SUMMARY
MicroRNAs (miRNAs) are small non-coding RNA molecules that regulate gene expression. Among these, members of the let-7 miRNA family control many cell fate determination genes to influence pluripotency, differentiation, and transformation. Lin28 is a specific, post-transcriptional inhibitor of let-7 biogenesis. We report crystal structures of mouse Lin28 in complex with sequences from let-7d, let-7-f1, and let-7g precursors. The two folded domains of Lin28 recognize two distinct regions of the RNA and are sufficient for inhibition of let-7 in vivo. We also show by NMR spectroscopy that the linker connecting the two folded domains is flexible, accommodating Lin28 binding to diverse let-7 family members. Protein-RNA complex formation imposes specific conformations on both components that could affect downstream recognition by other processing factors. Our data provide a molecular explanation for Lin28 specificity and a model for how it regulates let-7.
INTRODUCTION
Since the discovery of the first human microRNAs (miRNAs) about a decade ago, examples of miRNA regulation have been found for virtually every cellular process (Kim et al., 2009; Krol et al., 2010). Precursors of miRNAs undergo a series of processing steps after transcription to generate an active product. In this canonical pathway, a newly transcribed primary miRNA (pri-miRNA) with at least one hairpin structure is cleaved within the nucleus by an RNAseIII enzyme, Drosha, that acts in complex with DGCR8. The resulting pre-miRNA is exported to the cytoplasm, where another RNAseIII, Dicer, removes the “terminal loop region”, or pre-element (preE), to yield the mature miRNA (Figure 1A). Mechanisms of transcriptional control have been analyzed for many miRNAs, but the recent identification of post-transcriptional regulators of miRNA biogenesis now provides a way to investigate the molecular details of miRNA maturation and regulation (Davis-Dusenbery and Hata, 2010; Siomi and Siomi, 2010).
Figure 1.
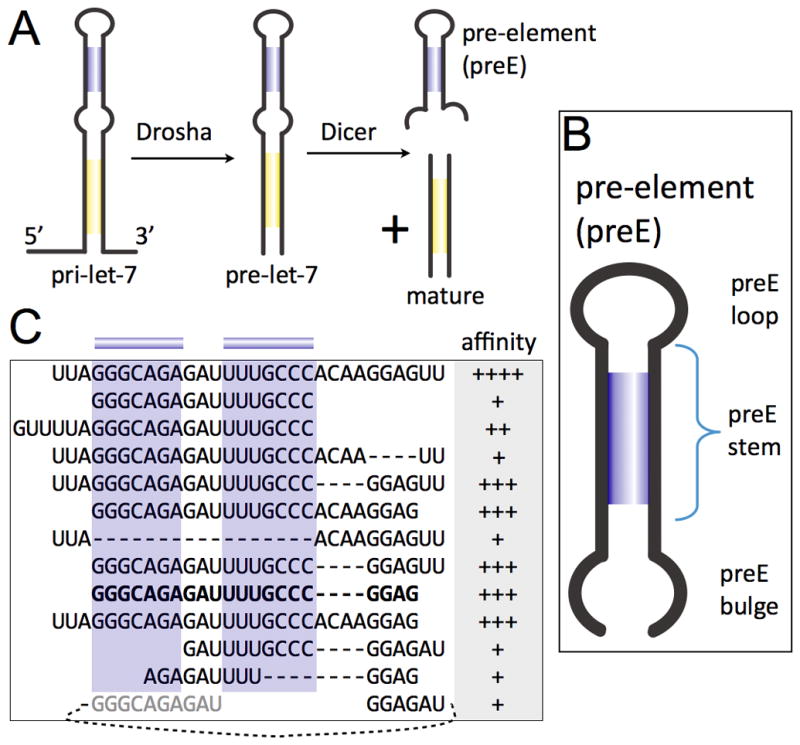
Mapping of Lin28 binding sites on pre-let-7. (A) Processing steps in canonical miRNA biogenesis. (B) Architecture of pre-elements (preEs). (C) Fragments of preE-let-7d tested on EMSA for association with Lin28. Affinity is indicated by Kd ranges: ++++, 0.2–1.5μM; +++, 1.5–3μM; ++, 3–15μM; +, >15μM. Predicted stem is highlighted in blue. Minimal fragment (preEM) identified is bolded. See also Figure S1.
The let-7 family of miRNAs regulates many factors that control cell fate decisions, including oncogenes (c-Myc, Ras, HMGA-2) and cell cycle factors (CyclinD1, D2) (Büssing et al., 2008; Viswanathan and Daley, 2010). Deregulation of let-7 influences tumorigenicity of breast cancer stem cells (Yu et al., 2007a). Moreover, IL-6 is a target of let-7, thereby bridging the inflammation and cell-transformation signaling pathways (Iliopoulos et al., 2009). There are several let-7 family members in mammals, with similar mature regions but divergent sequences in the preE removed by Dicer (Figure 1A). Despite low sequence identity, most preEs in let-7 are predicted to contain conserved structural elements (stem, bulge, and loop) that may be important for regulation of pre-miRNAs (Figure 1B).
Lin28, originally discovered as a heterochronic gene regulating developmental timing in worms (Moss et al., 1997), blocks let-7 biogenesis (Heo et al., 2008; Lehrbach et al., 2009; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). Its effects on gene expression are profound enough to make Lin28 one of the four factors sufficient to reprogram human somatic cells into induced pluripotent stem (iPS) cells (Yu et al., 2007b). Lin28 is activated in many human tumors (~15%) and appears to be associated with less differentiated cancers (Viswanathan et al., 2009). Studies with patient samples show correlation between over-expression or mutation of Lin28 with ovarian cancer (Peng et al., 2010; Permuth-Wey et al., 2011) and colon cancer (King et al., 2011). Variations in Lin28 have also been linked to developmental traits such as height and timing of puberty onset in humans and mice (Lettre et al., 2008; Lu et al., 2009; Ong et al., 2009; Perry et al., 2009; Sulem et al., 2009; Zhu et al., 2010).
Because it is one of few specific inhibitors of miRNA maturation to be discovered thus far, understanding Lin28 activity provides an avenue for investigating the mechanisms of miRNA biogenesis and regulation. Lin28 contains two well-known nucleic acid interaction domains—a cold shock domain (CSD) and two tandem Cys-Cys-His-Cys (CCHC)-type zinc-binding motifs (CCHCx2). Mammals have two paralogs, Lin28a and Lin28b, with different physiological expression patterns but similar behavior in vitro (Guo et al., 2006; Heo et al., 2008; Viswanathan et al., 2008; Yang and Moss, 2003). Lin28 binds precursor forms of let-7 miRNAs and can inhibit both pri-let-7 processing by Drosha (Newman et al., 2008; Viswanathan et al., 2008) and pre-let-7 processing by Dicer (Heo et al., 2008; Lehrbach et al., 2009; Rybak et al., 2008). Furthermore, Lin28 can recruit a terminal uridylyl transferase (TUTase) that adds uridine to the 3′ end of pre-miRNA to increase decay (Hagan et al., 2009; Heo et al., 2009; Lehrbach et al., 2009). Although parts of the preE segment are dispensable for pri-miRNA processing by Drosha (Han et al., 2006), point mutations in the preE can disrupt interactions with Lin28 (Heo et al., 2009; Lehrbach et al., 2009; Newman et al., 2008; Piskounova et al., 2008), thereby de-repressing Drosha-mediated processing (Newman et al., 2008). Sequence variability among preEs in let-7 (Figure S1A) has hindered interpretation of these results and extension of the conclusions to other let-7s, highlighting the need for an atomic-level view of divergent Lin28:let-7 complexes.
We present here high-resolution crystal structures of mouse Lin28a in complex with three preE constructs of let-7d, let-7f-1, and let-7g. These structures provide a direct view of a protein interacting with the terminal loop region of a miRNA. We identify sequence-specific interactions between Lin28 and let-7 precursors that give direct structural evidence for the role of preEs in miRNA regulation. The Lin28 CSD and the CCHC “zinc knuckles” make extensive contacts with the preE elements in two distinct regions. We also describe NMR studies and biochemical assays showing that the linker between the CSD and CCHCx2 regions introduces flexibility to accommodate variable preE sequences and lengths while preserving the joint contribution of the two interaction sites to overall affinity. We show that both the terminal and linker regions outside of the folded domains are not essential for blocking let-7 in vivo. Mutagenesis of preE fragments and full-length pre-miRNA molecules confirms our conclusions from the structure concerning specificity of Lin28, and allows us to predict how Lin28 recognizes other let-7s. Complex formation induces in both Lin28 and preE-let-7 a specific conformation that can affect recognition by downstream factors such as Drosha, Dicer and TUTase, and changes in the CCHCx2 region are particularly detrimental to Lin28 activity in vivo.
RESULTS
Two discrete binding sites in pre-let-7 for Lin28 binding
As a first step to understanding how pre-let-7 is recognized by Lin28, we tested a series of deletions in pre-let-7d for binding to the protein. Pre-let-7d has a relatively high affinity for Lin28 both in vivo and in vitro (Hagan et al., 2009; Heo et al., 2009; Newman et al., 2008), and secondary structure predictions indicate that it has the most stable preE-stem among mouse pre-let-7s, without interrupting bulges (Markham and Zuker, 2005). We focused our analysis on the preE, as mutagenesis studies had indicated its importance in direct association with Lin28 (Heo et al., 2009; Newman et al., 2008; Piskounova et al., 2008; Rybak et al., 2008). We observed that an isolated preE segment, containing none of the mature-region nucleotides, can bind Lin28 and that two distinct regions are critical for binding to Lin28, thereby defining a minimal preE-let-7d (preEM-let-7d) sufficient for high-affinity binding (Figure 1C, S1B-C). The first required region includes the preE-stem and the preE-loop; truncating the stem reduces binding. The other is the GGAG motif, which occurs at the 3′ end of the preE bulge. Although overall preE sequence conservation is low, even within the preE stem and loop, the GGAG tetranucleotide element is well conserved throughout the let-7 family (Figure S1A). Our mapping results suggest that the GGAG element provides an independent binding site, as deleting the neighboring nucleotides, thereby altering the distance to the CSD binding site, does not abolish Lin28 binding. The presence of two independent binding sites explains how diverse preE-let-7s containing variable linker sequences can all bind Lin28 with high specificity and affinity.
Domains of Lin28 tethered to each other are sufficient for inhibiting let-7 processing
Lin28 has two folded regions, CSD and CCHCx2, connected by a positively charged linker of ~15 amino acids, with extensions of ~30 residues at both the amino and carboxy termini. Mutagenesis studies have implicated both folded domains in repression of let-7 (Heo et al., 2009; Piskounova et al., 2008). The region C-terminal to the CCHCx2 domain also promotes translation of certain mRNA targets (Jin et al., 2011; Peng et al., 2011; Qiu et al., 2009). Using limited proteolysis and electrophoretic mobility shift assay (EMSA), we analyzed a series of truncation constructs of Lin28 to identify the essential region for interaction with preE-let-7. Both the N- and C-terminal regions can be removed without affecting affinity for RNA, but removal of either the CSD or the CCHCx2 abolishes high-affinity preE-let-7 binding (Figure S2A).
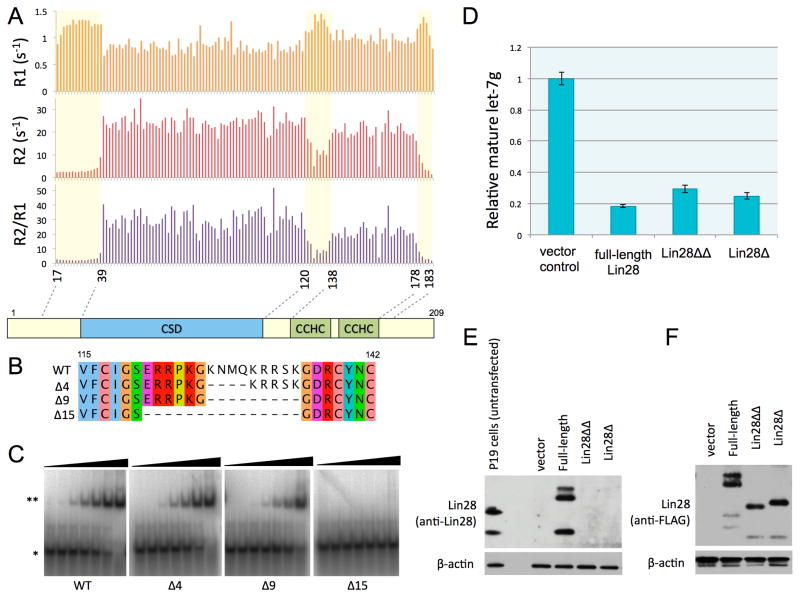
We used NMR spectroscopy to study the dynamics of Lin28:preEM-let-7d complexes in more detail (Figure 2A). We measured longitudinal (R1) and transverse (R2) relaxation rates to probe backbone dynamics. The R2/R1 ratio, which is a measure of correlation time, is an indicator of tumbling rate in solution. This ratio is similar for the folded domains but much lower for the terminal segments and the intervening linker, indicating more rapid motion in those regions. We conclude that the linker sequence lacks secondary structure, an inference consistent with absence of inter-residue backbone NOE crosspeaks in 15N-NOESY (Figure S2B). Comparing the Cα, Cβ, C′ chemical shifts to random coil chemical shifts also indicates that the linker region lacks secondary structure (Figure S2C). Deletion of up to 9 amino acids in the linker region supports binding to preE-let-7d or preE-let-7f-1, although further deletion prevents complex formation (Figure 2B-C, S2D). We conclude that a Lin28 fragment (31–187) with N- and C- terminal truncations and a 9-residue linker deletion (Lin28ΔΔ), is sufficient for binding to preE-let-7 in vitro.
Figure 2.
Linker between CSD and CCHCx2 is flexible. See also Figure S2. (A) Longitudinal (R1) and transverse (R2) relaxation rates and the ratio (R2/R1), plotted against the residue number. Relatively more dynamic regions are marked with a light yellow box. (B) Alignment of internal deletions in the linker, indicated with the number of amino acids deleted on left. (C) EMSAs with preE-let-7d as probe, mixed with increasing concentrations (0.005, 0.02, 0.08, 0.3, 1.2, 5, 20μM) of linker deletion constructs of Lin28(16–184): *, free probe; **, complex. (D) Quantitative RT-PCR results for in vivo levels of mature let-7g. Lin28Δ is truncated at both N and C termini. Lin28ΔΔ has both of the terminal extensions and the linker removed. The standard deviation is calculated from triplicate experiments; U6 RNA levels were used for normalization. (E) and (F) Western blots of Trizol bottom layer for transfections shown in (D). Anti-Lin28 antibodies do not recognize truncation constructs, so anti-FLAG was used in (F) to compare the relative expression levels of different Lin28 constructs.
To test whether Lin28ΔΔ can inhibit let-7 processing in cells, we compared the intracellular levels of processed mature let-7g when pri-let-7g is co-transfected with different Lin28 truncation constructs. Relative to vector alone, Lin28ΔΔ significantly reduces the level of mature let-7g, although not as much as the full-length Lin28 construct, probably due to slightly lower affinity (Figure 2C-D). Processing of pri-miR-122 or pri-miR-16 is not inhibited by either Lin28 construct (Figure S2E). Ectopically expressed Lin28 levels are similar to the endogenous levels observed in P19 cells and also among all Lin28 constructs (Figure 2E-F). The Lin28ΔΔ construct we have identified is therefore comparable to the full-length protein in its ability to inhibit processing in vivo as well as to bind let-7 precursors in vitro.
High resolution crystal structures of Lin28 with let-7 microRNAs
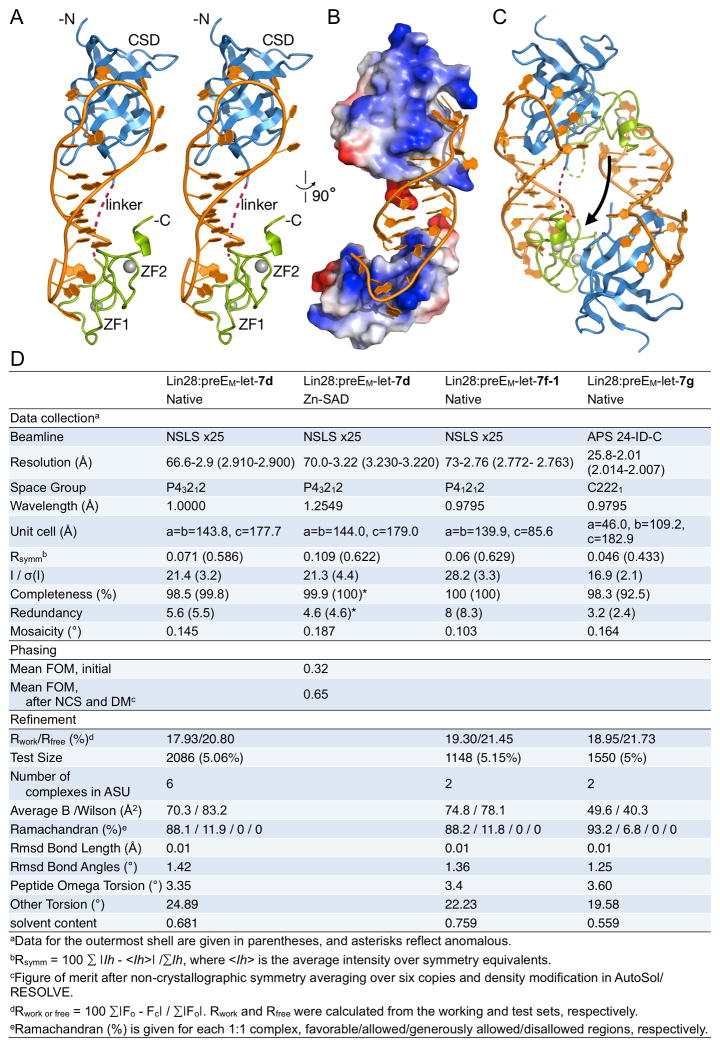
We determined crystal structures of Lin28ΔΔ in complex with preEM-let-7s derived from let-7d, let-7f-1, and let-7g, at resolutions 2.9Å, 2.8Å, and 2.0Å, respectively, from three different crystal forms (Figures 3, S3A). We used single-wavelength anomalous dispersion (SAD), with the bound zinc atoms as the anomalous scatterers, to determine the structure of the Lin28ΔΔ:preEM-let-7d complex; we determined the other structures by molecular replacement. While the overall architectures of the three complexes are similar (Lin28 Cα RMSD < 1.3Å), there are several local differences due to divergent RNA sequences (Figure S3B-C, and see CSD and CCHC sections below).
Figure 3.
Structure of Lin28:preEM-let-7d complex. Cartoon representations were colored by domain: blue, CSD; green, CCHCx2; grey, zinc; orange, RNA. (A) Stereo representation of the monomeric complex. Interdomain linker is represented by a purple dotted line. (B) Same complex in (A) represented with surface colored by electrostatic potential, and rotated. (C) Domain-swapped dimer. Arrow pointing from the domain-swapped to the biologically relevant CCHCx2 domain. Linker connecting swapped domains marked in green, dotted line. Linker connecting unswapped domains marked in purple, dotted line. See also Figure S3. (D) X-ray data collection and refinement statistics.
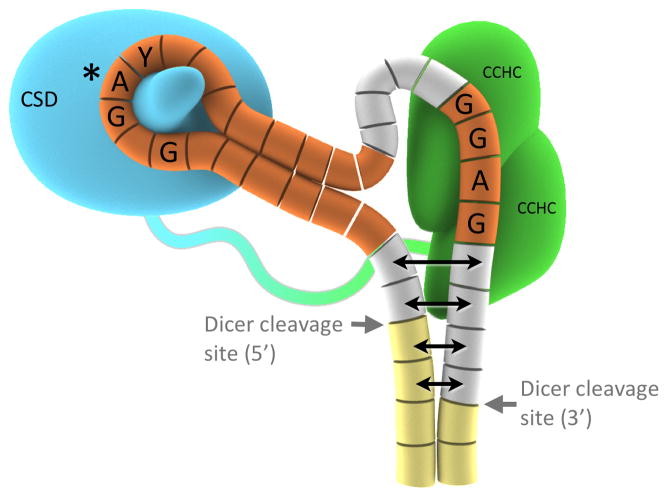
The structures reveal that the CSD and CCHCx2 domains of Lin28 interact with two distinct single-stranded regions of the RNA fragment (Figure 3A). The preE-loop encircles a protrusion of the CSD as a necktie would wrap around a collar, with the extensive contacts around the circle made possible by the presence of the preE-stem, which functions as the necktie’s knot. The CCHC zinc knuckles interact with the GGAG motif at the 3′ end, and several sequence-specific interactions shape the single stranded segment around the knuckles to introduce a distinctive kink in the RNA backbone. Positively charged surfaces on both domains interact with RNA throughout the complex (Figure 3B).
The shortened linker between CSD and CCHCx2 is the most variable region among the different complexes, as might have been expected from its flexibility (Figure S3D). In all three crystal forms, we observe a domain swap in which the Lin28 CSD interacts with the loop of one RNA molecule and the CCHCx2 interacts with the GGAG of a second RNA (Figure 3C). That is, each Lin28 monomer in the crystal interacts with distinct elements of two separate preEM-let-7d molecules. In sedimentation equilibrium ultracentrifugation experiments under more physiological conditions, we observe only monomeric complexes of Lin28:preE-let-7d, with or without internal deletions in the Lin28 linker (Figure S3E). An unswapped complex conformation can be modeled with a small rearrangement of the C-terminal extension of the CSD (residues 112–121) and a rotation of the 7-residue linker to span the 18–30Å distance between CSD and CCHCx2 on the same RNA (Figure 3A). Moreover, the longer, 16-residue linker in wildtype Lin28 would accommodate even longer RNA substrates, including pre-let-7d without internal deletions. The monomeric model is also consistent with our observation that high affinity RNA binding by Lin28 requires both Lin28-binding sites on the same molecule (Figure 1C). As all biochemical evidence points to a monomeric complex in solution, we restrict our description to a 1:1 complex, with CSD and CCHCx2 bound in cis to a single RNA.
Specific binding of preE-let-7 with CSD
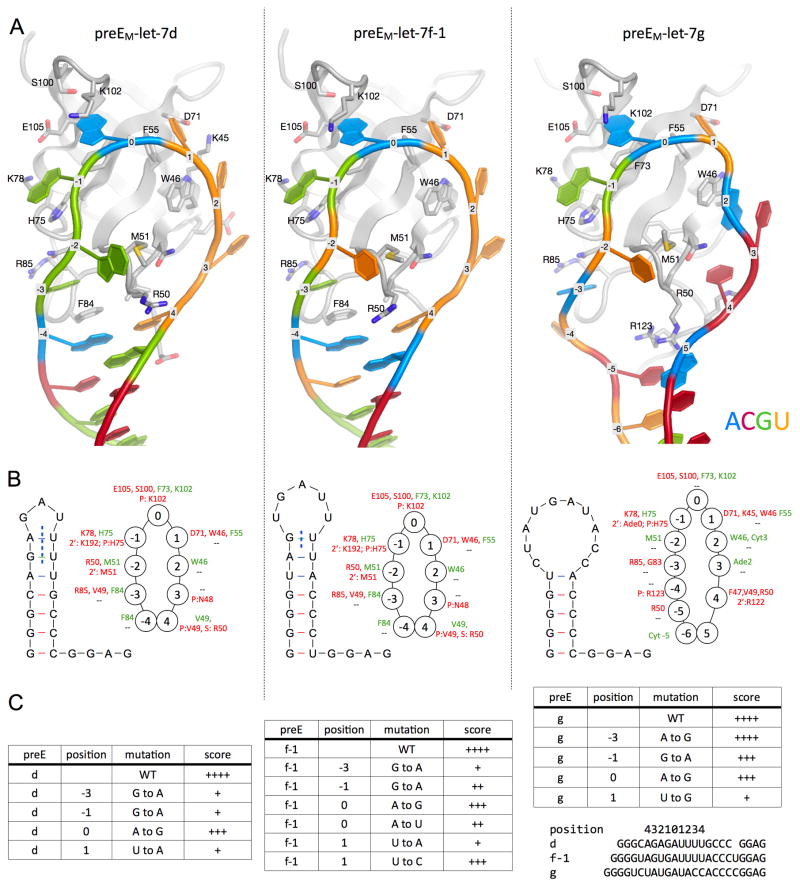
A detailed analysis of the contacts between the CSD and the preE-let-7 stem-loops suggests that specificity relies on both the sequence and the conformation of the RNA. Most of the direct contacts lie in a ≥9-nucleotide segment that includes the preE-loop (Figure 4, S4A-B). As the loop wraps around the CSD, the bases project and make a number of π-stacking interactions with aromatic side chains. Complementary to the Velcro-like effects of the hydrophobic interactions, hydrogen bonding and steric exclusion create nucleotide preferences and enhance specificity. From inspection of the binding pocket of each nucleotide, we can imagine an ideal RNA substrate for the CSD of Lin28. To simplify the discussion, we define the middle position of preE-let-7d that docks into the pocket lined by Phe73 and Lys102 as the “center”, or position 0. Purines are preferred at positions 0 and −1, near the tip of the loop so that the bulky bases can reach the protein. Position 1, on the other hand, is limited to a pyrimidine, as Lys45 and Asp71 impose steric hindrance. A deeper pocket at position −3 makes a purine more favorable, because a larger ring is necessary to stack over Phe84 (in d and f-1) and also to make favorable contacts with the Lin28 backbone (in all three). The hydrogen bonding networks around −3, −1 and 0 are specific for G, G and A, respectively.
Figure 4.
CSD:RNA interactions. See also Figure S4. (A) Close-up view of CSD (backbone as grey cartoon; key residues also shown with sticks and labels) interacting with the preEM-loops as labeled, shown in the same orientation. RNA is colored by base identity (Azure, Ade; Crimson, Cyt; Green, Gua; Umber, Ura), and marked by position number on backbone as defined in text. (B) Schematic drawing of predicted structures of preEM sequences used for crystallization. Some of the predicted base pairs are broken (blue vertical dotted line) in the complex structures. Rings of circles show the protein:RNA interactions at each nucleotide position, marked with interacting residues (green, hydrophobic or π-stacking; red, H-bond). Top line is for base contacts and bottom line is for sugar or phosphate interactions at each position. (C) Comparison of Lin28 affinity for various preEM-let-7 mutants, described by the parent let-7, position of the mutation, and the base identity. Accompanying gels are shown in Figure S4.
We evaluated the effect of several point mutations in the co-crystallized preEs at positions where specific interactions are observed in the structures (Figure 4C, S4C). Most of the mutant probes have lower affinity for Lin28 than wild-type. Although Gua is strongly preferred over Ade at positions −3 and −1, substitution of Ade0 with a Gua is not as deleterious. Ade replaces Gua−3 in the Lin28:let-7g complex, and as a result some favorable hydrogen bonds are absent in comparison to other structures. Due to the small size of the pocket, a pyrimidine is strongly preferred at position 1. Some of the previously reported mutations of preE-let-7g include a transversion (purine to pyrimidine) at position 0 (Newman et al., 2008) and changes in the preE-stem that disrupt base pairing (Piskounova et al., 2008). While our studies focused on mouse Lin28a, the observed effects of preEM point mutations on complex stability are equivalent for human Lin28a and Lin28b (Figure S4C).
Comparing the structure of Lin28 bound to the divergent preE-let-7g with those of the preE-let-7d and -7f-1 complexes illustrates how the CSD accommodates variability in substrate RNAs. The short preE-loops in let-7d and -f-1 require that base pairs be broken to fit around the CSD. In order to tighten the longer loop in preE-let-7g (Figure 4B), Arg50 moves in to mimic a base, pairing with Cyt−5 and stacking against Ade5. The other extra bases have π-stacking interactions: two with the side chains of Arg122 and Arg123 at the amino-terminal end of the inter-domain linker, and Ade2 and Cyt3 with each other. A closed RNA loop appears to be important to maintain full contact with the CSD, perhaps explaining the more extensive interactions here than in other CSD:RNA complex structures (Frazão et al., 2006; Max et al., 2006, 2007) (Figure S4D).
Interactions of zinc knuckles with preE-let-7
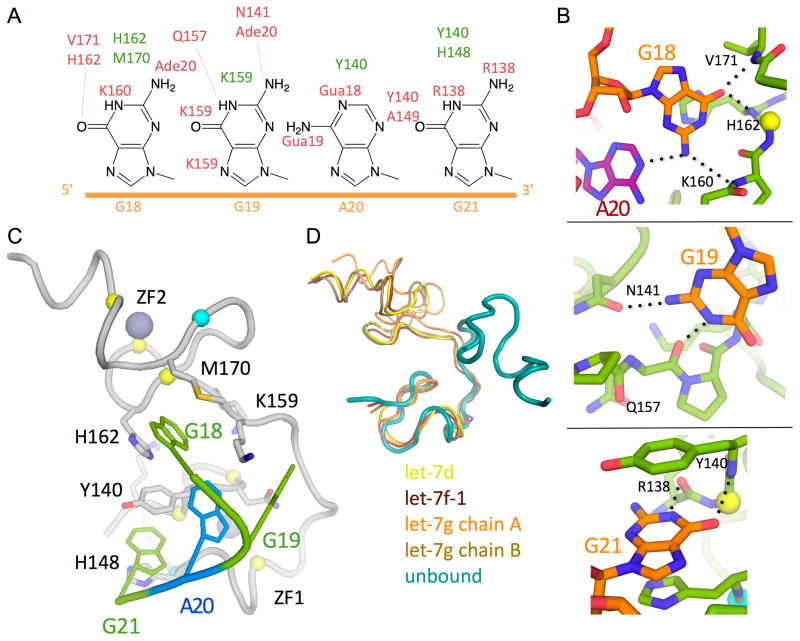
The CCHC knuckles maximize favorable interactions with a small number of nucleotides by making many contacts with the bases (Figure 5A-C). The intimate interaction between GGAG and CCHCx2 produces a distinctive kink in the RNA backbone. Most of the protein atoms participating in the extensive hydrogen bonding network lie in relatively rigid regions of the protein, such as adjacent to zinc-coordinating residues or in a proline-rich linker, thereby imposing a specific, rigid conformation on the 3′ end of the RNA (Figure 5C, S5A-B). Ring stacking and hydrophobic interactions with side chains of the CCHCx2 further stabilize the particular conformation by aligning the bases. One of the key residues is Y140, which establishes the kinked conformation by sandwiching between the last two bases (AG) and interacting with H162, which braces the first (G). Although the adenine base does not have as many polar contacts with Lin28, it packs closely against the first Gua and makes a hydrogen bond that assists in bending the RNA backbone. The resulting conformation of the ssRNA resembles that of the so-called “K-turn”, which often participates in specific protein-RNA interactions (Klein et al., 2001).
Figure 5.
CCHCx2:RNA interactions. See also Figure S5. (A) Schematic drawing of GGAG, and atoms making contact are marked with amino acid/nucleotide numbers (green, hydrophobic or π-stacking; red, H-bond). (B) Close-up view of base interactions. H-bonds are marked with dashed lines. (C) View of GGAG interactions with CCHCx2. Lin28 is represented with grey cartoon, and GGAG are colored by sequence (Green, Gua; Azure, Ade). Zinc (large grey spheres)-coordinating residues are represented with small spheres at Cα positions (yellow, Cys; cyan, His). (D) Comparison of the CCHCx2 region of Lin28 in different states by superimposition of the first CCHC motif (unbound, PDB code =2CQF).
The CCHCx2 regions from all our structures align well with each other, except for slight differences, due to crystal contacts, in one of the two non-crystallographic copies of preEM-let-7g (Figure S5C-D). When compared with the conformation seen in the solution structure of an isolated Lin28 zinc-knuckle fragment (PDB 2CQF), however, there is a large rearrangement of the inter-knuckle joint in Lin28 (Figure 5D). Therefore, association of CCHCx2 with GGAG imposes specific conformational constraints on both the RNA and the protein; this reciprocal effect may be functionally important for regulation.
Two NMR structures of CCHC motifs from HIV NCp1 have been determined previously, in which the knuckles bind a tetraloop of sequence GGAG or GGUG in two stem loops (SL2 and SL3) of the ψ-site (Amarasinghe et al., 2000; De Guzman et al., 1998). The conformation of the GGAG motif in complex with Lin28 is very different from its conformation in complex with HIV NCp1, indicating that the conformation we observe is specific to Lin28 (Figure S5E-F).
Lin28 interactions with full-length pre-let-7
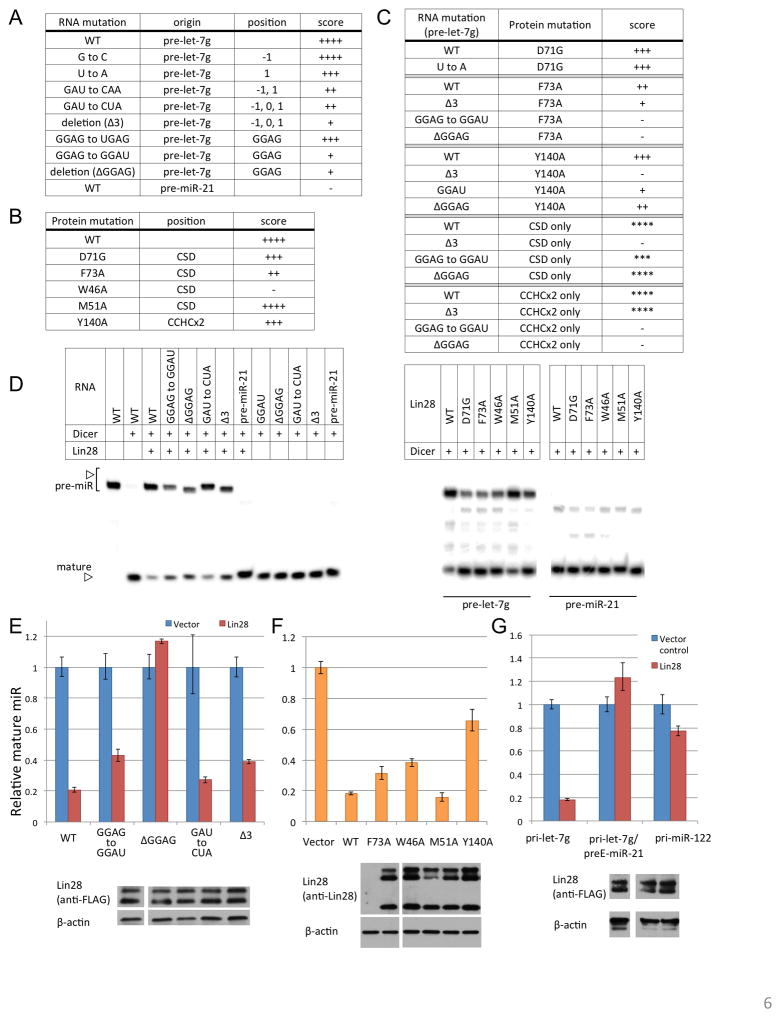
To test our conclusions from the model provided by the crystal structures, and to verify that the truncations and deletions we have made for crystallization do not affect specificity, we generated mutant forms of full-length Lin28 and pre-let-7g. Alteration of the key binding sites of CSD (near position 0) or CCHCx2 (GGAG) in pre-let-7g reduces affinity, consistent with the mutagenesis studies with preE fragments (Figure 6A, S6A). In addition, mutation of RNA-contacting residues in CSD and CCHCx2 also interferes with complex formation, especially when aromatic side chains are replaced with Ala (Figure 6B, S6B). We then conducted binding assays using combinations of protein and RNA mutants (Figure 6C, S6C). The D71 side chain, which is near nucleotide position 1, limits the size of the pocket and restricts it to pyrimidine rings. Presumably due to the additional free space provided by a glycine, a D71G mutant no longer discriminates against a purine at position 1 (Figure 6C, D71G block).
Figure 6. Structure validation with full-length molecules.
(A-C) Results of EMSA with various RNA mutants (full-length pre-miRNA background) combined with protein mutants (full-length Lin28, except for the isolated-domain experiments, designated "CSD or CCHCx2 only"). Affinity indicated by Kd ranges: ++++, 0.13–0.26μM; +++, 0.26–0.52μM; ++, 0.52–1μM; +,1–2μM; −, >2μM. For isolated domains, scores relative to wildtype are given by asterisks, as protein concentrations are higher than with full-length Lin28. See also Figure S6. (D) In vitro Dicer processing assay with different pre-miR and protein combinations. (E–G) In vivo processing assay as described in Figure 2D. Full-length Lin28 was co-transfected with pri-let-7g with indicated mutations (E), or with the entire preE region swapped with preE of an unrelated miR-21(G)(Piskounova et al., 2008), or pri-miR-122 (G). Wildtype pri-let-7g was co-transfected with full-length Lin28 constructs with indicated mutations in (F). Error bars indicate standard deviation calculated from triplicate measurements. Immunoblots are shown to compare expression levels of Lin28 in each panel.
The bipartite character of the Lin28:let-7 interactions implies that one should observe strong synergy when combining a mutation in one of the two let-7 interaction sites with a mutation in the Lin28 domain that recognizes the other let-7 interaction site. Indeed, a CSD mutation (F73A) has much greater effect on binding with RNA bearing a mutation in the GGAG motif (to GGAU or deletion) than it does on binding with RNA bearing a preE-loop mutation near the CSD binding site (Figure 6C, F73A block). Similarly, for binding with a mutated CCHCx2 (Y140A), GGAG mutations are not as detrimental as a CSD binding-site mutation (Figure 6C, Y140A block). We have also tested binding of individual domains of Lin28 to various pre-let-7g mutants (Figure 6C, CSD and CCHCx2 blocks). Neither isolated domain binds to let-7 as specifically or tightly as does full-length Lin28. Nevertheless, RNA mutations at each binding site affect only the affinity of the corresponding domain, consistent with our model. In summary, the results of all these mutational studies are all consistent with the conclusion that Lin28 binds full-length pre-let-7 in the same way as does the truncated form present in our crystals.
The GGAG motif is conserved among let-7s not only in its sequence but also in its proximal position with respect to the Dicer site in the context of the full pre-let-7 molecule. The last G is 4 bases from the Dicer cleavage site on the 3′ strand, and only 2 bases from the position at which complementarity to the mature strand begins. Using previously determined structures of Dicer and the proposed location of the cut site (Du et al., 2008; Macrae et al., 2006), we have modeled how a Lin28:pre-let-7 complex would interact with Dicer (Figure S6D). Because their binding sites on RNA are close together and because Lin28 bends the RNA backbone, Lin28, especially its CCHCx2, may hinder Dicer directly. To test whether binding of Lin28 with pre-let-7g is sufficient to inhibit Dicer processing, we used different mutants in an in vitro Dicer assay (Figure 6D). The mutations that disrupt association between Lin28 and pre-let-7 lead to increased Dicer cleavage, compared with wildtype control. Our data are thus consistent with a direct effect of Lin28 on Dicer processing of pre-let-7.
We also tested the effect of the described mutations on in vivo processing of let-7 (Figure 6E-G). Mutations that affect CSD binding de-repress processing of pri-let-7g only modestly, perhaps because the presence of other cellular factors partially compensate for the affinity change (<10 fold). Altering the CCHCx2:GGAG interaction—by changes in RNA or protein—is more detrimental to Lin28 activity. Levels of mature let-7 in our in vivo assay depend on both complex formation between Lin28 and let-7 precursors and downstream effects of Lin28, such as hindering Drosha and Dicer while recruiting TUTase. Our results indicate that although both CSD and CCHCx2 contribute to affinity and specificity for let-7 precursors, the CCHCx2:GGAG interaction is more critical for the effector function of Lin28.
DISCUSSION
Deciphering Lin28 specificity
The structural and biochemical studies presented here reveal how Lin28 recognizes let-7 precursors and allow us to postulate how Lin28 might bind diverse pre-let-7s. We propose a preferred sequence consensus for CSD binding: NGNGA0YNNN (Y=pyrimidine; N=any base). The sequences and distances between the CSD binding site and the CCHCx2-binding GGAG motif are variable, but the two sites can be identified in many of the preE-let-7 sequences (Figure S7A). In cases where no significant preE-stem structure is predicted (e.g., in let-7a-2 or let-7c-1), the nearby mature region with its stable double-stranded helix may aid in closing the loop around the CSD. Loss of one or a few favorable interactions in other preE-let-7s might not completely exclude the RNAs from binding to Lin28, but rather result in differences in affinity that could affect the sensitivity of particular let-7s to Lin28 regulation in vivo. Indeed, understanding Lin28 specificity from preE-let-7d and preE-let-7f-1 allowed us to crystallize the preE-let-7g complex, which binds to Lin28 in an energetically less stable conformation (Figure S7B-C).
The sequence of the linker between CSD and CCHCx2 has a strong net positive charge, probably to interact with the negatively charged RNA sugar-phosphate backbone, or to compensate for any unpaired bases, as seen in the case of preE-let-7g complex. Evolutionary conservation of the electrostatic property suggest that the linker does play some role, even though its sequence is not crucial for binding specificity. The length of the linker varies in some organisms, and shorter linkers occur in those with only one copy of let-7 containing a shorter preE sequence. Longer, more flexible linkers might have evolved in higher eukaryotes to recognize longer and divergent let-7 precursors. Our preE-let-7g complex structure illustrates how the linker can adapt to different RNA substrates; Arg122 and Arg123 at the amino terminal end of the inter-domain linker stack against extra bases near the ds-ss junction (Figure S5B).
The GGAG tetranucleotide motif is well conserved among the members of the let-7 family within a particular species. In evolutionarily distant organisms such as worms and fruit flies, however, other sequences (such as GGUG or AUCA) are found in place of GGAG, perhaps due to co-evolution of RNA and protein. Although not included in the crystal structure, the two nucleotides following GGAG are A and U in most let-7 sequences. In the context of full-length molecules, there may be more contacts between the bulge near GGAG and CCHCx2. The importance of the GGAG motif has been explored previously, by introducing a GGAG motif into an unrelated RNA sequence, miR-16, to generate a chimeric pre-miRNA that has gained affinity for Lin28 (Heo et al., 2009). From our binding experiments and structural data, the GGAG motif alone cannot confer robust binding with Lin28, and shifting its position by a base or two relative to the CSD binding site does not affect Lin28 binding significantly. In the case of the chimeric RNA with miR-16, its preE also coincidentally contains a sequence similar to the preferred CSD binding site (UAAGAUUCU vs NGNGAYNN), at the 5′ side of the GGAG motif, explaining why this chimera could bind Lin28. Our structural and biochemical data thus provide a molecular explanation for Lin28 specificity, making it possible to investigate further its role in let-7 biogenesis as well as its function in binding various mRNA targets (Jin et al., 2011; Peng et al., 2011; Qiu et al., 2009).
Implications for miRNA regulatory mechanisms
Although Drosha and Dicer are known to cut at opposite ends of the mature miRNA, there are still major questions regarding how they recognize their target and how the cleavage can be regulated. Our structures of Lin28:preE-let-7 complexes combined with known structural data for Dicer have allowed us to postulate how the Lin28 binding event itself can inhibit processing of pre-let-7 in at least two ways (Figure S6D). First, Lin28 might act as a “wedge” to melt part of the double-stranded mature region as it bends GGAG and situates itself in a particular conformation on one of the strands. As a result, Dicer might be unable to recognize its substrate properly. Second, given the location of CCHCx2 binding site, the volume of CCHCx2, and the location of its N-terminus from which the interdomain linker would have to traverse to CSD, Lin28 is likely to clash with the Dicer dsRNA binding domains and also mask one of the cleavage sites.
The role of the preE in Drosha processing is less clear, especially since the Drosha cleavage site is at the opposite end of the mature region from preE. Nonetheless, the direct association of Lin28 with the preE shows that the observed effects of both the preE modifications and Lin28 on Drosha activity are probably linked (Michlewski et al., 2008; Zeng, 2003; Zeng and Cullen, 2005; Zeng et al., 2005; Zhang and Zeng, 2010). Other small RNA-binding proteins such as hnRNP-A1 and KSRP have been proposed to modify Drosha processing by binding to the preE region (Michlewski and Cáceres, 2010; Michlewski et al., 2008). Rather than being a mere by-product of miRNA processing, preE is clearly a critical handle for regulatory factors such as Lin28.
Our mutagenesis studies strongly suggest that the GGAG:CCHCx2 region has an important functional role in regulating let-7, in addition to contributing to the specificity and tightness of complex formation. Our in vitro binding results show that the observed strong effect of mutations in the CCHCx2:let-7 interface cannot be attributed to the overall affinity of the molecules alone. As the GGAG motif is closer to the mature sequence, mutations that lead to lower occupancy at this site—regardless of association of CSD with preE—may be more directly linked to hindrance of processing enzymes. Moreover, the specific conformation of CCHCx2:GGAG induced by complex formation, as observed in our crystals, is probably important for recruiting downstream factor(s) such as TUTase. The critical role of Y140 of CCHCx2 in determining the RNA conformation is described in Results, and a uracil base (in GGAU mutant) would not be large enough to stack against Y140 efficiently in the observed conformation. Transition mutations in GGAG sequence might also result in slightly different conformations, without greatly reducing complex formation. Some of these mutations (to GAGG or AAGG) maintain their affinity for Lin28, but can obliterate uridylation by TUTase (Heo et al., 2009). That is, the CSD provides a larger contact and contributes more strongly than CCHxCx2 to let-7 affinity, but the latter domain has additional effector functions.
Conclusion
The structures of the three Lin28:preE-let-7 complexes we have determined show a bipartite interaction of Lin28 with its let-7 family partners (Figure 7). The CSD inserts into the loop at one end of the central stem-loop structure in preE-let-7, and the CCHCx2 module recognizes a GGAG motif at the other end. The linker between CSD and CCHCx2 is flexible, to accommodate variable sequences and lengths among Lin28-regulated let-7 family members without compromising affinity or specificity. This molecular organization explains several conserved features of preE-let-7s: first, a minimum loop length of 9-nucleotides, with a preferred sequence of NGNGAYNNN; second, a stem-like structure that closes the loop into a circle; and third, a GGAG motif close to the 3′ end of the preE. The model provided by our crystal structures provides a mechanistic explanation for the inhibitory effect of Lin28 on miRNA processing by Dicer; it further suggests that the CCHCx2:GGAG part of the complex directly influences downstream factor(s) important for let-7 regulation. These structural details will be useful for developing therapies that target the Lin28:pre-let-7 complex and its effects on let-7 processing.
Figure 7.
Schematic model for Lin28 domains binding to two distinct regions of preE-let-7 (f-1 was used for the model figure). For Lin28: Blue, CSD; Green, CCHCx2; Blue-green loop, protein linker. For pre-let-7 depicted as array of cylinders: Yellow, mature region; Orange, bases included in the crystallization construct; Grey, parts of preE not included in the crystal. Potential partial melting of dsRNA near Dicer sites is represented with double-headed arrows; it is uncertain how far the effect would carry. From structural models, interactions with other preE-let-7 sequences can be postulated as shown in Figure S7.
Experimental Procedures
More details are provided in Extended Experimental Procedures.
Protein purification
Lin28 constructs derived from mouse Lin28a were purified after overexpression in E.coli, using Nickel affinity, cation exchange, and size exclusion chromatography.
Electrophoretic Mobility Shift Assay
For preE probes, RNA oligonucleotides were synthesized (IDT), and full pre-miR probes were purified by PAGE after in vitro transcription followed by double ribozyme cleavage, as detailed in (Walker et al., 2003). RNAs were radiolabeled with ATP[γ-32P] using T4 polynucleotide kinase, incubated with protein in a buffer containing 20mM Tris 7.5, 100mM NaCl, 10mM DTT, 50μM ZnCl2, 15μg/μL yeast tRNA, and 1U/μL RNAse inhibitor.
NMR spectroscopy
All NMR samples were prepared as 0.5mM Lin28:preEM-let-7d complexes. Sequence-specific chemical shifts for backbone atoms were determined for 157 residues (out of 166 total, including 13 prolines), using the TROSY versions of HNCA, HN(CO)CA, HNCACB, HN(CO)CACB, HNCO, and HN(CA)CO, using a 15N, 13C and 85% 2H-labeled protein combined with unlabeled RNA. Experiments were conducted at 30°C on Bruker spectrometers equipped with cryogenic probes, operating at 1H frequencies of 600MHz (sequence assignment and relaxation experiments) or 750MHz (NOESYs).
Crystallography
Crystals of all three complexes were produced by vapor diffusion, using reservoir solution containing 0.6M NaH2PO4, 1.4M K2HPO4 and 5% glycerol for preEM-let-7d and preEM-let-7f-1 complexes, and 0.1M Tris pH 8.0, 32% w/v PEG 4000, and 0.2M sodium acetate for preEM-let-7g complex. Experimental phases were obtained for preEM-let-7d complex by anomalous scattering from zinc atoms (SAD), and the structures of preEM-let-7f-1 and preEM-let-7g complexes were solved by molecular replacement with Lin28:pre-let-7d as search model.
Dicer in vitro processing assay
Dicer expression construct (Addgene plasmid 19873) and purification are described as in (Landthaler et al., 2008), and radiolabeled pre-miR constructs were prepared similarly to EMSA probes. Dicer assays were carried out as described in (De and Macrae, 2011), using a buffer containing 20mM Tris 7.5, 5% glycerol, 3.2mM MgCl2, 5mM DTT, 50mM NaCl, and 100μM ZnCl2.
MicroRNA in vivo processing assay
Ability of Lin28 constructs to block let-7 processing in cells was compared as outlined in (Viswanathan et al., 2008). Briefly, pri-let-7g was co-transfected with FLAG-tagged Lin28 constructs (25ng unless otherwise noted) or vector control into 293T cells (12well) using lipofectamine. Total RNA was isolated using TriZol reagent, treated with DNAse I, and quantitative RT-PCR was used with miRNA-specific stem-loop primers as previously described (Wan et al., 2010). Relative levels of mature miRNAs were analyzed by ΔΔCt method, and normalized by U6 snRNA levels.
Supplementary Material
Acknowledgments
The authors declare no competing financial interests. We thank Stephen C Harrison for discussion and critical review of the manuscript, members of the Chou Lab for discussion about NMR experiments, Janet Iwasa for help with design of Figure 7, George Q Daley for the pri-miR constructs for in vivo processing assays, Thomas Tuschl for Dicer expression construct, Katarzyna Szatkowski for technical assistance, and Elizabeth O’Day for participation during the early stages of the project. This work is based upon research conducted at the Advanced Photon Source (Northeastern Collaborative Access Team beamlines) and Brookhaven National Laboratory (beamline X25). This work was partially supported by Center for Molecular and Cellular Dynamics at Harvard Medical School, NIH grant 5U54GM094608 and by postdoctoral fellowships to Y.N. from the Damon Runyon Cancer Research Foundation (DRG-#1953-07) and the Charles A. King Trust, N.A., Bank of America, Co-Trustee.
Footnotes
AUTHOR CONTRIBUTIONS
Y.N. performed experiments. C.C. provided technical assistance with experiments. R.G. provided the Lin28 clone, and participated in discussions. J.C. helped with NMR spectroscopy. Y.N. and P.S. designed and interpreted experiments and prepared the manuscript.
ACCESSION NUMBERS
Coordinates and structure factors for the structures of Lin28:preEM-let-7d, Lin28:preEM-let-7f-1, and Lin28:preEM-let-7g complexes have been deposited with the Protein Data Bank under accession codes 3TRZ, 3TS0, and 3TS2.
Supplemental Information includes 7 figures and Extended Experimental Procedures, and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. Journal of Molecular Biology. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- Büssing I, Slack F, Groβhans H. let-7 microRNAs in development, stem cells and cancer. Trends in molecular medicine. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Davis-Dusenbery BN, Hata A. MicroRNA in Cancer: The Involvement of Aberrant MicroRNA Biogenesis Regulatory Pathways. Genes & Cancer. 2010;1:1100–1114. doi: 10.1177/1947601910396213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science (New York, NY) 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- De N, Macrae IJ. Purification and Assembly of Human Argonaute, Dicer, and TRBP Complexes. Methods in molecular biology (Clifton, NJ) 2011;725:107–119. doi: 10.1007/978-1-61779-046-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Lee JK, Tjhen R, Stroud RM, James TL. Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2008;105:2391–2396. doi: 10.1073/pnas.0711506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazão C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature Structural & Molecular Biology. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Molecular Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Jing W, Lei X-X, Feng C, Peng S, Boris-Lawrie K, Huang Y. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- King C, Cuatrecasas M, Castells A, Sepulveda A, Lee JS, Rustgi AK. Lin28b promotes colon cancer progression and metastasis. Cancer research. 2011 doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. The EMBO Journal. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA (New York, NY) 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nature Structural & Molecular Biology. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nature genetics. 2008;40:584. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Katsaros D, Shaverdashvili K, Qian B, Wu Y, de la Longrais IAR, Preti M, Menato G, Yu H. Pluripotent factor lin-28 and its homologue lin-28b in epithelial ovarian cancer and their associations with disease outcomes and expression of let-7a and IGF-II. European journal of cancer (Oxford, England : 1990) 2009;45:2212–2218. doi: 10.1016/j.ejca.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science (New York, NY) 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Research. 2005;33:W577–581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max KEA, Zeeb M, Bienert R, Balbach J, Heinemann U. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. Journal of Molecular Biology. 2006;360:702–714. doi: 10.1016/j.jmb.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Max KEA, Zeeb M, Bienert R, Balbach J, Heinemann U. Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. The FEBS journal. 2007;274:1265–1279. doi: 10.1111/j.1742-4658.2007.05672.x. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nature Structural & Molecular Biology. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Molecular Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA (New York, NY) 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Li S, Zhao JH, Luan Jaa, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nature genetics. 2009;41:729. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. STEM CELLS. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29:2153–2159. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- Permuth-Wey J, Kim D, Tsai Y-Y, Lin H-Y, Chen YA, Barnholtz-Sloan J, Birrer MJ, Bloom G, Chanock SJ, Chen Z, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer research. 2011 doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JRB, Stolk L, Franceschini N, Lunetta KL, Zhai G, Mcardle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nature genetics. 2009;41:648. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, Lapierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. The Journal of biological chemistry. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Research. 2009 doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl E, Nitsch R, Wulczyn F. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature Cell Biology. 2008 doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. Posttranscriptional Regulation of MicroRNA Biogenesis in Animals. Molecular Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nature genetics. 2009;41:734. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science (New York, NY) 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nature genetics. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SC, Avis JM, Conn GL. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Research. 2003;31:e82. doi: 10.1093/nar/gng082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, En Lim Q, Too HP. High-performance quantification of mature microRNAs by real-time RT-PCR using deoxyuridine-incorporated oligonucleotides and hemi-nested primers. RNA (New York, NY) 2010;16:1436–1445. doi: 10.1261/rna.2001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene expression patterns : GEP. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007a;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, NY) 2007b;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zeng Y. Sequence requirements for micro RNA processing and function in human cells. RNA (New York, NY) 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. The Journal of biological chemistry. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. The EMBO Journal. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Research. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nature genetics. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.