05, for each panel).
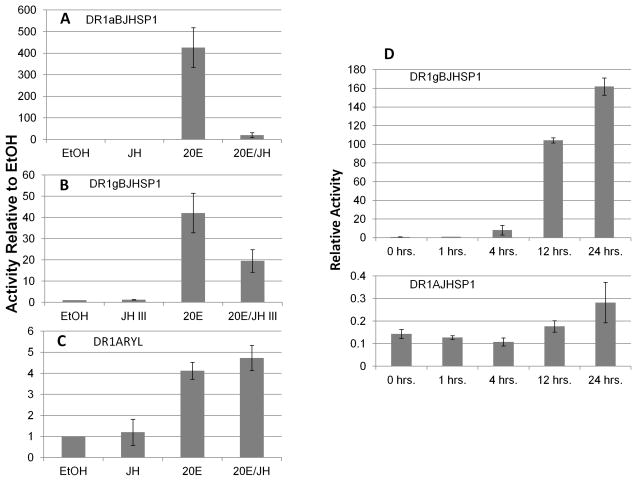
In view of previous reports that these genes respond in vivo to 20E (Manohar et al., 2010), we placed the DR1 enhancer (an established EcR/USP heterodimer binding site) 5′ to each promoter and assessed hormone responsiveness. The DR1AJHSP1 construct (with two tandem DR1 copies) remained unresponsive to any hormone regimen (Fig. 2C), verified over a 24 h time course (Fig. 3D). However, the DR1 enhancer imparted a marked transcriptional activation of the BJHSP1 core promoter by 20E, with just one copy of the enhancer (ca. 40 fold, Fig. 3B). Increasing the number of the DR1 enhancer to four copies yielded an even greater increase in transcriptional induction (ca. 400 fold, Fig. 3B). In addition, the induction was evident within 2 h of treatment, consistent with a direct induction of the promoter by the EcR/USP heterodimer through the DR1 binding motif (not shown).
Figure 3.
Panels A and B) Transcriptional activities of the BJHSP1 core promoter with DR1 enhancer (four tandem copies, “DR1g”, 24 h hormone treatment, or a single copy, “DR1a,” placed immediately 5′ to the BJHSP1core. Panel C) Transcriptional activities of the ARYL core promoter with a DR1 enhancer (two tandem copies) placed immediately 5′ to the core promoter (24 h hormone treatment). Shown are means and standard error of reported transcriptional activities relative to ethanol (“EtOH”) carrier control treatment, which is shown as value of 1.0. Experimental treatments are 1 μM 20-hydroxy ecdysone (“20E”), 100 μM juvenile hormone III (“JH III”), or a combined treatment of the two hormones at these concentrations (“20E/JH”). For either DR1-enhanced BJHSP1 promoter construct, ANOVA was significant (p < 0.05); 20E significantly induced transcriptional activity (p< 0.01 for both DR1aBJHSP1 and DR1gBJHSP1); JH III alone had no significant effect (p>0.05), but the cotreatment of JH III with 20E significantly suppressed the reported activity below that of 20E alone (p<0.01 DR1aBJHSP1; p<0.02 DR1gBJHSP1). The DR1ARYL construct also showed a significant treatment effect (ANOVA, p < 0.05). However, in contrast to the DR1-enhanced BJHSP1 constructs, the 20E-induced activity of the DR1-enhanced ARYL core promoter (p < 0.01) was not significantly affected by cotreatment with JH III (p> 0.05). Panel D) Over a 24 h time course, the DR1BJHSP1 construct exhibited more than a 100 fold increase in transcriptional activity in response to 1 μM 20E, in comparison to the lack of response to 20E of the DR1AJHSP1 construct (note the much different scales for the two Y-axes in panel D).
3.3 Contribution of Core Promoter Motifs
The above results showed that the presence of the DR1 motif enabled the BJHSP1 core promoter segment to be activated by 20E, however, the AJHSP1 core promoter fragment remained unresponsive even in the presence of the same DR1 enhancer motif. Hence, the difference in 20E response appeared to reside to information in the core promoter segment, rather than in DR1 enhancer motif. Inspection of the sequence of the four related hexamerin genes shows the region from the TATA box to the transcription start point of the BJHSP1, BJHSP2 and ARYL genes share several motifs, that are not conserved with AJHSP1 (Fig. 1). This conservation of sequences prompted our mutational testing of the contribution of these motifs to the observed 20E-modulation (through a DR1) of the BJHSP1 promoter.
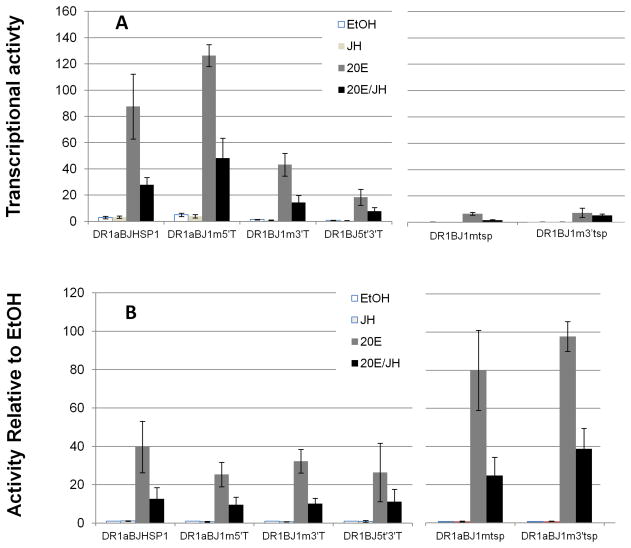
Immediately 3′ to the TATA box (TATAAA), the BJHSP1, BJHSP2 and ARYL core promoters conserve the sequence AAGG(G/C)G, while the AJSHP1 core promoter is very dissimilar. Immediately 5′ to the TATA box, the BJHSP1, BJHSP2 and ARYL core promoters conserve an “AG” motif, while again the AJHSP1 gene is dissimilar. Transversion mutation of these two motifs did not each significantly change the level of 20E-induction of transcriptional activity (p > 0.05, Fig. 4A). However, doubly-mutating both of these 3′-to TATA and 5′-to-TATA motifs did reduce the level of 20E-induced transcriptional activity (p < 0.05, Fig. 4).
Figure 4.
DR1g-enhanced transcriptional activities of the wild type and several mutants of the BJHSP1 core promoter after 24 h of hormone treatment. Panel A) Normalized transcriptional activity of the indicated mutations (see Fig. 1). Panel B) Transcriptional activity of Panel A expressed instead as relative to EtOH control (= value of 1). Shown are means and standard error of the experimental treatments of 1 μM 20-hydroxy ecdysone (“20E”), 100 μM juvenile hormone III (“JH III”), or a combined treatment of the two hormones at these concentrations (“20E/JH”). Each of the single motif mutations retained a significant effect of cotreatment of JH III with 20E compared to the transcription induction with 20E only (Panel B, p < 0.01 each).
The other area of strong conservation in the core promoter sequences is around the transcription start point. The sequence at this location for the BJHSP1, BJHSP2 and ARYL genes is conserved as ACAGT, with the first base being the actual transcription start point (Jones et al. 1996). In contrast, the sequence at the AJHSP1 transcription start point is different at two bases (underlined): ACTGGC. We tested the functional significance of this difference by mutating this transcription start motif of the BJHSP1 promoter to be that of the AJHSP1 promoter. This mutation very severely reduced the ability of 20E to induce transcriptional activity of the DR1-enhanced BJHSP1 (p < 0.05, Fig. 4). Hence, the transcription start site sequence itself of the AJSHP1 promoter cannot enable the marked 20E-inducibility of a core promoter (of BJHSP1) that otherwise is strongly 20E sensitive.
Finally, we noted that the 5′ untranslated region, from the transcription start site to the ATG translation initiation codon, is longer in the BJHSP1, BJHSP2 and ARYL genes. That region also contains a GTGG motif in common for these three genes, that is not present in the region between the transcription start point and the ATG codon of the AJHSP1 gene. When that GTGG was mutationally altered for the DR1BJHSP1 construct, it also severely reduced the 20E-inducibility (p < 0.05, Fig. 4A).
3.4 JH III Suppression of 20E-Activation
The constituitive activities of the core promoters of AJHSP1 or BJHSP1 were not affected by JH III (p > 0.05), nor did the presence of the DR1 motif confer positive or negative sensitivity of either to JH III alone (p > 0.05, Fig. 2, 3). However, the 20E induction of DR1-enhanced BJHSP1 promoter construct was sensitive to being suppressed by the cotreatment with JH III (p < 0.05, Fig. 3A, B). The suppression of the 20E activation of the BJHSP1 was greater in the presence of four DR1 enhancers (Fig. 3A) than with a single DR1 enhancer (p < 0.05, Fig. 3B).
The ARYL promoter that is similar in sequence to the BJHSP1 from the TATA box to the initiator (Fig. 1), is made 20E-inducible by the presence of a DR1, but that induced activity is not suppressed by cotreatment with JH III (Fig. 3C). Hence, the presence of the DR1 is insufficient in and of itself to confer vulnerability for its 20E-induced activity to be suppressed by cotreatment with JH III (a similar hormone response pattern was seen when an IR1 motif was used instead of an IR1 motif, unp. obs.). Further, as mentioned above, mutation of the motifs in BJHSP1 that are in common to the BJHSP1 and ARYL promoters at immediately 5′ and 3′ to the TATA box, and 3′ to the initiator (Fig. 1) did reduce the level of 20E activation of DR1BJHSP1 (Fig. 4A). However, those mutations did not eliminate the sensitivity of the remaining induced activity to being suppressed by cotreatment with JH III (Fig. 4B).
3.5 Location of Sequences for JH Modulation of 20E in DR1BJHSP1/−109
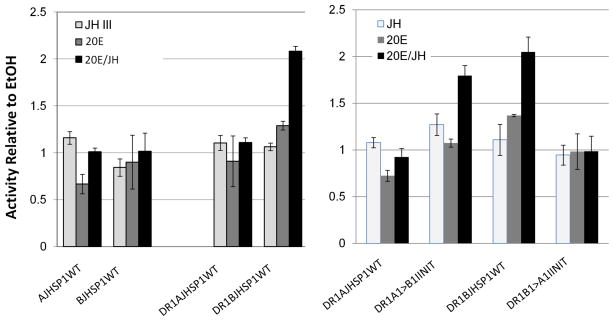
The above results indicated that the conserved sequences at or between the TATA box and initiator are not necessary for the JH III effect to suppress 20E-induced activity. Nor is the DR1 motif in and of itself sufficient for the JH III suppression of 20E-induced transcription. Hence, we reasoned that it could be that motifs in the BJHSP1 construct between the TATA box and the 5′ upstream boundary (at −109) contain regulatory information for JH suppression of 20E response. Thus, we made a BJHSP1 promoter construct truncated at 9 bases 5′ to the TATA (position −41), enhanced by a pair of tandem DR1 motifs placed immediately 5′ to the core promoter segment (Fig. 1). As a control, we created the corresponding construct for the AJHSP1 core promoter. The DR1-enhanced “−41” AJHSP1 core promoter retained its 20E and JH insensitivity (Fig. 5). The DR1 enhanced “−41” BJHSP1, while much reduced in overall transcriptional activity, and insensitive to JH alone, remarkably this time possessed a property for activity in presence of 20E to become significantly increased by JH III, not suppressed by JH III (p < 0.05, Fig. 5). Interestingly, when the initiator of the DR1-enhanced “−41” BJHSP1 core promoter was mutationally made to instead possess the initiator of the unresponsive AJHSP1, the DR1BJHSP1 became similarly transcriptionally inactive (Fig. 5, P > 0.05). Conversely, when the DR1-enhanced “−41” AJHSP1 core promoter was made to possess the common initiator of the ARYL/BJHSP1 promoters, it newly gained the property of the DR1”−41” BJHSP1, by exhibiting the greatest activity in the presence of both 20E and JH III (Fig. 5, p < 0.05). Hence, when the sequence between −109 and −41 of the BJHSP1 gene was not present, the initiator sequence of the DR1-enhanced BJHSP1 core promoter imparted information that the highest transcriptional induction would in the combined 20E/JH III treatment.
Figure 5.
Normalized transcriptional activities of the wild type (WT) AJHSP1and BJHSP1 core promoter, each trimmed to include only the region from −41 to the first base of the translation start codon, and enhanced (or not, as indicated) by two copies of a 5′-placed DR1 motif. Also as indicated, the initiator of AJHSP1 was replaced with the five bases of the BJHSP1 initiator (DR1A1>B1INIT), and the initiator for the BJHSP1 was changed to be that of AJHSP1 (DR1B1>A1INIT). Shown relative to the EtOH control are means and standard error of the experimental treatments of 48 h exposure to 1 μM 20-hydroxy ecdysone (“20E”), 100 μM juvenile hormone III (“JH III”), or a combined treatment of the two hormones at these concentrations (“20E/JH”). Experiment-wise ANOVA was significant at p < 0.01. The wild type AJHSP1 core promoter, enhanced by DR1 (DR1AJHSP1WT), was not significantly affected by hormone treatment (p > 0.05), while the DR1BJHSP1WT activity was significantly induced by cotreatment with 20E and JH III, as compared to treatment with only 20E (p<0.05). However, when the initiator motif of the two promoters were exchanged (INIT), the DR1BJHSP1 promoter lost hormone responsiveness (p>0.05), while the DR1AJHSP1 promoter showed significantly more activity by cotreatment with 20E and JH III, than the treatment with 20E alone (p < 0.05).
4. Discussion
There has been much research on the properties of various EcR/USP heterodimer binding sites. Such studies typically use very heterologous core promoters (e.g., bacterial thymidine kinase promoter enhanced with eukaryotic nuclear hormone receptor binding sites). Additional research has been done on physical parameters affecting 20E-mediated EcR association with USP, often using chimeric receptor constructs (part bacterial, part eukaryotic). While these approaches have much utility in addressing particular aspects of ligand-mediated nuclear hormone receptor action, there are questions that these techniques do not experimentally reach. For example, such approaches do not assess whether information is present in the sequence of the core promoter, that can control the nature of the promoter response to nuclear receptor-mediated hormone regulation. In addition, transcriptional cofactors that are necessary for a particular direction of ligand-regulated nuclear receptor action at a particular promoter may not be present in highly heterologous cell transfection systems. In the present study, we have tested specific questions on ligand-mediated transcriptional regulation using an established EcR/USP heterodimer binding site (DR1; Antoniewski et al., 1996; Vogtlii et al., 1998; Fang et al., 2005) to probe regulatory information in the promoter sequence of several related noctuid genes, using a noctuid cell line environment.
4.1 DR1 is necessary but not sufficient for 20E response
As a starting control, none of the three natural AJHSP1, BJHSP1 and ARYl promoter constructs responded to 20E treatment. The DR1 enhanced BJHSP1 ‘−109’ promoter construct was transcriptionally activated by 20E, much more so from four DR1 motifs than from a single DR1 motif (as would be anticipated, Antoniewski et al., 1996; Fang et al., 2005; Heneghan et al., 2006). The DR1 also enhanced transcription by the ARYL promoter. However, the presence of the same DR1 motif did not enable 20E activation of the core promoter of the related AJHSP1. Hence, an enhancer binding site for EcR/USP (DR1) was necessary, but was not sufficient, to cause even a related core promoter to be transcriptionally activated by 20E.
4.2 Sequence immediately 5′ to TATA necessary but not sufficient for DR1-transducing 20E response
When sequences immediately 5′ to the TATA box (to −109) were present, even just a single DR1 motif enabled a nearly 40-fold activation of transcription by 20E treatment. When the sequence between −109 and −41 was removed, then even two tandem DR1 motifs were not able to effectuate a significant increase in transcription over the EtOH control (p > 0.05). Further, when this 5′ sequence out to −109 was present, but without the presence of DR1, there was no significant response of the promoter to 20E treatment (p > 0.05). These results indicate that a sequence(s) within −109 to −41 is necessary for the DR1 motif to be able to transduce the promoter response to 20E. We infer that this necessary sequence is not itself a conventional EcR/USP heterodimer binding site because no such site is discerning within that region.
4.3 DR1 is not sufficient for JH suppression of 20E response
Although treatment with JH III alone did not significantly regulate any of the DR1-enhanced core promoters, cotreatment of JH III with 20E did cause suppression in the level of 20E-induced activity. The degree to which JH III suppressed the 20E activity was greater when more DR1 motifs were present (Fig. 3A, B). The phenomenon of synergistic increase in transcriptional activation with increasing number of nuclear hormone receptor binding sites has been reported for a number of single hormone systems, e.g., the homodimerizing class of vertebrate steroid receptors (Heneghan et al. 2006). In addition, synergistic increase in response to 20E in transfection reporter assay by the insect heterodimer EcR/USP has also been associated with an increase in number of synthetic heterodimer binding sites (Antonieweski et al., 1996). Similarly, a greater number of direct repeat binding sites yielded a greater transcriptional activation of the JH esterase core promoter by JH III alone (Xu et al., 2002). So far as we are aware, the present data are the first report, for the insect two-hormone system, that the level of JH suppression of 20E-induced transcription can be made a function of the number of DR1 motifs located at the regulated core promoter.
4.4 Regulatory Information Contained in Transcription Initiator Motif
When the region of the BJHSP1 core promoter from −41 to the translation start codon was enhanced by DR1, it responded to cotreatment with JH III by increasing transcription significantly above the level occurring in the presence of 20E alone (p < 0.05). When the DR1 was missing from this construct, no such effect of JH III and 20E treatment was observed. Hence, sequence between −41 to the translation start codon of the BJHSP1 core promoter was the target of the DR1 modulatory effect. We noted that the related AJHSP1 core promoter, from its −41 to translation start codon, did not respond to cotreatment with JH III and 20E even when the same DR1 was placed immediately 5′ to the AJSHP1 core promoter. We further noted that the five bases starting from the transcription start point (+1 to +5) were absolutely conserved in the BJHSP1, BJHSP2 and ARYL promoters (ACAGT), but that the motif for AJHSP1 was uniquely different (ACTGG). We hypothesized that the initiator motif for BJHSP1 might contain the information that enabled the DR1 BJHSP1 “−41” construct to exhibit hormone response that the AJHSP1 (“−41”) construct did not exhibit. To test this hypothesis, we mutationally swapped the BJHSP1 initiator motif to the AJHSP1, and vice versa. Intriguingly, the BJHSP1 construct then exhibited the nonresponsiveness previously exhibited by the AJHSP1 construct, while the AJHSP1 then exhibited the hormonal response previously shown by the BJHSP1 promoter (Fig. 5). These results suggest that the initiator motif contains regulatory information that communicates with the transcription-enhancing mechanism of the DR1. So far as we are aware, these results are the first to report juvenile hormone-response regulatory information to be contained in the transcription initiation motif.
4.5 Caution When Using Heterologous ‘Reporter Core Promoters’
It is a common approach in the study of hormone action to test a putative ligand’s interaction with a nuclear hormone receptor by a transfection assay, in which a nuclear hormone receptor binding site is placed in tandem with a minimal heterologous promoter. Such an approach has even commonly extended to testing hypotheses on eukaryotic nuclear hormone receptor action using a bacterial minimal core promoter as the assay reporter (e.g., Solomin et al., 1998; Raucy ad Lasker, 2010). It is sometimes asserted that if a putative nuclear receptor ligand fails to activate transcription of the minimal heterologous promoter, it can be inferred as disproving that the putative ligand naturally interacts in vivo with that nuclear hormone receptor. Our studies here and reported previously have shown that the same DR1 enhancer, in the same cell line (Sf9), either (a) increases minimal core promoter activity in response to JH III alone (JH esterase, Xu et al., 2002), or (b) increases response to 20E but suppresses 20E-activation when cotreated with JH III, (c) has no response to JH III alone but increases minimal core promoter activity in response to cotreatment with both JH III and 20E (−41″ “BJHSP1), or (d) increases response to 20E alone but does not respond to JH III alone or together with 20E (ARYL), or (d) has no response to either JH III alone, or JH III in cotreatment with 20E (AJHSP1). Hence, our results provide a caution to the use of heterologous minimal core promoters as the ‘litmus test’ for action of a potential ligand through a nuclear receptor that binds to the 5′-placed enhancer in such plasmid constructs. A lack of transcriptional activation by hormone treatment may simply be the result of an inappropriate core promoter, rather than a lack of cognancy between the test ligand and target nuclear receptor.
5. Conclusions
Our use here of related core promoters has enabled discernment of specific features of DR1, core promoter, and immediately proximal sequences in controlling the nature of the transcriptional response to single and multiple hormones. The molecular mechanism by which JH and 20E signaling become integrated into modulation of a transcriptional response is still an active area of research, though precise molecular models have not yet been within reach (Konopova and Jindra, 2007; Bitra and Palli, 2009; Abdou et al. 2011; Li et al., 2011; Zhang et al., 2011). Whether the JH III applied in the present study is acting by directly binding to USP (Jones et al., 2006), or by binding to the bHLH/PAS factor “MET” (Miura et al. 2005) that in turn is in complex with the EcR/USP heterodimer (Bitra and Palli, 2009), or is binding directly to both USP and MET, remains to be ascertained. While the system here is one performance in a cell line of a model enhancer synthetically placed with the core promoters, this system offers a characterized nuclear hormone binding site, with differential modulatory effects on defined and closely related promoter sequences. This ‘tight’ model system offers much potential in furthering our understanding of the specific DNA sequences, and the factors binding to them, that enable these two important hormones act to regulate particular genes during insect larval development.
Acknowledgments
This research was supported in part by grants from the National Science Foundation (1052142) and the National Institutes of Health (GM075248-04).
Abbreviations
- 20E
20-hydroxy ecdysone
- JH III
juvenile hormone III
- AJHSP1
acidic juvenile hormone suppressible protein 1
- BJHSP1
basic juvenile hormone suppressible protein 1
- BJHSHP2
basic juvenile hormone suppressible protein 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdou MA, He Q, Wen D, Zyaan O, Wang J, Xu J, Baumann AA, Joseph J, Wilson TG, Li S, Wang J. Drosophila Met and Gce are partially redundant in transducing juvenile hormone action. Insect Biochem Mol Biol. 2011 Sep 29; doi: 10.1016/j.ibmb.2011.09.003. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Antoniewski C, Mugat B, Delbac F, Lepesant JA. Direct repeats bind the EcR/USP receptor and mediate ecdysteroid responses in Drosophila melanogaster. Mol Cell Biol. 1996;16:2977–86. doi: 10.1128/mcb.16.6.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger EM, Goudie K, Klieger L, Berger M, DeCato R. The juvenile hormone analogue, methoprene, inhibits ecdysterone induction of small heat shock protein gene expression. Dev Biol. 1992;151:410–8. doi: 10.1016/0012-1606(92)90181-f. [DOI] [PubMed] [Google Scholar]
- Berger EM, Dubrovsky EB. Juvenile hormone molecular actions and interactions during development of Drosophila melanogaster. Vitam Horm. 2005;73:175–215. doi: 10.1016/S0083-6729(05)73006-5. [DOI] [PubMed] [Google Scholar]
- Bitra K, Palli SR. Interaction of proteins involved in ecdysone and juvenile hormone signal transduction. Arch Insect Biochem Physiol. 2009;70:90–105. doi: 10.1002/arch.20281. [DOI] [PubMed] [Google Scholar]
- Calléja C, Messaddeq N, Chapellier B, Yang H, Krezel W, Li M, Metzger D, Mascrez B, Ohta K, Kagechika H, Endo Y, Mark M, Ghyselinck NB, Chambon P. Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev. 2006;20:1525–38. doi: 10.1101/gad.368706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke B, Hegstrom CD, Villeneuve LS, Breedlove SM. Sexual differentiation of the vertebrate brain: principles and mechanisms. Front Neuroendocrinol. 1998;19:323–62. doi: 10.1006/frne.1998.0171. [DOI] [PubMed] [Google Scholar]
- D’Avino PP, Crispi S, Cherbas L, Cherbas P, Furia M. The moulting hormone ecdysone is able to recognize target elements composed of direct repeats. Mol Cell Endocrinol. 1995;113:1–9. doi: 10.1016/0303-7207(95)03584-t. [DOI] [PubMed] [Google Scholar]
- Fang F, Xu Y, Jones D, Jones G. Interactions of ultraspiracle with ecdysone receptor in the transduction of ecdysone- and juvenile hormone-signaling. FEBS J. 2005;272:1577–89. doi: 10.1111/j.1742-4658.2005.04578.x. [DOI] [PubMed] [Google Scholar]
- Grazzini E, Guillon G, Mouillac B, Zingg HH. Inhibition of oxytocin receptor function by direct binding of progesterone. Nature. 1998;392:509–12. doi: 10.1038/33176. [DOI] [PubMed] [Google Scholar]
- Hayes TB. Sex determination and primary sex differentiation in amphibians: genetic and developmental mechanisms. J Exp Zool. 1998;281:373–99. [PubMed] [Google Scholar]
- Heneghan AF, Connaghan-Jones KD, Miura MT, Bain DL. Cooperative DNA binding by the B-isoform of human progesterone receptor: thermodynamic analysis reveals strongly favorable and unfavorable contributions to assembly. Biochemistry. 2006;45:3285–96. doi: 10.1021/bi052046g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich VC. The ecdysteroid receptor. In: Gilbert LJ, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 3. Oxford: Elsevier/Pergamon; 2005. pp. 243–282. [Google Scholar]
- Hoopes BC, LeBlanc JF, Hawley DK. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J Mol Biol. 1998;277:1015–3. doi: 10.1006/jmbi.1998.1651. [DOI] [PubMed] [Google Scholar]
- Hunter J, Kassam A, Winrow CJ, Rachubinski RA, Capone JP. Crosstalk between the thyroid hormone and peroxisome proliferator-activated receptors in regulating peroxisome proliferator-responsive genes. Mol Cell Endocrinol. 1996;116:213–21. doi: 10.1016/0303-7207(95)03717-9. [DOI] [PubMed] [Google Scholar]
- Jones G, Manczak M, Schelling D, Turner H, Jones D. Transcription of the juvenile hormone esterase gene under the control of both an initiator and AT-rich motif. Biochem J. 1998;335:79–84. doi: 10.1042/bj3350079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Manczak M, Horn M. Hormonal regulation and properties of a new group of basic hemolymph proteins expressed during insect metamorphosis. J Biol Chem. 1993;268:1284–91. [PubMed] [Google Scholar]
- Jones G, O’Mahony P, Chang S, Schachtschabel U. Identification of regulatory sequences of juvenile hormone-sensitive and -insensitive serum protein-encoding genes. Gene. 1996;173:209–14. doi: 10.1016/0378-1119(96)00100-x. [DOI] [PubMed] [Google Scholar]
- Jones G, Jones D, Teal P, Sapa A, Wozniak M. The retinoid-X receptor ortholog, ultraspiracle, binds with nanomolar affinity to an endogenous morphogenetic ligand. FEBS J. 2006;273:4983–96. doi: 10.1111/j.1742-4658.2006.05498.x. [DOI] [PubMed] [Google Scholar]
- Jones D, Jones G, Teal P, Hammac C, Messmer L, Osborne K, Belgacem YH, Martin JR. Suppressed production of methyl farnesoid hormones yields developmental defects and lethality in Drosophila larvae. Gen Comp Endocrinol. 2010;165:244–54. doi: 10.1016/j.ygcen.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Jones D, Li X, Tang L, Ye L, Teal P, Riddiford L, Sandifer C, Borovsky D, Martin JR. Activities of natural methyl farnesoids on pupariation and metamorphosis of Drosophila melanogaster. J Insect Physiol. 2010;56:1456–64. doi: 10.1016/j.jinsphys.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Jones G, Wozniak M, Jones D. Newly identified, basic hemolymph proteins from noctuid species. Biochem Biophys Res Commun. 1987;146:265–9. doi: 10.1016/0006-291x(87)90720-0. [DOI] [PubMed] [Google Scholar]
- Kakizaki S, Karami S, Negishi M. Retinoic acids repress constitutive active receptor-mediated induction by 1,4-bis[2-(3,5-Dichloropyridyloxy)]benzene of the Cyp2b10 gene in mouse primary hepatocytes. Drug Metab Dispos. 2002;30:208–11. doi: 10.1124/dmd.30.2.208. [DOI] [PubMed] [Google Scholar]
- Kethidi DR, Perera SC, Zheng S, Feng QL, Krell P, Retnakaran A, Palli SR. Identification and characterization of a juvenile hormone (JH) response region in the JH esterase gene from the spruce budworm, Choristoneura fumiferana. J Biol Chem. 2004;279:19634–42. doi: 10.1074/jbc.M311647200. [DOI] [PubMed] [Google Scholar]
- Kim J, Parvin JD, Shykind BM, Sharp PA. A negative cofactor containing Dr1/p19 modulates transcription with TFIIA in a promoter-specific fashion. J Biol Chem. 1996;271:18405–18415. doi: 10.1074/jbc.271.31.18405. [DOI] [PubMed] [Google Scholar]
- Konopova B, Jindra M. Juvenile hormone resistance gene Methoprene-tolerant controls entry into metamorphosis in the beetle Tribolium castaneum. Proc Natl Acad Sci USA. 2007;104:10488–93. doi: 10.1073/pnas.0703719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Robinson GE, Palli SR. Identification and characterization of a juvenile hormone response element and its binding proteins. J Biol Chem. 282:37605–17. doi: 10.1074/jbc.M704595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mead EA, Zhu J. Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc Natl Acad Sci USA. 2001;108:638–43. doi: 10.1073/pnas.1013914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sheng Z, Liu H, Wen D, He Q, Wang S, Shao W, Jiang RJ, An S, Sun Y, Bendena WG, Wang J, Gilbert LI, Wilson TG, Song Q, Li S. Juvenile hormone counteracts the bHLH-PAS transcription factors MET and GCE to prevent caspase-dependent programmed cell death in Drosophila. Development. 2009;136:2015–25. doi: 10.1242/dev.033712. [DOI] [PubMed] [Google Scholar]
- Maden M. Role and distribution of retinoic acid during CNS development. Int Rev Cytol. 2001;209:1–77. doi: 10.1016/s0074-7696(01)09010-6. Review. [DOI] [PubMed] [Google Scholar]
- Manohar D, Gullipalli D, Dutta-Gupta A. Ecdysteroid-mediated expression of hexamerin (arylphorin) in the rice moth, Corcyra cephalonica. J Insect Physiol. 2010;56:1224–31. doi: 10.1016/j.jinsphys.2010.03.026. [DOI] [PubMed] [Google Scholar]
- Meyer T, Carlstedt-Duke J, Starr DB. A weak TATA box is a prerequisite for glucocorticoid-dependent repression of the osteocalcin gene. J Biol Chem. 1997;272:30709–14. doi: 10.1074/jbc.272.49.30709. [DOI] [PubMed] [Google Scholar]
- Miura K, Oda M, Makita S, Chinzei Y. Characterization of the Drosophila Methoprene -tolerant gene product. Juvenile hormone binding and ligand-dependent gene regulation. FEBS J. 2005;272:1169–78. doi: 10.1111/j.1742-4658.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Henrich VC. Arthropod nuclear receptors and their role in molting. FEBS J 2009. 2009;276:6128–57. doi: 10.1111/j.1742-4658.2009.07347.x. [DOI] [PubMed] [Google Scholar]
- Raucy JL, Lasker JM. Current in vitro high throughput screening approaches to assess nuclear receptor activation. Curr Drug Metab. 2010;11:806–14. doi: 10.2174/138920010794328896. [DOI] [PubMed] [Google Scholar]
- Riddiford LM. Juvenile hormone: The status of its “status quo” action. Arch Insect Biochem Physiol. 1996;32:271–86. doi: 10.1002/(SICI)1520-6327(1996)32:3/4<271::AID-ARCH2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Riddiford LM, Truman JW, Mirth CK, Shen YC. A role for juvenile hormone in the prepupal development of Drosophila melanogaster. Development. 2010;137:1117–26. doi: 10.1242/dev.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala G. Progesterone signaling and mammary gland morphogenesis. J Mammary Gland Biol Neoplasia. 1999;4:89–104. doi: 10.1023/a:1018760721173. [DOI] [PubMed] [Google Scholar]
- Smith SM, Dickman ED, Power SC, Lancman J. Retinoids and their receptors in vertebrate embryogenesis. J Nutr. 1998;128:467S–470S. doi: 10.1093/jn/128.2.467S. [DOI] [PubMed] [Google Scholar]
- Solomin L, Johansson CB, Zetterström RH, Bissonnette RP, Heyman RA, Olson L, Lendahl U, Frisén J, Perlmann T. Retinoid-X receptor signalling in the developing spinal cord. Nature. 1998;395:398–402. doi: 10.1038/26515. [DOI] [PubMed] [Google Scholar]
- Steinera B, Pfister-Wilhelma R, Grossniklaus-Bürgina C, Remboldb H, Treiblmayrc K, Lanzrein B. Titres of juvenile hormone I, II and III in Spodoptera littoralis (Noctuidae) from the egg to the pupal moult and their modification by the egg–larval parasitoid Chelonus inanitus (Braconidae) J Insect Physiol. 1999;45:401–413. doi: 10.1016/s0022-1910(98)00139-5. [DOI] [PubMed] [Google Scholar]
- Truman JW, Riddiford LM. Endocrine insights into the evolution of metamorphosis in insects. Annu Rev Entomol. 2002;47:467–500. doi: 10.1146/annurev.ento.47.091201.145230. [DOI] [PubMed] [Google Scholar]
- Truman JW, Riddiford LM. The morphostatic actions of juvenile hormone. Insect Biochem Mol Biol. 2007;37:761–70. doi: 10.1016/j.ibmb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Vasudevan N, Koibuchi N, Chin WW, Pfaff DW. Differential crosstalk between estrogen receptor (ER)alpha and ERbeta and the thyroid hormone receptor isoforms results in flexible regulation of the consensus ERE. Mol Brain Res. 2001a;95:9–1. doi: 10.1016/s0169-328x(01)00165-6. [DOI] [PubMed] [Google Scholar]
- Vasudevan N, Zhu YS, Daniel S, Koibuchi N, Chin WW, Pfaff DJ. Crosstalk between oestrogen receptors and thyroid hormone receptor isoforms results in differential regulation of the preproenkephalin gene. Neuroendocrinology. 2001b;13:779–90. doi: 10.1046/j.1365-2826.2001.00693.x. [DOI] [PubMed] [Google Scholar]
- Vogtli M, Elke C, Imhof MO, Lezzi M. High level transactivation by the ecdysone receptor complex at the core recognition motif. Nucleic Acids Res. 1998;26:2407–14. doi: 10.1093/nar/26.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, LeBlanc GA. Interactions of methyl farnesoate and related compounds with a crustacean retinoid X receptor. Mol Cell Endocrinol. 2009;309:109–16. doi: 10.1016/j.mce.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Willis JH. Morphogenetic action of insect hormones. Annu Rev Entomol. 1974;19:97–115. doi: 10.1146/annurev.en.19.010174.000525. [DOI] [PubMed] [Google Scholar]
- Xu Y, Fang F, Chu Y, Jones D, Jones G. Activation of transcription through the ligand-binding pocket of the orphan nuclear receptor ultraspiracle. Eur J Biochem. 2002;269:6026–36. doi: 10.1046/j.1432-1033.2002.03293.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Sheng Z, Sui Y, Palli SR. Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J Biol Chem. 2011;286:8437–47. doi: 10.1074/jbc.M110.191684. [DOI] [PMC free article] [PubMed] [Google Scholar]