SUMMARY
As nascent polypeptides exit ribosomes, they are engaged by a series of processing, targeting and folding factors. Here we present a selective ribosome profiling strategy that enables global monitoring of when these factors engage polypeptides in the complex cellular environment. Studies of the Escherichia coli chaperone Trigger Factor (TF) reveal that, while TF can interact with many polypeptides, β-barrel outer membrane proteins are the most prominent substrates. Loss of TF leads to broad outer membrane defects and premature, cotranslational protein translocation. While in vitro studies suggested that TF is prebound to ribosomes waiting for polypeptides to emerge from the exit channel, we find that in vivo TF engages ribosomes only after ~100 amino acids are translated. Moreover, excess TF interferes with cotrantslational removal of the N-terminal formyl methionine. Our studies support a triaging model in which proper protein biogenesis relies on the fine-tuned, sequential engagement of processing, targeting ad folding factors.
INTRODUCTION
Co-translational events play a critical role in determining the fate of polypeptides. Indeed as soon as a nascent chain emerges from the ribosomal exit tunnel, it is acted upon by a series of processing enzymes, targeting factors and molecular chaperones (Kramer et al., 2009). The ribosome serves as a platform for the regulated association of these various factors. Yet we have only a limited understanding of the spatial and kinetic coordination of these events.
In bacteria, the exit tunnel of the large ribosomal subunit can accommodate an extended peptide of ~30 amino acids (Ban et al., 2000). Shortly after the peptide exits this tunnel, the formyl group of the N-terminal formylmethionine is removed by a ribosome-bound peptide deformylase (PDF) (Bingel-Erlenmeyer et al., 2008), after which the methionine can be cleaved by methionine aminopeptidase (MAP) (Ball and Kaesberg, 1973). In addition, many nascent polypeptides interact with the ribosome-associated chaperone trigger factor (TF), which is thought to assist in cotranslational folding. Alternatively, the signal recognition particle (SRP) or the ATPase SecA can interact with nascent chains harboring an N-terminal signal sequence in order to target them for translocation across the cytoplasmic membrane (Huber et al., 2011; Ullers et al., 2003). The chaperone SecB also associates with nascent secretion substrates (Randall and Hardy, 2002).
Ribosome-associated chaperones play critical roles in both prokaryotes (Kramer et al., 2009) and eukaryotes (Albanèse et al., 2006; Hundley et al., 2005). Of these, TF is the best characterized in terms of the molecular details of its action (Hoffmann et al., 2010). The ability of TF to promote folding of newly synthesized proteins depends on its association with ribosomal protein L23, which is situated on the surface of the ribosome near the polypeptide exit channel (Kramer et al., 2002). The ribosome binding activity of TF has been extensively characterized in vitro. Although TF binds to non-translating ribosomes with a KD of ~1 μM (Patzelt et al., 2002) and with a mean residence time of 10 to 15 sec (Kaiser et al., 2006), the presence of nascent substrates can increase this affinity up to 30-fold (Rutkowska et al., 2008). In addition, structural analyses of TF in complex with ribosomes suggest that TF forms a protective dome over the tunnel exit (Ferbitz et al., 2004) that could shield nascent chains from degradation by proteases (Hoffmann et al., 2006; Tomic et al., 2006) or improve the efficiency of folding by reducing the speed of folding (Agashe et al., 2004).
By contrast, many aspects of the mechanism of action of TF in vivo are unknown. For example, how TF aids in the folding of proteins remains unresolved. Likewise, it is unclear whether TF interacts with all nascent chains or only a specific subset, and, although TF can interact with relatively short nascent chains in vitro (Merz et al., 2008), it is unknown when TF begins to associate with them in vivo. Furthermore, the interplay of TF with other chaperones, targeting factors and enzymes remains unclear. Finally, despite extensive studies, the phenotypic cost to cells lacking TF has not been apparent unless combined with the loss of the DnaK chaperone (Deuerling et al., 1999; Teter et al., 1999).
To enable the systematic and quantitative analysis of proteins in prokaryotes, we have developed a strategy for monitoring bacterial translation through ribosome profiling (deep sequencing of ribosome protected mRNA fragments) (Ingolia et al., 2009). Furthermore, by combining ribosome profiling with a procedure to affinity purify ribosomes whose nascent chains are bound by TF, we quantitatively defined when TF engages its substrates. Analysis of these data revealed several fundamental features of TF action, including a role for TF in the biogenesis of β-barrel outer membrane proteins. Additionally, we found that in contrast to in vitro studies, full recruitment of TF is delayed until the peptide is ~100 amino acids in length, providing a protected window during which other processing and targeting factors have preferential access to the nascent chain. More generally, the approach developed here enables the comprehensive and quantitative analysis of co-translationally acting factors involved in the maturation and folding of newly synthesized polypeptides.
RESULTS AND DISCUSSION
Ribosome Profiling in Escherichia coli
Dramatic advances in DNA sequencing technology (Bentley et al., 2008) have made it possible to sequence bacterial genomes rapidly and at low costs. This has led to an enormous increase in our understanding of the genetic diversity of the prokaryotic world. However, our ability to systematically identify the proteins encoded within these genomes or monitor their rates of production has lagged far behind. Eukaryotic ribosome profiling experiments (Guo et al., 2010; Ingolia et al., 2009) have provided the means to (1) experimentally define open reading frames (ORFs) in an unbiased manner including those that play a regulatory role in translation (or are too small to be identified by other approaches) rather than leading to production of stable proteins; (2) comprehensively evaluate protein production rates for each gene under different environmental conditions and (3) measure the variability of rates in translation within genes that arise from ribosome pausing at specific positions along the message. We sought to extend this approach to prokaryotes to enable the analysis of both translation and co-translational processes that promote the maturation of nascent polypeptides. Although we focused on E. coli, our method provides a general tool for decoding proteomes and monitoring rates of protein production in other bacteria.
Development of bacterial ribosome profiling
Ribosome profiling requires four distinct steps: (1) generation of cell extracts, in which ribosomes have been halted along the mRNA that they are translating; (2) treatment of polysomes with nuclease to remove regions of the message not protected by the ribosome; (3) conversion of these RNA fragments into double stranded DNA copies and (4) analysis of these fragments by high throughput sequencing.
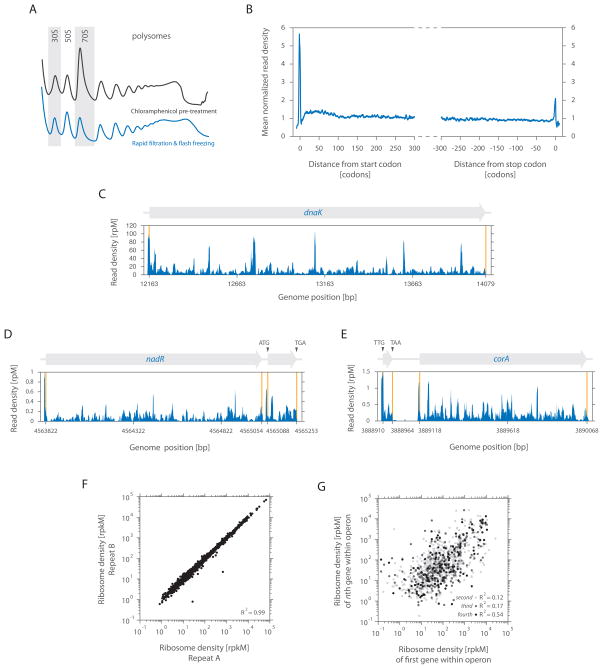
We developed two alternative approaches to capture the cellular state of translation in E. coli. For the first, we pre-treated exponentially growing cells with chloramphenicol to arrest translating ribosomes. For the second, we collected the cells by fast filtration of the culture. For each case, cells were rapidly frozen in liquid nitrogen and lysed in a frozen state, preventing continued elongation during sample preparation. Both approaches allowed extraction of intact polysomes (Figure 1A), although modest differences in polysome profiles were seen between them. Although rapid filtration is essential for robust analysis of ribosome pausing (see below), chloramphenicol pre-treatment is especially useful in cases where rapid recovery of cells is difficult.
Figure 1. Characterizing prokaryotic translation by ribosome profiling of bacterial cells.
(A) Translating ribosomes were extracted from cells (MC4100) either pre-treated with chloramphenicol (black trace) or collected by rapid filtration (blue trace). Polysomes were resolved by 10 to 55% (w/v) sucrose density gradients.
(B) Meta-gene analysis of ribosome density as a function of position from fast filtered cells. Genes were aligned from either their start (left panel) or stop (right panel) codon and averaged across them [See Extended Experimental Procedures].
(C) Ribosome density of dnaK as a function of position. The density in reads per million (rpM) was corrected for the total number of reads that aligned to its coding sequence.
(D) Example of a novel ORF starting at a non-canonical UUG codon.
(E) Example of a newly identified canonical ORF.
(F) Quantifying gene expression levels by ribosome profiling from fast filtered cells. Ribosome densities of two independent replicates were plotted for comparison. The density in reads per kilobase million (rpkM) is a measure of overall translation along each gene [See Extended Experimental Procedures].
(G) The ribosome density of the first gene in an operon was compared with the ribosome density of either the second, third or fourth gene in the same operon as indicated.
After digestion with micrococcal nuclease (MNase), ribosome-protected mRNA footprints were isolated through the collection of monosomes either using a sucrose gradient or by pelleting them through a sucrose cushion. Protected mRNA regions that are derived from other ribosomal complexes (such as disomes) can be distinguished from monosomal footprints based on the size of the protected fragments using PAGE purification. Finally, we converted RNA fragments into a sequenceable DNA library using a previously described method (Ingolia, 2010), except that 3′ ends were ligated with a defined linker rather than being polyadenylated. Following conversion, each footprint was identified by deep sequencing and mapped to its genomic position.
Meta-gene analysis
Focusing on the top ~2000 highly expressed genes (out of 4084 annotated), we analyzed the average ribosome density across these ORFs using cells harvested by rapid filtration. A strong peak was seen over the initiation codon whose density was ~5.5-fold greater than those within the body of the message (Figure 1B, left panel). A less pronounced peak (~2-fold greater density) was observed over the termination codon (Figure 1B, right panel). The elevated ribosome density at the beginning and end of coding sequences presumably reflects the slower kinetics of translation initiation and termination when compared with the average rate of elongation. There was also a modest (~1.3-fold) excess in density over the first 50 to 100 codons. This is similar in span but of much smaller magnitude to the ~3-fold excess density seen at the 5′ end of yeast messages (Ingolia et al., 2009).
Examination of the ribosome occupancy profile of individual genes revealed that the density of ribosome footprints varies substantially across individual messages (Figure 1C), resulting from local differences in the rate of elongation as the ribosome moves down a message. For example, dnaK had a median read density of 7.4 reads per million, yet five peaks exceeded this median by more than ten fold and most likely represent prominent ribosome pausing sites (Figure 1C). This observed variability was highly reproducible ([R2] = 0.92 for dnaK) and thus likely represents an intrinsic feature of the translation of individual messages. Ribosome pausing regulates the synthesis (Morris and Geballe, 2000), folding (Zhang et al., 2009), and localization of certain proteins (Mariappan et al., 2010). However, the difficulties in identifying pause sites have limited previous analyses to a small number of examples. Our data provide a critical resource for understanding the causes and biological roles of such pauses.
Defining open reading frames
Ribosome profiling provides a direct readout of the regions being translated along any mRNA, allowing the experimental definition of protein boundaries and thus the identification of novel ORFs. Although the E. coli genome has been extensively annotated, we identified a number of short ORFs, including a well-expressed 55 residue protein (Figure 1D) and an upstream uORF with a near cognate (UUG) initiation codon preceding corA (Figure 1E). uORFs can regulate the expression of downstream genes in the same operon (Tenson and Ehrenberg, 2002), but their identification has been challenging. Thus ribosome profiling provides a general tool for identifying and monitoring production of these species under many environmental conditions independent of their size or stability.
Global analysis of gene expression
Ribosome profiling provides a high precision tool for monitoring translation rates as evidenced by density of ribosomes on messages. Under optimal growth conditions (Luria broth, mid-log phase, 37°C), ~75% of known ORFs were quantifiable. Such measures are highly reproducible ([R2] = 0.99), with rates of translation spanning five orders in magnitude (Figure 1F). This measure of protein expression is expected to be a far better predictor of protein levels than measures of mRNA levels as it captures both transcriptional and translational control (Ingolia et al., 2009). This point is illustrated by analyzing the translation rates of polycistronic messages. Despite being encoded on the same mRNA, the expression levels of genes in the same operon are only modestly correlated with one another (Figure 1G). This finding argues that translational control plays an important role in determining the overall rate of protein production in E. coli.
Investigation of TF-nascent chain interactions by selective ribosome profiling
We next sought to extend our technique to selectively profile ribosomes by enriching for ribosomes bound by factors that act on nascent chains. In general, selective ribosome profiling depends on the efficient enrichment of a well-defined population of ribosomes (Figure 2A). Here we focused on monitoring monosomes that were engaged by TF, predominantly through its association with the nascent chain.
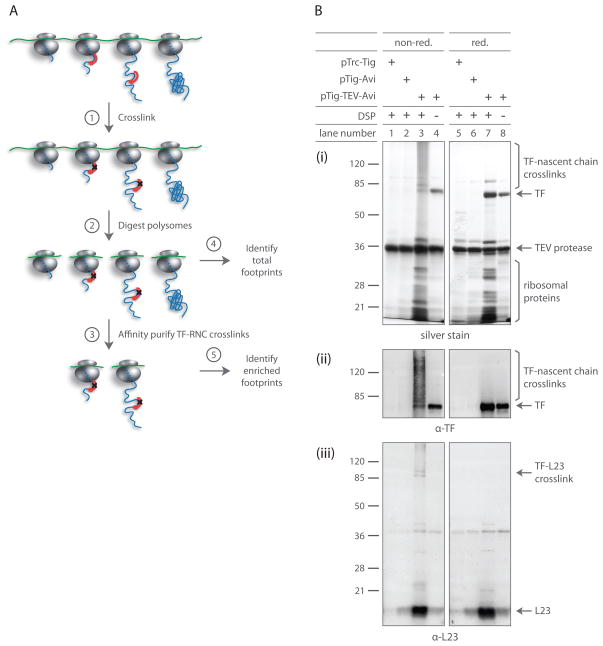
Figure 2. TF crosslinked RNCs can be isolated with high specificity.
(A) Schematic for affinity purifying TF crosslinked RNCs: ➀ cells expressing epitope-tagged TF are harvested at mid-log phase, cryogenically lysed and chemically crosslinked. ➁ Polysomes digested with MNase yield footprint-containing monosomes. ➂ Digested monosomes are forced through a sucrose cushion, separating free TF molecules from those crosslinked to RNCs. TF crosslinked RNCs are affinity purified and eluted by cleaving TF with TEV protease. ➃ mRNA footprint fragments derived from all monosomes and ➄ those enriched through affinity purification are cloned into a cDNA library for deep sequencing analysis.
(B) Gel analysis of DSP crosslinking and affinity purification. Δtig::kan cells expressing specified TF variants were harvested by centrifugation. Following cryogenic lysis, lysates were crosslinked with DSP as they thawed. TF-RNCs were affinity purified and eluted through TEV protease cleavage. Eluates were analyzed under both reducing and non-reducing conditions. Gels were either silver stained (i) or immunoblotted using antisera specific for TF (ii) or L23 (iii).
To facilitate the purification of TF-bound ribosomes, we fused a tag consisting of a TEV protease-cleavable AviTag, which is biotinylated by an endogenous biotin ligase, to the C-terminus of the protein. The tagged TF appears to be fully functional both in vitro and in vivo. The tag neither altered the affinity of TF for the ribosome nor interfered with TF’s ability to aid refolding of chemically denatured GAPDH (data not shown). Furthermore, expression of tagged TF at wild-type levels complemented the synthetic lethal phenotype seen for the Δtig ΔdnaK double knockout (Figures S1A and S1B) and the chemical sensitivities of Δtig cells (see below) (Figures S5A and S5B).
To stabilize the transient association of TF with ribosome-nascent chain complexes (TF-RNCs), we crosslinked TF to nascent polypeptides using the thiol-cleavable crosslinker DSP (dithiobis succinimidyl propionate), which reacts with primary amines in lysine side chains and N-termini. In order to capture physiologically relevant substrate interactions, frozen lysates were directly thawed in the presence of DSP. Lysates were subsequently treated with MNase to generate monosomes prior to affinity purification in order to avoid the co-purification of unbound ribosomes tethered through the polysomal mRNAs. The ribosomal fraction was separated from uncrosslinked TF by ultracentrifugation through a high salt sucrose cushion, followed by affinity purification and elution of TF-RNCs.
Our analysis of crosslinking products demonstrates that we specifically enriched for ribosomes whose nascent chains were engaged by TF. Robust recovery of ribosomes strictly depended on DSP crosslinking (Figure 2B; lanes 4 and 8), the presence of an AviTag (Figure 2B; lanes 1 and 5) and a TEV protease cleavage site (Figure 2B; lanes 2 and 6). Importantly, ribosome recovery also depended on the ability of TF to bind ribosomes (Figure S1C), indicating that TF is unable to directly engage nascent chains without docking to L23. Crosslinking of TF by DSP gave rise to products of diverse molecular weight, representing nascent chains of various lengths (Figure 2B; lane 3, i and ii). However, we observed only negligible crosslinking of TF to L23, since L23 migrated almost exclusively as a single band under both non-reducing (Figure 2B; lane 3, iii) and reducing conditions (Figure 2B; lane 7, iii). Likewise, we did not observe significant levels of crosslinking between TF and ribosomal proteins L24 or L29 (data not shown), which were suggested to come in close proximity to ribosome-bound TF (Baram et al., 2005; Schlünzen et al., 2005). These observations argue that the purified TF-RNCs were captured predominantly on the basis of the interaction of TF with nascent chains.
Features of TF engagement to nascent chains
We next compared the density of ribosome footprints across individual genes for the affinity purified TF-RNCs to the total pool of ribosomes. The ratio of these values provides a position specific measure of the propensity of TF to engage nascent chains. We performed a meta-gene analysis to determine the average enrichment efficiency as a function of polypeptide length. Since DSP specifically crosslinks TF to nascent chains, the minimal length at which nascent chains engage TF is expected to exceed the 30 amino acids needed to traverse the ribosomal exit tunnel. Indeed, ribosomes within this region were poorly captured by affinity purification (Figure 3A). However, this N-terminal depletion extended well beyond the minimal length needed for the nascent chain to emerge from the ribosome, indicating that effective TF binding requires substantial extension of the polypeptide outside of the tunnel exit.
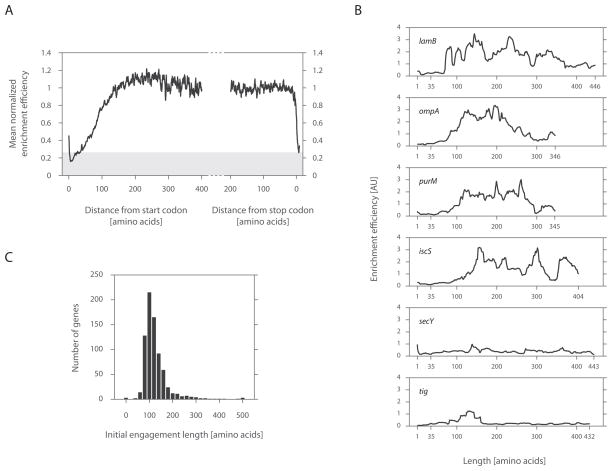
Figure 3. TF interaction propensity as a function of nascent chain length.
(A) Meta-gene enrichment efficiency derived as a function of ribosome position. Meta-gene ribosome densities (described in Figure 1B) were each computed for footprints derived from TF enriched RNCs and those from the total monosome pool. Ratios between these profiles were taken along indicated positions. Background signal is shaded in gray, corresponding to the enrichment efficiency at codon 30, a length that should be inaccessible to soluble factors.
(B) Individual enrichment efficiencies plotted as a function of nascent chain length. Characteristic examples of cytoplasmic (IscS and PurM), inner membrane (SecY) and outer membrane (LamB and OmpF) proteins are shown.
(C) A histogram of the initial position, at which TF engages nascent chains.
By examining individual profiles, we found that enrichment efficiency was low near the N-terminus and typically rose sharply thereafter (Figure 3B). The position of this rise (indicative of the first TF binding) varied between different genes. Thus to identify the initial point at which TF engaged each polypeptide, we measured the position at which each profile first crossed an empirically derived threshold. This threshold was chosen to be well above background but still able to capture these early binding events (Figure 3C and Extended Experimental Procedures). The median length at which TF first engaged a polypeptide was 112 amino acids, with half of all nascent chains being bound within ±20 amino acids of this position. TF was depleted at the beginning of translation in virtually all nascent chains (Figures 3B and 3C).
We also observed from the meta-gene analysis that the intensity of TF engagement leveled off after ~135 residues (Figure 3A). If multiple TF molecules were bound per nascent chain, the likelihood of purifying crosslinked TF-RNCs should increase in proportion to the length of the nascent chain. Yet this was not the case, suggesting that nascent chains are generally not engaged by multiple TF molecules. Consistent with this view, we observed varying levels of TF engagement for each polypeptide (Figure 3B), with periods of TF binding interrupted by regions with little detection of TF association over background, suggesting that TF cycles on and off the nascent chain during synthesis.
TF recruitment by the ribosome occurs concurrently with nascent chain binding
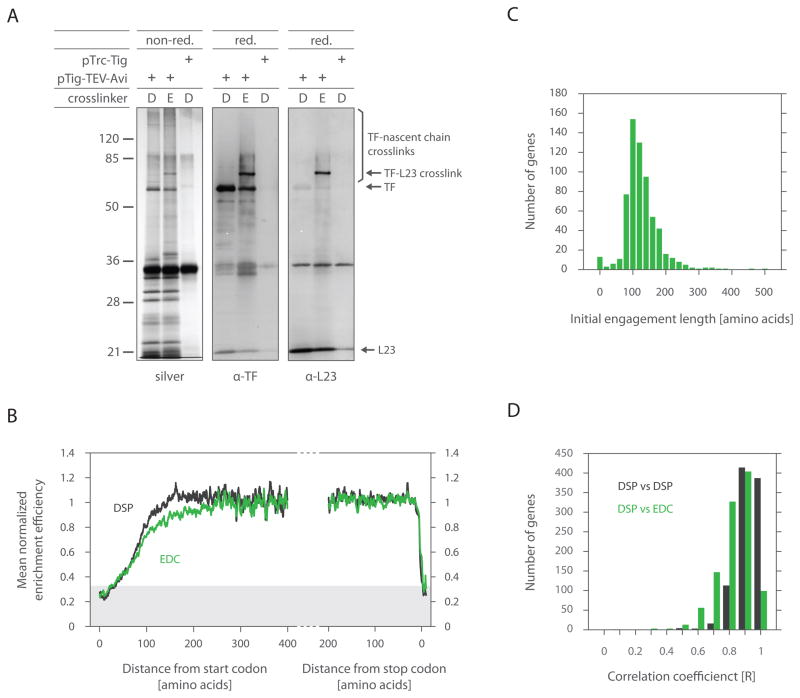
The depletion of TF in the beginning of translation could arise either because TF is not bound to the ribosome or because it is present on the ribosome but not engaged with the nascent chain. To discriminate between these possibilities, we used the crosslinker EDC (1-ethyl-3-[3-dimethylaminopropyl] carbodiimide), which couples carboxyl groups to primary amines. In contrast to DSP, EDC covalently linked TF not only to nascent chains, but also to the ribosome at L23 (Figures 4A and S2). This ability of EDC, unlike DSP, presumably reflects the availability of carboxyl groups in L23 that are in close proximity to TF. Remarkably, the TF enrichment efficiency was highly similar between DSP and EDC at both the meta-gene (Figure 4B) and single gene (Figures 4C and 4D) levels, suggesting that robust ribosome binding occurs concurrently with nascent chain engagement.
Figure 4. Ribosome recruitment of TF occurs at the same time as nascent chain binding.
(A) Gel analysis of DSP and EDC crosslinking and affinity purification. Δtig::kan cells were processed as before (Figure 2), but treated with DSP (D) or EDC (E).
Resulting eluates were resolved by SDS-PAGE under both reducing (red.) and non-reducing (non-red.) conditions. EDC crosslinks are irreversible under reducing conditions unlike DSP. Gels from non-reduced samples were silver stained (i), while reduced samples were immunoblotted using antisera specific either for TF (ii) or L23 (iii).
(B) The same test was performed as in Figure 3A, except cells were harvested by rapid filtration followed by fast freezing. Cryogenically pulverized cells were crosslinked with either EDC or DSP.
(C) The same test was performed as in Figure 3C, except cells were harvested by filtration (as in Figure 4B) and TF-RNCs were stabilized by EDC crosslinking.
(D) Gene by gene correlation of TF binding profiles for DSP replicates and DSP compared to EDC.
Nascent chain N-termini are resistant to surveillance by TF as they emerge from the ribosomal exit tunnel
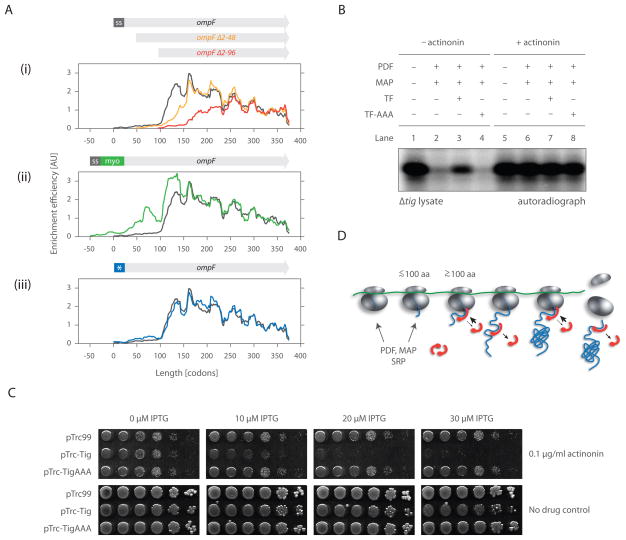
In principle, the observed delay in TF recruitment to ribosomes until ~100 amino acids have been synthesized could result from a general paucity of binding sites robustly recognized by TF. Alternatively, the observed depletion could be an intrinsic feature of translation in vivo that disfavors TF recruitment to the N-termini of nascent chains even if TF recognition sites are present. To discriminate between these possibilities, we used the selective ribosome profiling approach to monitor TF engagement in cells expressing variants of a TF substrate (see below) altered at their N-termini. Specifically, we constructed a series of OmpF variants in which 48 or 96 residues had been truncated from the N-terminus, 50 residues derived from human myoglobin had been added following the signal sequence, or charged residues (N5D, V9E, A13D, and V16E) had been introduced in the signal sequence. These variants were expressed from a plasmid at levels similar (within a factor of two) to endogenous ompF expression (Figure S3). Selective ribosome profiling experiments were then performed to determine when TF engaged the different OmpF variants.
Our results establish that the distance from the N-terminus is the critical determinant of TF recruitment. For each of the truncations and the insertion mutant, there was a complete lack of TF recruitment prior to translation of 50 amino acids, and the synthesis of at least ~100 amino acids was required for the full engagement of TF, even though TF binding sites were present earlier (Figure 5A, i and ii). Following the initial binding event, the pattern of TF engagement along the nascent chain mirrored the binding pattern seen for wild-type OmpF. The signal sequence mutant had no discernible effect on TF binding (Figure 5A, iii), further emphasizing that initial TF engagement depends on the position along the nascent chain, rather than sequence composition of the residues near the N-terminus.
Figure 5. TF does not engage the N-terminal end of nascent chains in vivo as they emerge from the ribosome.
(A) Individual enrichment efficiencies of ompF variants compared with wild-type ompF.
(B) Cell-free coupled transcription/translation reactions initiated by non-stalled barnase. 5 μM each of either TF or TF-AAA (deficient in ribosome binding) and 2 μM each of both PDF and MAP were supplemented prior to translation initiation where indicated. Extracts treated with actinonin (lanes 5–8), an inhibitor of PDF, were used to assess overall levels of barnase synthesis. Reactions were quenched by TCA precipitation and visualized using SDS-PAGE and autoradiography.
(C) TF overexpression can interfere with N-terminal processing. MC4100 ΔacrA::kan cells transformed with pTrc99 (empty vector), pTrc-Tig or pTrc-TigAAA were spotted as 1:10 serial dilutions on LB plates containing 100 μg/ml of ampicillin and indicated concentrations of IPTG and actinonin. 10 μM IPTG induces TF from pTrc-Tig near endogenous levels (Kramer et al., 2004), thus overall expression of TF is increased roughly two-fold (from both the plasmid and endogenous locus).
(D) A model for the dynamic binding of trigger factor to ribosomes and nascent chains. Interaction between TF and the ribosome is limited early on translation (i.e. before the nascent chain emerges from the exit tunnel). TF fully engages the ribosome and nascent chain not before ~100 amino acids are translated. After release from the nascent chain, TF can re-bind, but each polypeptide is, on average, bound by only one, maximal two TF molecules at a time. Following translation termination, TF may stay associated with the released polypeptide, guiding further folding steps.
The delay in TF recruitment to the ribosome—until well after the polypeptide emerges from the exit tunnel—contrasts with the current view, mainly drawn from in vitro data, that TF is prebound to the ribosome and waits for the nascent chain to emerge (Patzelt et al., 2002). It is presently unclear what prevents TF from associating with shorter polypeptides in vivo. Although TF is thought to be in excess of ribosomes (Patzelt et al., 2002), the fraction of TF molecules available for nascent chain and ribosome binding at steady state conditions is not known. For example, TF has recently been suggested to have an additional ribosome-independent function in the assembly of oligomeric complexes (Martinez-Hackert and Hendrickson, 2009). As a result, fewer TF molecules would be available to interact with ribosomes. This could drive TF to associate preferentially with translating ribosomes exposing longer nascent chains. Indeed, such RNCs have been demonstrated in vitro to exhibit higher association rates for TF binding (Rutkowska et al., 2008).
Regardless of the mechanism, this delayed association of TF to RNCs may provide a window for other ribosome-associated nascent chain interacting factors, such as PDF and MAP, to act on the emerging polypeptide. To investigate the interplay of TF with PDF and MAP, we developed an in vitro assay for examining the action of these processing enzymes. We monitored the synthesis of the TF model substrate barnase (which has only one methionine residue at the initiation codon) by following 35S-methionine incorporation in a translation-competent Δtig extract devoid of PDF and MAP activity. In the absence of both enzymes, we detected a pronounced band corresponding to full-length, non-processed barnase (Figure 5B; lane 1). In their presence, however, the radioactive signal dramatically decreased (Figure 5B; lane 2), indicating that the N-terminal methionine was both deformylated and cleaved. Addition of excess TF prior to translation initiation (Figure 5B; lane 3), but not of the TF mutant impaired in ribosome binding (Figure 5B; lane 4) partially restored the radioactive signal, indicating that ribosome-bound TF interferes with N-terminal processing. Similarly, even a modest (~2-fold) overexpression of TF resulted in increased sensitivity to the PDF inhibitor actinonin (Figure 5C). These results suggest that TF can be driven to engage the N-termini of nascent chains, but that premature engagement of TF interferes with the removal of the N-terminal fMet residue from nascent chains, i.e. the essential N-terminal processing carried out by PDF and MAP. More broadly, our data suggest a model in which initial binding of TF to RNCs is determined by the length of the polypeptide, providing access for other factors. After the initial engagement event, TF can repeatedly bind to and release from the nascent chain and may stay associated with it even after translation has terminated (Figure 5D).
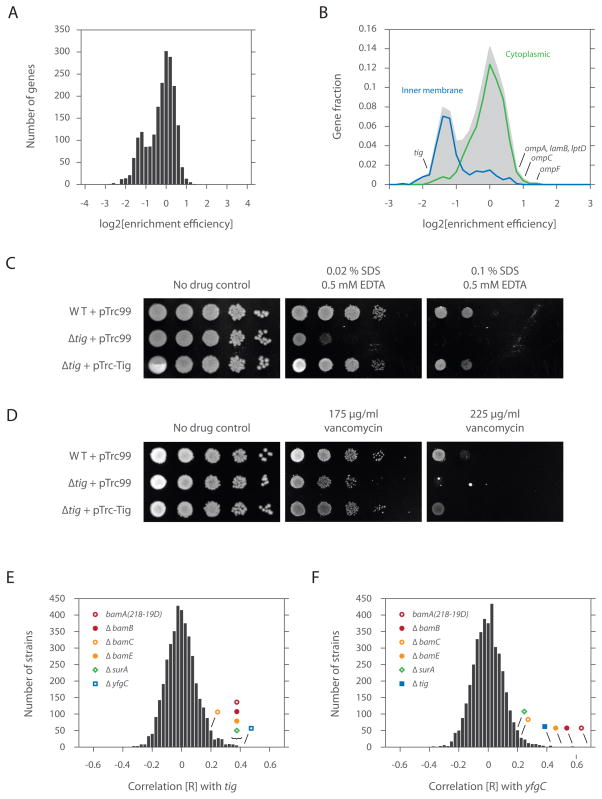
TF interacts with cytoplasmic proteins, but shows strong preference for outer membrane β-barrel proteins
To characterize the substrate specificity of TF, we determined the overall enrichment efficiency for each gene—defined as the sum of the enriched footprint density divided by the sum of the total footprint density (excluding the N-terminal region not engaged by TF). This analysis revealed an apparent bimodal distribution, with a major subset comprised of nascent chains robustly interacting with TF and a minor subset showing modest engagement to the chaperone (Figure 6A). While most nascent chains actively engaged by TF were localized to the cytoplasm (P = 5.2 × 10−16, rank sum test), those poorly engaged by the chaperone were generally targeted for the inner membrane (P = 3.2 × 10−44) (Figure 6B). This division of the TF interactome along cellular localization is consistent with the view that SRP outcompetes TF for binding to nascent inner membrane proteins (Eisner et al., 2006; Ullers et al., 2006; Ullers et al., 2003).
Figure 6. TF chaperones outer membrane β-barrel protein biosynthesis.
(A) A histogram of the overall enrichment efficiency (defined as the ratio of the enriched ribosome footprint density to the total ribosome footprint density). Nascent chains that interact well with TF show positive log values while those that interact poorly with the chaperone show negative log values.
(B) A histogram comprising the overall enrichment efficiency of each nascent chain for those with known GO (gene ontology) annotations based on cellular localization (i.e. cytoplasm, GO=0005737; inner membrane, GO=0019866; outer membrane, GO=0009279). The number of genes was represented as a fraction of the total, with the shaded area reflecting the total number.
(C) Growth analyses of cells expressing or lacking TF. 1:10 serial dilutions (horizontal dimension) of indicated strains (vertical dimension) were spotted on LB plates containing 10 μM IPTG, 50 μg/ml of ampicillin and specified levels of SDS/EDTA.
(D) Same as Figure 6C, but dilutions were spotted on LB plates containing 10 μM IPTG, 50 μg/ml of ampicillin and specified levels of vancomycin.
(E) Chemical sensitivities of BW25113 Δtig cells correlated with those of more than 3900 bacterial mutants (Nichols et al., 2011) and represented as a histogram of correlation [R] values.
(F) Same as Figure 6E, but for BW25113 ΔyfgC cells.
Strikingly, outer membrane β-barrel proteins were among the strongest TF interactors (Figure 6B). For example, five of the best characterized outer membrane proteins (OMPs) (LamB, LptD, OmpA, OmpC and OmpF) were among the top 25 most strongly enriched polypeptides (out of the ~2000 proteins examined). In addition, LamB, OmpA, OmpC and OmpF were at least an order of magnitude more strongly expressed than the other proteins that were highly enriched in the TF pull downs. Thus they accounted for the large majority of polypeptides fluxing through TF among this enriched group. Interestingly, TF was originally identified on the basis of its ability to promote insertion of chemically denatured proOmpA into membrane vesicles (Crooke and Wickner, 1987), although the physiological significance of this was unclear (Guthrie and Wickner, 1990).
Loss of TF function mimics the loss of outer membrane chaperones
The enrichment of OMPs among the cotranslational substrates of TF suggested that TF could play a role in OMP biogenesis. Defects in the biogenesis of outer membrane β-barrel proteins (including porins) often disrupt outer membrane integrity, leading to increased sensitivity to SDS/EDTA and vancomycin (Hagan et al., 2010). In support of the notion that TF plays a role in OMP biogenesis, Δtig cells displayed increased sensitivity to both SDS/EDTA (Figures 6C and S4) and vancomycin (Figures 6D and S4) in a manner that was rescued by wild-type TF or tagged TF expressed from plasmid (Figures S5A and S5B). Additionally, the activity of σE, which controls the envelope stress response, decreased by ~2-fold (Figure S6). As σE monitors the protein flux and integrity of folding in the outer membrane, the lowered activity of σE could reflect altered delivery of OMPs to the periplasm (Mecsas et al., 1993).
To further examine the phenotypic consequences of the loss of TF function, we compared the chemical sensitivities of Δtig cells to ~4000 deletion strains that were exposed to more than 300 conditions in a large scale chemical genetic screen (Nichols et al., 2011). Because mutations in functionally related genes have closely related chemical sensitivities (Hillenmeyer et al., 2008), examining the correlation across these sensitivities can reveal relationships among either unknown or seemingly unrelated genes. The chemical sensitivities of Δtig cells correlate highly with those carrying mutations in bamA (P = 1.5 × 10−11), bamB (P = 8.0 × 10−12), bamE (P = 8.2 × 10−11), surA (P = 5.8 × 10−11) and yfgC (P = 6.0 × 10−15) (Figure 6E). bamA, bamB and bamE encode for three of five components of the β-barrel assembly machinery, which together with SurA (a periplasmic chaperone) mediates the insertion of β-barrel proteins into the outer membrane (Hagan et al., 2010), strongly implicating a role for TF in OMP biogenesis. yfgC encodes for a predicted periplasmic peptidase and although its function is unknown, the chemical genetic data suggests that it also plays a role in OMP biogenesis. Indeed, when we queried the chemical sensitivities of ΔyfgC cells against all other strains, we found that these correlations were among the highest with components of the Bam complex and TF (Figure 6F).
The phenotypic link between SurA/Bam(s) and TF is particularly remarkable in light of the structural similarity between SurA and TF. SurA and TF have no apparent sequence similarity, yet their chaperone domains possess the same fold (Merz et al., 2006). Moreover, the chaperone domain of both proteins is followed by cis/trans peptidyl prolyl isomerase domain(s). Although the functional significance of these observations was not evident at the time, we now suggest the possibility that there was a primordial chaperone for β-barrel proteins, which underwent a gene duplication event, such that one copy gained (or retained) a ribosome-binding domain (TF), while the other (SurA) gained an N-terminal signal sequence, targeting it to the periplasm. This could then give rise to a pathway capable of chaperoning β-barrel proteins from their synthesis by the ribosome to their insertion into the outer membrane.
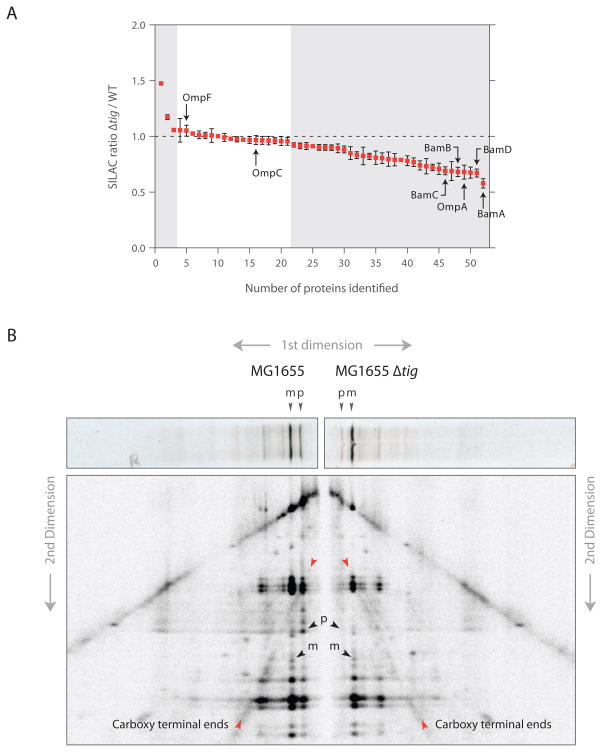
To provide quantitative data on the outer membrane defect caused by TF deletion, we analyzed the protein content of the outer membrane of wild-type and Δtig cells using SILAC (stable isotope labeling with amino acids in cell culture) combined with mass spectrometric analysis. We found that ~60% of all detected proteins in the outer membrane fraction showed a significant decrease in their steady-state levels in Δtig cells compared to wild-type cells whereas almost no proteins were found in higher amounts upon TF deletion (Figure 7A). OmpA and components of the Bam complex were among the most prominently disenriched proteins. Levels of OmpC and OmpF remained unaltered, which may result from compensatory mechanisms that are known to regulate their transcription so as to maintain proper OmpC/OmpF levels in the outer membrane. Interestingly, when we also analyzed the soluble fraction of proteins by SILAC, we found that SecA is one of the most strongly induced proteins in the Δtig mutant (increased by ~40%, P = 2.6 × 10−4), suggesting that Δtig cells have a mild translocation defect (Riggs et al., 1988).
Figure 7. TF absence causes a broad reduction in outer membrane protein levels and shifts the mode of translocation.
(A) Quantifcation of proteins from isolated outer membranes using SILAC. The SILAC ratio (Δtig/wild-type) was calculated for all outer membrane proteins identified with at least three peptides.
(B) 2D gel assay for monitoring translocation of newly synthesized LamB. Wild-type and Δtig::kan cells were pulse-labeled with 35S-methionine for 30 s and quenched using 5% TCA. LamB chains were immunoprecipitated and resolved by 12% SDS-PAGE (first dimension). Gel slices were excised, digested in gel with V8 protease and resolved by 15% SDS-PAGE (second dimension). Red arrows highlight C-terminal fragments that converge either to the precursor (p) as seen for wild-type cells or mature (m) form as seen for Δtig cells (black arrows).
The impact of loss of TF on OMP translocation
Taken together, the above observations indicate that an important function of TF is to chaperone β-barrel OMPs and/or modulate their export to the periplasm by the translocation system. Consistent with previous studies (Lee and Bernstein, 2002; Ullers et al., 2007), we observed that Δtig mutants accumulated less full-length precursors in pulse-labeling experiments for all exported proteins we examined (LamB, MBP, OmpA, and OmpF), suggesting that signal sequence processing (and therefore protein translocation) is more cotranslational in strains lacking TF. To test for this directly, we used an established two-dimensional gel assay (Josefsson and Randall, 1981) to determine if the timing of translocation with respect to nascent polypeptide length was affected in a Δtig mutant. 35S-labeled LamB was immunoprecipitated and resolved by size using SDS-PAGE. The protein was then subjected to partial proteolysis in gel with a site-specific protease (V8), with the proteolytic fragments separated by size in a second dimension. For wild-type cells, the prominent C-terminally derived fragments converged on the pro-LamB form, implying that a substantial fraction of the polypeptides were exported only after completion of protein synthesis (Figures 7B and S7B for a similar analysis of MBP in which the N-terminal fragments are more prominent). However, for Δtig cells, the C-terminal fragments converged on the mature form, indicating that there was significantly more cotranslational export of LamB. This switch in translocation mechanism helps explain the suppression of the translocation defect of a secB mutant by mutations in the tig gene (Ullers et al., 2007). In addition, the mechanism of translocation can affect the folding of exported proteins in the periplasm (Kadokura and Beckwith, 2009), which can partially explain the apparent OMP defect of Δtig mutants. More generally, our findings underscore the challenge in attempts to define the function of components in highly redundant systems by following the phenotypes resulting from the loss of these components.
Perspective
Here we present a strategy for the quantitative analysis of translation in bacteria using ribosome profiling. This technique provides a critical tool for decoding unknown bacterial proteomes, quantitatively monitoring translation rates and exploring the various mechanisms for regulating translation. Additionally, we present an approach to selectively profile ribosomes, which enabled us to query the substrates of the co-translationally acting chaperone TF. Our studies revealed that recruitment of TF is delayed until well after the polypeptide has emerged from the ribosome exit channel. This delay is likely to be critical in allowing other factors to engage nascent chains. In support of this notion, excess TF prevents N-terminal processing both in vitro and in vivo. Selective ribosome profiling of other factors should help decipher the logic underlying the coordinated action of the various ribosome-associated processing enzymes, targeting factors and molecular chaperones that ensure the efficient biogenesis of proteins in vivo.
The value of selective ribosome profiling is also illustrated by the identification of OMPs as critical targets of TF. Defining the function of TF has been challenging because of its redundancy with other chaperone systems, which masks the phenotypic consequence of loss of TF. By contrast, our approach can monitor the natural flux of TF substrates in unperturbed cells, which revealed that abundantly expressed OMPs were consistently among the most prominent substrates of TF. This was complemented by analysis of a comprehensive chemical genetic screen, which showed that the chemical sensitivities of the loss of TF closely resembled that seen with the loss of the OMP chaperone machinery, as well as global mass spectrometry analysis. We anticipate that this combination of quantitative phenotypic loss of function analysis and high resolution monitoring of chaperone action in unperturbed cells will be key to elucidating the in vivo function of chaperone networks.
EXPERIMENTAL PROCEDURES
General ribosome profiling
Bacterial cells were grown in LB media at 37°C to an OD600 of 0.4 to 0.5. Cells were harvested either by pre-treatment with chloramphenicol to a final concentration of 100 μg/ml or by rapid filtration. Collected cells were flash frozen in liquid nitrogen and cryogenically pulverized by mixer milling (Retsch). Pulverized cells were thawed and clarified by centrifugation. Resulting lysates were digested with MNase, quenched with EGTA and resolved by sucrose density gradients. Ribosome-protected mRNA footprints were processed as previously described (Ingolia, 2010) and deep sequenced by Illumina GA II or HiSeq2000.
Selective ribosome profiling
Bacterial cells were collected and pulverized as with the general approach, but cells were thawed directly in the presence of 2.5 mM DSP or 20 mM EDC pH 5.8 (Pierce). To quench the crosslinking reactions, lysates were brought up to 100 mM Tris pH 8.0 for DSP or 100 mM Tris pH 8.0, 250 mM glycine and 4 mM NaHCO3 for EDC. Crosslinked lysates were digested with MNase and resolved by sucrose density gradients or cushions. Ribosome pellets derived from sucrose cushions were resuspended in buffer containing 50 mM Tris pH 7.0, 200 mM NaCl, 10 mM MgCl2, 1 mM chloramphenicol, 1 mM PMSF, 0.4% Triton X-100, 0.1% NP-40 and incubated overnight on ice. The resuspended ribosome mixture was affinity purified with 220 μl of 50% Strep-Tactin sepharose (IBA, Germany) and thoroughly washed. TF-RNCs were next eluted by TEV protease treatment at room temperature for 1 hr. Typical yields ranged between 50 to 100 μg of RNA as determined by Nanodrop (Thermo Scientific). Ribosome-protected mRNA fragments were isolated and converted to a cDNA library for identification.
Supplementary Material
Acknowledgments
We thank N. Ingolia for help with ribosome profiling and data analysis; C. Chu for help with sequencing; S. Rouskin for developing a method for ligation; G. Brar and J. Dunn for developing an rRNA subtraction strategy; N. Reifenberger and B. Zachmann-Brand for help with cloning and crosslinking experiments; Thomas Silhavy for providing antibodies; Thomas Ruppert and the mass spectrometry facility at the ZMBH for experimental support; Yves Cully for graphical assistance; and members of the Bukau, Gross and Weissman labs for insightful discussions and critical readings of the manuscript. This work was supported by the SFB638 and FOR967 of the Deutsche Forschungsgemeinschaft (B.B. and G.K.); the Peter und Traudl Engelhorn-Stiftung and EMBO (A.B.); and the NIH (P01 AG10770) and the Howard Hughes Medical Institute (J.S.W.).
Footnotes
Supplemental Information includes Extended Experimental Procedures and seven figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell. 2004;117:199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- Albanèse V, Yam AYW, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Ball LA, Kaesberg P. Cleavage of the N-terminal formylmethionine residue from a bacteriophage coat protein in vitro. J Mol Biol. 1973;79:531–537. doi: 10.1016/0022-2836(73)90404-x. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Baram D, Pyetan E, Sittner A, Auerbach-Nevo T, Bashan A, Yonath A. Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action. Proc Natl Acad Sci USA. 2005;102:12017–12022. doi: 10.1073/pnas.0505581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingel-Erlenmeyer R, Kohler R, Kramer G, Sandikci A, Antolić S, Maier T, Schaffitzel C, Wiedmann B, Bukau B, Ban N. A peptide deformylase-ribosome complex reveals mechanism of nascent chain processing. Nature. 2008;452:108–111. doi: 10.1038/nature06683. [DOI] [PubMed] [Google Scholar]
- Crooke E, Wickner W. Trigger factor: a soluble protein that folds pro-OmpA into a membrane-assembly-competent form. Proc Natl Acad Sci USA. 1987;84:5216–5220. doi: 10.1073/pnas.84.15.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- Eisner G, Moser M, Schäfer U, Beck K, Müller M. Alternate recruitment of signal recognition particle and trigger factor to the signal sequence of a growing nascent polypeptide. J Biol Chem. 2006;281:7172–7179. doi: 10.1074/jbc.M511388200. [DOI] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–596. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie B, Wickner W. Trigger factor depletion or overproduction causes defective cell division but does not block protein export. J Bacteriol. 1990;172:5555–5562. doi: 10.1128/jb.172.10.5555-5562.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan CL, Silhavy TJ, Kahne DE. β-Barrel Membrane Protein Assembly by the Bam Complex. Annu Rev Biochem. 2010 doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Bukau B, Kramer G. Structure and function of the molecular chaperone Trigger Factor. Biochim Biophys Acta. 2010;1803:650–661. doi: 10.1016/j.bbamcr.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Merz F, Rutkowska A, Zachmann-Brand B, Deuerling E, Bukau B. Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J Biol Chem. 2006;281:6539–6545. doi: 10.1074/jbc.M512345200. [DOI] [PubMed] [Google Scholar]
- Huber D, Rajagopalan N, Preissler S, Rocco MA, Merz F, Kramer G, Bukau B. SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol Cell. 2011;41:343–353. doi: 10.1016/j.molcel.2010.12.028. [DOI] [PubMed] [Google Scholar]
- Hundley HA, Walter W, Bairstow S, Craig EA. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science. 2005;308:1032–1034. doi: 10.1126/science.1109247. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Meth Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson LG, Randall LL. Different exported proteins in E. coli show differences in the temporal mode of processing in vivo. Cell. 1981;25:151–157. doi: 10.1016/0092-8674(81)90239-7. [DOI] [PubMed] [Google Scholar]
- Kadokura H, Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138:1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–460. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwülbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- Kramer G, Rutkowska A, Wegrzyn RD, Patzelt H, Kurz TA, Merz F, Rauch T, Vorderwülbecke S, Deuerling E, Bukau B. Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains. J Bacteriol. 2004;186:3777–3784. doi: 10.1128/JB.186.12.3777-3784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Bernstein HD. Trigger factor retards protein export in Escherichia coli. J Biol Chem. 2002;277:43527–43535. doi: 10.1074/jbc.M205950200. [DOI] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hackert E, Hendrickson WA. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell. 2009;138:923–934. doi: 10.1016/j.cell.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, et al. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J. 2008;27:1622–1632. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz F, Hoffmann A, Rutkowska A, Zachmann-Brand B, Bukau B, Deuerling E. The C-terminal domain of Escherichia coli trigger factor represents the central module of its chaperone activity. J Biol Chem. 2006;281:31963–31971. doi: 10.1074/jbc.M605164200. [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzelt H, Kramer G, Rauch T, Schönfeld HJ, Bukau B, Deuerling E. Three-state equilibrium of Escherichia coli trigger factor. Biol Chem. 2002;383:1611–1619. doi: 10.1515/BC.2002.182. [DOI] [PubMed] [Google Scholar]
- Randall LL, Hardy SJS. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci. 2002;59:1617–1623. doi: 10.1007/PL00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs PD, Derman AI, Beckwith J. A mutation affecting the regulation of a secA-lacZ fusion defines a new sec gene. Genetics. 1988;118:571–579. doi: 10.1093/genetics/118.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowska A, Mayer MP, Hoffmann A, Merz F, Zachmann-Brand B, Schaffitzel C, Ban N, Deuerling E, Bukau B. Dynamics of trigger factor interaction with translating ribosomes. J Biol Chem. 2008;283:4124–4132. doi: 10.1074/jbc.M708294200. [DOI] [PubMed] [Google Scholar]
- Schlünzen F, Wilson DN, Tian P, Harms JM, McInnes SJ, Hansen HAS, Albrecht R, Buerger J, Wilbanks SM, Fucini P. The binding mode of the trigger factor on the ribosome: implications for protein folding and SRP interaction. Structure. 2005;13:1685–1694. doi: 10.1016/j.str.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999;97:755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- Tomic S, Johnson AE, Hartl FU, Etchells SA. Exploring the capacity of trigger factor to function as a shield for ribosome bound polypeptide chains. FEBS Lett. 2006;580:72–76. doi: 10.1016/j.febslet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- Ullers RS, Ang D, Schwager F, Georgopoulos C, Genevaux P. Trigger Factor can antagonize both SecB and DnaK/DnaJ chaperone functions in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:3101–3106. doi: 10.1073/pnas.0608232104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullers RS, Houben ENG, Brunner J, Oudega B, Harms N, Luirink J. Sequence-specific interactions of nascent Escherichia coli polypeptides with trigger factor and signal recognition particle. J Biol Chem. 2006;281:13999–14005. doi: 10.1074/jbc.M600638200. [DOI] [PubMed] [Google Scholar]
- Ullers RS, Houben ENG, Raine A, ten Hagen-Jongman CM, Ehrenberg M, Brunner J, Oudega B, Harms N, Luirink J. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J Cell Biol. 2003;161:679–684. doi: 10.1083/jcb.200302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.