Abstract
Aggregation-prone proteins often misfold into multiple distinct amyloid conformations dictating their different physiological impacts. Although amyloid formation is triggered by a transient nucleus, the mechanism of how an initial nucleus is formed and allows the protein to form a specific amyloid conformation remains unclear. Here we show that, prior to fiber formation, the prion domain (Sup35NM, consisting of residues 1-254) of yeast prion Sup35, the [PSI+] protein determinant, forms oligomers in a temperature-dependent, reversible manner. Mutational and biophysical analyses revealed that “nonnative” aromatic interactions outside of the amyloid core drive oligomer formation by bringing different Sup35NM monomers together, which specifically leads to the formation of highly infectious strain conformations with more limited amyloid cores. Thus, transient nonnative interactions in the initial nucleus play pivotal roles in determining the diversity of amyloid conformations and resulting prion strain phenotypes.
A ubiquitous feature of protein misfolding is that the same polypeptide can adopt structurally distinct fibrillar β-sheet-rich aggregates termed amyloid fibers with the specific misfolded conformation dictating its physiological impact1-3. A striking example of this is the prion strain phenomenon, in which prion particles composed of the same protein cause distinct heritable states4. In the yeast prion [PSI+], the prion state results from self-propagating β-sheet-rich aggregates (amyloid) of the translation termination factor Sup35 (5-7). Inactivation of Sup35 due to the aggregation causes a nonsense suppression phenotype that can be readily monitored using an ade1 nonsense reporter gene whereas in non-prion states [psi−] colonies require adenine and accumulate a red pigment. Strikingly, like mammalian prions4, [PSI+] exhibits a range of heritable strain variants: a variety of strain variants showing distinct color phenotypes from dark pink to white exist depending on the degree of Sup35 inactivation8,9. Synthetic Sup35NM prions created at different temperatures adopt distinct conformations and introduction of the different prion conformations into yeast leads to readily distinguishable and stably inherited differences in the resulting [PSI+] strains. Specifically, Sup35NM fibers formed at 4°C (Sc4) and 37°C (Sc37) induced strong and weak [PSI+] strains, respectively10. This study established that the ability of Sup35 to misfold into multiple amyloid conformations constitutes the physical foundation of the strain phenomenon.
The self-propagating nature of amyloids implies that an initial nucleus formed in the very early phase of protein fibrillation may encode key structural information that determines the conformation of the final amyloid state. Owing to the transient nature of the initial nucleus, however, our understanding of the interactions that form the initial nucleus and how such species allow proteins to overcome the strong kinetic barrier to form a specific amyloid conformation is highly limited11-15. The abilities to create different infectious conformations of Sup35NM in a test tube and introduce them in yeast10,16 make the [PSI+] system uniquely well suited for investigating the mechanism by which a single polypeptide (Sup35NM) under the different polymerization conditions forms initial nuclei and misfolds into self-propagating distinct amyloid conformations.
In this study, we revealed that nonnative interactions outside of the amyloid core drive oligomer formation of Sup35NM, which specifically leads to the formation of highly infectious amyloid conformations. Our findings established that the initial nucleus plays pivotal roles in determining the diversity of prion strain conformations and resulting strain phenotypes.
RESULTS
Oligomers are formed in a temperature-dependent manner
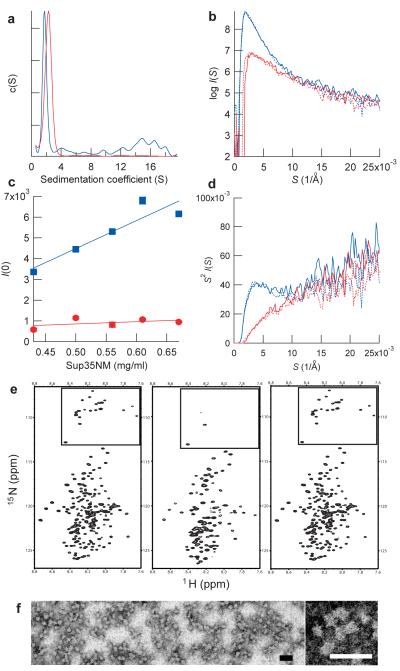
We first examined secondary structures of Sup35NM in 5mM potassium phosphate buffer containing 150mM sodium chloride (pH7.4) by circular dichroism (CD) spectroscopy. At both 4°C and 37°C, prior to polymerization, Sup35NM showed CD spectra typical of disordered proteins although, consistent with an earlier report17, the 37°C form showed evidence for modest structure (Supplementary Fig. 1). Under the same condition, we next investigated the oligomeric state of Sup35NM at 4°C and 37°C by analytical ultracentrifugation (AUC). Remarkably, we found that at 4°C Sup35NM forms oligomers whose sedimentation coefficients are between 4S and 20S, whereas at 37°C Sup35NM was solely in a monomeric state (Fig. 1a) as previously observed for Sup35NM at 25°C (18). The AUC analysis suggests oligomers are formed by association of 4-5 Sup35NM monomers (Supplementary Fig. 2) and that under the present concentration (10μM) roughly half of the protein forms oligomers with the remainder being monomeric.
Figure 1. Sup35NM forms oligomers in a temperature-dependent, reversible manner.
(a) Sedimentation coefficient distributions of Sup35NM at 4°C (blue) and 37°C (red).
(b) SAXS profile of Sup35NM protein. SAXS profile was first measured at 4°C (blue solid line) and the temperature was increased to 37°C (red solid line), returned to 4 °C (blue broken line) and finally shifted again to 37°C (red broken line).
(c) Concentration-dependent changes in scattering intensities of Sup35NM protein at 4°C. Maximum scattering intensities of Sup35NM protein at 4°C (blue) and 37 °C (red) were plotted against the concentration of Sup35NM.
(d) Kratky plot of the SAXS profile shown in b. The peak at 4 Å−1 for Sup35NM at 4°C indicates the presence of globular structures20.
(e) HSQC NMR spectra of Sup35NM protein. Spectra were first measured at 25 °C (left), the temperature was decreased to 10 °C (center), and finally returned to 25 °C (right). The boxed area shows signals from glycines in Sup35NM.
(f), Images of Sup35NM oligomer by transmission electron microscopy (left). A zoomed view of Sup35NM oligomer (right). Scale bar, 50 nm.
We further characterized the oligomeric species, by measuring small angle X-ray scattering (SAXS) with synchrotron X-ray radiation, which is highly sensitive to detection of soluble oligomers19. At 37°C, Sup35NM monomer has a radius of gyration (Rg) of 36.4±1.4 Å (Fig. 1b), which is consistent with an extended monomeric structure17. In contrast, at 4°C Sup35NM showed increased scattering intensities with Rg of 79.2±1.4 Å (Fig. 1b), confirming the presence of oligomers under these conditions (Fig. 1a). Interestingly, the oligomer formation was completely reversible in a temperature-dependent manner as the SAXS profile of Sup35NM monomer at 37°C was indistinguishable from that of Sup35NM following repeated temperature shifts between 4°C and 37°C (Fig. 1b). We also found a concentration-dependent increase in the scattering intensity for Sup35NM at 4°C but not 37°C (Fig. 1c). A Kratky plot of the SAXS profile of Sup35NM at 4°C showed a peak at S (= 2sinθ/λ) = 4.0±0.2 Å−1 (Fig. 1d), suggesting that the Sup35NM oligomer formed at 4°C has a globular structure20.
The temperature-dependent reversible formation of Sup35NM oligomer was readily monitored by NMR spectroscopy (Fig. 1e). A large fraction of the peaks in the hetero-nuclear single quantum coherence (HSQC) spectrum of Sup35NM disappeared at low temperature due to the slow tumbling rate of oligomeric species but those signals were quantitatively restored by increasing temperature. The formation of Sup35NM oligomer was further confirmed by the analysis with transmission electron microscopy (TEM), which showed spherical particles with an average diameter of 15-20nm (Fig. 1f). Taken together, these results established that as has been seen with other amyloid forming proteins21-26 and consistent with the earlier observations of Serio and co-workers15, Sup35NM forms oligomers prior to amyloid formation but the oligomer formation is seen only at low temperatures.
Oligomers are on pathway to Sc4 amyloid formation
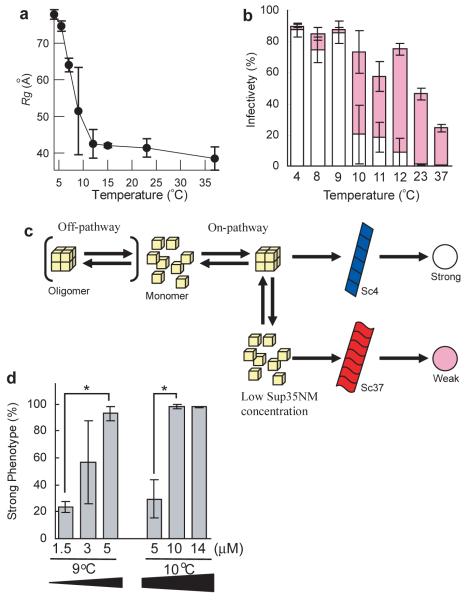
We next explored whether the Sup35NM oligomers played an important kinetic role in determining the prion strain conformation that results from a polymerization reaction. First, we searched for the highest temperature at which oligomers are formed. SAXS analysis (Fig. 2a) revealed that Sup35NM oligomers begin to be detected below 9°C. We then investigated how temperature influenced the final prion strain conformation Sup35NM adopted, using the protein infection assay to introduce Sup35NM amyloids formed at various temperatures into [psi−] yeast10. The temperature dependence to accumulation of the oligomeric species prior to polymerization precisely mirrored formation of the Sc4 prion conformation that leads to the induction of strong [PSI+] phenotypes: Sup35NM amyloids formed below 9°C and above 10°C induced strong (white) and weak (pink) [PSI+] strains, which are readily distinguishable by their color phenotypes on rich media, respectively (Fig. 2b). If the oligomer is on pathway to formation of the Sc4 conformation, we expect that increasing the Sup35NM oligomer by increasing the Sup35NM concentration would favor formation of the Sc4 strain conformation (Fig. 2c). Accordingly, we found that increasing Sup35NM concentration from 5μM to 14μM at 10°C increased the fraction of strong (Sc4) [PSI+] strains, while the decrease of Sup35NM concentration from 5μM to 1.5μM at 9°C reduced the proportion of strong [PSI+] strains (Fig. 2d). Taken together, these studies indicate that the Sup35NM oligomer formed at low temperatures is an intermediate that specifically facilitates conversion to the Sc4 strain conformation.
Figure 2. Low temperature Sup35NM oligomers are on pathway to formation of Sc4 amyloid fibers.
(a) Temperature-dependent changes in the amounts of Sup35NM oligomer. Rg values determined by the Guinier approximation are plotted against temperature.
(b) Relationships between levels of Sup35NM oligomer and prion strain phenotypes. Sup35NM amyloids formed at various temperatures were introduced into [psi−] yeast and resulting degree of prion infectivity as well as a fraction of strong (white bar) and weak (pink bar) [PSI+] phenotypes was determined. The infection experiments were performed more than twice and fractions (%) of the strain phenotypes were calculated using more than 100 colonies in each experiment.
(c) Schematic representation of on-pathway or off-pathway Sup35NM oligomer in the fiber formation.
(d) Effects of changes in concentration of Sup35NM protein on the ratio of strong to weak [PSI+] strain phenotypes. Asterisk denotes P<0.05.
Distinct residues control nucleation and amyloid growth
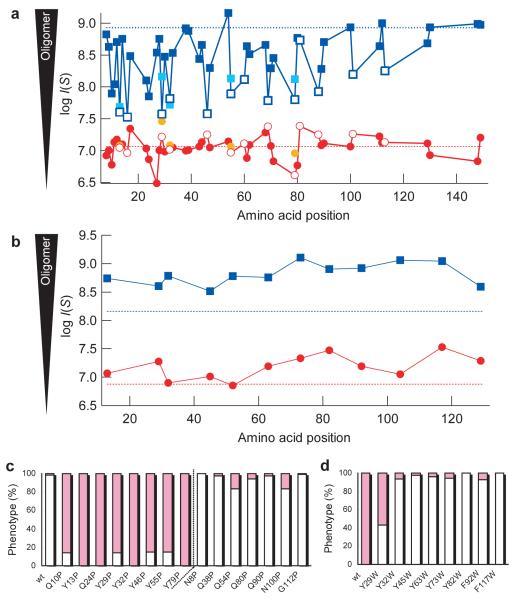
We next examined the structural properties of the Sup35NM oligomer by monitoring the effects of a variety of proline mutants on oligomer formation at 4°C. Earlier amide hydrogen exchange and proline scanning studies indicated the amyloid core of Sc4 conformation was largely restricted to the first 35 amino acids whereas there is a dramatic expansion (near doubling) of the amyloid core in the Sc37 conformation27. Interestingly, in marked contrast to the effects on growth of the Sc4 fibers, mutations across the entire prion domain decreased oligomer formation, indicating that amino acid residues involved in the oligomer formation extend far beyond the Sc4 amyloid core (Fig. 3a). In particular, replacement of most tyrosines, which are located throughout the prion domain with proline, strongly decreased oligomer formation and disfavored formation of the Sc4 strain conformation as assessed by the prion infection assay (Fig. 3a and c). The amount of oligomer in the mutants agreed well with the fraction of the Sc4 strain conformation (Fig. 3a and c). In the infection assay, the primary sequence of Sup35 between the donor (proline mutants) and the recipient (wild-type) is different. However, we revealed that wild-type Sup35NM protein could be readily recruited into the seeds of proline mutants and infection of [psi−] yeast with the resulting wild-type amyloids also led to similar fractions of strong [PSI+] strains (Supplementary Fig. 3). Thus, the in vitro generated strain conformations determined by the amount of oligomer, rather than the incompatibility of Sup35 sequence, would be responsible for the observed change in prion strength (Fig. 3c). These results confirmed the roles of the oligomer in formation of the Sc4 strain conformation (Fig. 2b).
Figure 3. Distinct amino acid regions in the prion domain of Sup35NM are involved in nucleation and amyloid growth.
(a) Effects of mutations on oligomer formation. Maximum SAXS intensities of Sup35NM proline mutants (4°C, blue; 37°C, red) and leucine mutants (4°C, cyan; 37°C, orange) were plotted against amino acid position of the mutations. The dotted lines denote the intensity of wild-type Sup35NM at 4°C (upper, blue) and 37°C (lower, red). Tyrosine residues are highlighted by open symbols.
(b) Effects of tryptophan mutations on oligomer formation. Maximum SAXS intensities of Sup35NM tryptophan mutants (4°C, blue; 37°C, red) were plotted against amino acid position of the mutations. The dotted lines indicate intensities of wild type Sup35NM at 4°C (upper, blue) and 37°C (lower, red).
(c,d) Effects of proline (c) or tryptophan (d) mutants on prion strain phenotypes. Fractions of strong (white bar) and weak (pink bar) [PSI+] phenotypes were determined. Fibers of Sup35NM proline and tryptophan mutants were formed at 8°C and 11°C, respectively, and used for amyloid infection.
We also substituted five tyrosine residues (Tyr13, 29, 32, 55 or 79), that we had found to be highly sensitive to proline substitution, with leucine which has an increased hydrophobicity but lacks an aromatic ring. These mutations also decreased oligomer formation (Fig. 3a), indicating that the aromatic ring, rather than solely the hydrophobicity, in tyrosine drives oligomer formation. The critical role of the aromatic rings in oligomer formation was confirmed by the increase in amounts of oligomer as well as the fraction of strong [PSI+] variants following amyloid infection, when tyrosine residues were replaced with tryptophan (Fig. 3b and d).
The NMR analysis further supports the proposal that the oligomer structure extends well beyond the 35 amino acid amyloid core found in Sc4 as the 21 glycine residues that are found throughout the 123 amino acid N-terminal domain became less mobile upon oligomer formation. Thus, they are not observed in low temperature HSQC spectra whereas the two glycine residues in the charged middle domain are observable, and therefore highly mobile, in both high and low temperature spectra (Fig. 1e). Taken together, these results revealed that interactions driving formation of the initial nucleus (oligomer) are distinct from those involved in the final amyloid structure, emphasizing that nucleation and amyloid growth are different processes. These findings also help explain the previous observation that increasing the number of the tyrosine-rich oligo repeat sequences found in Sup35NM promotes de novo prion formation28.
Nonnative interactions drive formation of oligomers
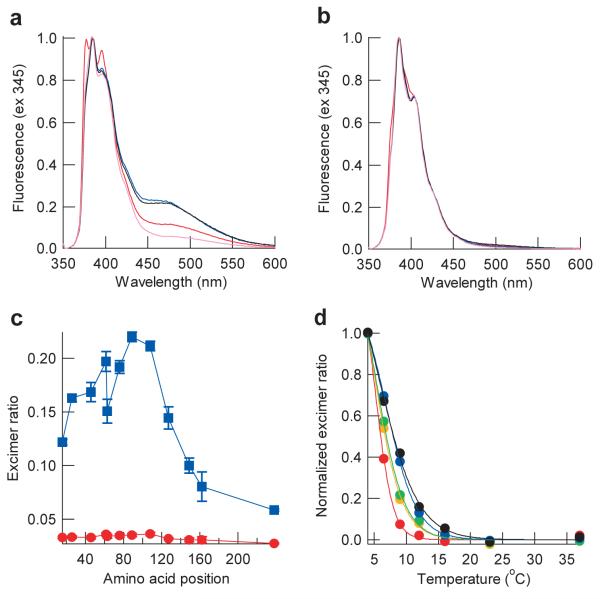
To further characterize the structure of Sup35NM oligomers, we prepared pyrene-labeled Sup35NM which allows us to detect excimer fluorescence when labeled residues from different Sup35NM molecules are in close proximity (~10Å)29. A variety of single cysteine mutants were labeled by pyrene and fluorescence spectra of the SupNM mutants were measured at various temperatures. As expected, excimer fluorescence of pyrene-labeled Sup35NM was observed at 4°C due to the oligomer formation but not at 37°C (Fig. 4a and b) and the fluorescence intensities at 4°C were dependent on the pyrene positions introduced into Sup35NM (Fig. 4c). Interestingly, pyrene placed C-terminal (specifically residues 62, 76, 89 and 108) to the Sc4 amyloid core showed the highest excimer signal (Fig. 4c). Moreover, these regions mediating intermolecular interactions in the oligomer are distinct from those observed previously to be involved in intermolecular contacts in the fiber using a similar pyrene strategy, which showed Sup35NM fibers adopted “head to head” and “tail to tail” interactions involving residues 10-38 and 85-120, respectively29,30. Our pyrene data are also consistent with the mutational data showing that the low temperature oligomer is stabilized by interactions that extend far beyond the Sc4 amyloid core (Fig. 3a). Furthermore, the excimer fluorescence intensity of pyrene-labeled Sup35NM mutants showed different dependence on temperature. Upon decrease in temperature from 37°C, excimer fluorescence was first detected for the C-terminal mutants, followed by the N-terminal mutants (Fig. 4d and Supplementary Fig. 4). Thus, the C-terminal residues in the prion domain are first assembled in the oligomer and this, in turn, facilitates the subsequent polymerization by the N-terminal part of the prion domain, underscoring the mechanistic and structural differences between the nucleation and amyloid growth processes.
Figure 4. Nonnative interactions outside of the Sc4 amyloid core drive oligomer formation of Sup35NM.
(a,b) Normalized excimer fluorescence spectra of pyrene-labeled Sup35NM mutants at 4°C (a) or 37°C (b). Shown are fluorescence spectra of Cys16 (red), Cys89 (blue), Cys108 (black), and Cys238 (pink) mutants.
c) The ratio of excimer fluorescence intensities (470nm) to non-excimer fluorescence intensities (385 nm). Normalized intensities of excimer fluorescence of the pyrene-labeled Sup35NM mutants at 4°C (blue) or 37°C (red) were plotted against amino acid position.
(d) Normalized intensities of excimer fluorescence are plotted against temperature for pyrene-labeled Sup35NM Cys16 (red), Cys26 (orange), Cys46 (yellow), Cys89 (blue) and Cys108 (black) mutants.
In agreement with this conclusion, several observations indicate that despite the roles of the oligomer in facilitating de novo formation of the Sc4 conformation, this species is not involved in growth of the Sc4 fiber. First, Sup35NM polymerization reactions carried out at 37°C, conditions where the oligomer is absent, but seeded with Sc4 fibers lead to formation of the Sc4 strain conformation (Supplementary Fig. 5). Second, the average diameter of the Sup35NM oligomers (15-20nm) (Fig. 1e) is larger than that of Sup35NM amyloid (~10nm)31,32 and we did not observe Sc4 fibers formed by aligned oligomers by TEM analysis32. Moreover, the reactivity of Sup35NM oligomer with an amyloid-specific dye, thioflavine T, was much lower than that of fibers (Supplementary Fig. 6). Finally, the Y46P mutant, which had previously been found to strongly interfere with the growth of Sc37 but not Sc4 fibers27, showed a precisely opposite preference during de novo fiber formation: Y46P strongly interfered with oligomer formation and favored formation of the Sc37 strain conformation (Fig. 3a) despite its strong defect in fiber growth of Sc37 conformation.
Thus, the oligomer plays a role of the scaffold where Sup35NM monomers bind and Sc4 amyloids begin to grow. Nonetheless, we found that initially formed oligomers do not seed monomeric Sup35NM (Supplementary Fig. 7), indicating that structural reorganization in the oligomer would be required for the oligomer to have the seeding activity. This result is consistent with the observation that there is a lag time in spontaneous formation of Sup35NM amyloids at 4°C (Supplementary Fig. 8a) despite the presence of substantial amounts of oligomers immediately after decreasing temperature to 4°C (Fig. 1b).
DISCUSSION
Owing to their transient nature, our mechanistic and structural understanding of the species that promote de novo fiber formation has lagged far beyond that of those involved in protofibril formation and amyloid growth27,29,33,34. Yet it is such species that play a central role in determining when a protein aggregates and what conformation it adopts during polymerization. One critical issue has been the oligomeric state of nuclei that precede formation of stable amyloid seeds. On the one hand, kinetic analysis of the concentration dependence of polymerization of relatively long stretches of polyglutamine suggested the nucleus was monomeric35. In addition, the earlier and present kinetic studies imply that de novo formation of Sup35NM fibers at 25°C (18) and 37°C (Supplementary Fig. 9), both of which lead to weak [PSI+] phenotypes, involves a very small (and potentially even monomeric) nucleus although it could be a larger one such as hexamer36. However, for many proteins, prior to polymerization, spectra of oligomeric species are ubiquitously present and have been suggested to facilitate seed formation15,24-26. Our present results regarding the formation mechanism of the two different strain conformations suggest unification between these two views as we found with Sup35 that both of the fibrillation pathways15,18 can occur, depending on temperature. While oligomeric species may not be required for amyloid conversion per se, such oligomeric species, driven by “nonnative” interactions distinct from those found in the amyloid form play a key role in determining which fiber conformation a protein adopts.
However, we could not rule out the possibility that other factors also regulate the strain conformations. For instance, strong prion conformation may simply get a selective advantage independent of the oligomer in the in vitro condition under which overall rates of prion formation increase. In addition, the oligomer may prevent formation of the weak (Sc37) strain conformation by hiding certain regions of the protein and therefore preventing formation of a larger amyloid core, rather than promoting formation of the strong (Sc4) strain conformation. Nonetheless, the oligomer is an on-pathway intermediate to the Sc4 amyloid formation and the amounts of oligomers correlated well with the fractions of strong [PSI+] phenotypes (Figs. 2 and 4). These results establish that the oligomer is closely involved in formation of the Sc4 strain conformation.
Despite the roles of the oligomer in formation of the strong strain conformation, the oligomer does not accelerate formation of Sc4 amyloid. Our kinetics analysis on spontaneous amyloid formation of proline and tryptophan mutants revealed that some mutants which inhibited or promoted oligomer formation showed kinetic profiles similar to wild-type Sup35NM (Supplementary Fig. 8). Therefore, the amount of oligomer does not necessarily correlate with the rate of de novo amyloid formation. This, in turn, suggests that structural reorganization in the oligomer after its initial formation is a rate-determining step.
Why then would oligomerization driven by nonnative interactions favor formation of specific fiber forms? A key distinction between the Sc4 conformation, favored by oligomers, from the Sc37 conformation is that the amyloid core in Sc4 is confined to roughly the first 35 amino acids whereas it is dramatically expanded in Sc37 (27). It is expected that formation of the very small nucleus which has a potential to lead to the weak strain conformation with an extended amyloid core (such as Sc37) would not be entropically disfavorable. In contrast, there would be a strong entropic barrier to bringing more Sup35NM monomers together to form the minimum stable core in Sc4 amyloids. The oligomeric species we observe would then be ideally suited to overcome this entropic barrier, as aromatic interactions C-terminal to the amyloid core could drive oligomerization by enthalpic effects. The aromatic interactions, in turn, bring multiple N-terminal amyloid core regions in close proximity overcoming the entropic barrier while keeping them free to polymerize (Fig. 5). The resulting amyloid with a limited core has a high propensity to fragment and yields more seeds by chaperone-mediated fiber breakage37-39, and the resulting increase in seed number in turn leads to strong [PSI+] phenotypes40. A critical role of regions flanking the amyloid core in controlling how protein polymerizes has also been suggested by recent studies on CFP-fused polyglutamine peptides and a truncated form of huntingtin, the causative protein of Huntington disease41,42, although the mechanism of these effects have not been elucidated. One intriguing possibility is that, as we observed with Sup35, these flanking regions promote oligomerization which both facilitates protein polymerization and alters the conformation of the resulting amyloid forms.
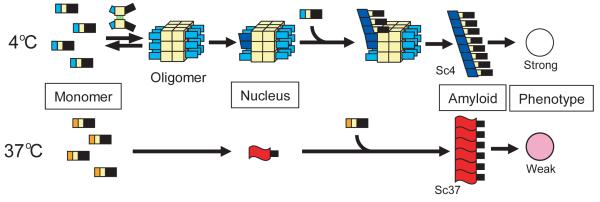
Figure 5. Global relationships between Sup35NM monomer, initial nucleus, amyloid conformation and prion strain phenotype.
Nonnative interactions (green lines) outside of the Sc4 amyloid core drive oligomer formation. This interaction brings Sup35NM monomers in close proximity while leaving the amyloid core regions free to interact, which eventually leads to highly infectious Sc4 strain conformations which have more limited amyloid cores. The nucleus in the fibrillation at 37°C could be larger than monomer36 but is still much smaller than the oligomer formed at 4°C.
In summary, we revealed that nonnative interactions in a transient nucleus drive oligomer formation, which specifically leads to the formation of highly infectious strain conformations with more limited amyloid cores. Thus, the initial nucleus plays pivotal roles in determining the conformation of final amyloid states and their physiological impacts. Our conclusion provides a basis of understanding how distinct amyloid conformations that dictate different physiological effects arise from a single polypeptide.
METHODS
Purification of Sup35NM protein and amyloid formation
Plasmids for bacterial expression of Sup35NM mutants were made using QuickChange site-directed mutagenesis (Stratagene). Nonlabeled or uniformly 15N labeled Sup35NM protein with a carboxy-terminal 7x-histidine tag was overexpressed in the bacterial strain BL21(DE3) and purified in a denatured condition as previously reported10,27. Single cysteine Sup35NM mutants were purified in the presence of 10mM 2-mercaptoethanol. The purified protein was passed through a 100KDa filter to remove pre-existing aggregates and stored at −80°C. Sup35NM amyloid was formed spontaneously or in the presence of 5-10% (mol/mol) indicated seeds at 4 or 37°C overnight as previously described10.
In vitro analysis of Sup35NM monomer and oligomers
Velocity AUC experiments were performed in a Beckman Optima XL-I analytical ultracentrifuge using an An60Ti rotor at 4 or 37°C. Velocity sedimentation was analyzed at a speed of 40,000rpm at a Sup35NM concentration of 10μM. Sedimentation data were collected at radial steps of 0.003cm and scanned every 9min. Data were analyzed with Sedfit using c(S) distribution method (http://www.analyticalultracentrifugation.com)18.
The measurement of SAXS was performed at the SAXS-dedicated beamline BL45XU (43) in SPring-8, operated by RIKEN, Japan. The sample-to-detector distance was 2.2m and the wavelength used was 0.9Å. The temperature of the sample (typically 18μM) was controlled by a high-precision peltier thermal controller (±0.05°C). Using an X-ray image intensifier and cooled CCD detector (XR-II+CCD)44, each scattering image was collected for 1sec during which no radiation damage was found. The radius of gyration (Rg) was determined by the Guinier approximation45. The Kratsky plot was calculated as previously reported46.
For NMR experiments, 15N-labeled Sup35NM protein in a native condition was passed through a 100KDa filter to remove pre-existing aggregates. 15N-HSQC spectra of 15N-labeled Sup35NM (300μM) in 5mM potassium phosphate buffer including 150mM sodium chloride and 10% (vol/vol) deuterium oxide were measured at 10 or 25°C on an 800MHz NMR spectrophotometer (Advance 800) equipped with cryoprobe (Brucker) as reported previosuly27.
For TEM analysis, an aliquot of 5μM Sup35NM wild-type solutions at 4°C was spread onto prechilled carbon-coated 400-mesh copper grids at 4°C and allowed to stand for 2min. The sample was fixed with 2% paraformaldehyde and 2.5% glutaraldehyde for 10min and washed with water, followed by staining with 2% phosphotungstic acid (pH7.5). Morphology of Sup35NM protein was investigated by electron microscopy (LEO).
For experiments for pyrene excimer fluorescence, a single cysteine mutant was labeled with pyrene maleimide in 6 M guanidine hydrochloride as recommended by Molecular Probes and purified with reverse phase HPLC chromatography, followed by lyophilization. The lyophilized protein was dissolved in 5mM phosphate buffer including 150mM sodium chloride and passed through a 100KDa filter to remove pre-existing aggregates. Spectra of excimer fluorescence of the labeled protein (5μM) were acquired with the excitation of 340nm and the emission of 345-600nm on a fluorescence spectrophotometer (FL-7000, Hitachi) equipped with a peltier thermal controller. The ratios of fluorescence intensities of pyrene excimer and monomer were determined by relative fluorescence intensities at 470 nm and 385nm, respectively. The labeling efficiency (>80%) was calculated using the extinction coefficient of pyrene (22,000 M−1cm−1 at 343nm)47.
Introduction of in vitro Sup35NM amyloid into yeast and determination of prion strain phenotypes
For amyloid infection, a [psi−][PIN+] 74D-694 yeast strain [MATa, his3, leu2, trp1, ura3; suppressible marker ade1-14(UGA)] was used and the infection procedure was described previously10. For the infection experiments with amyloids of Sup35NM mutants, amyloids of the proline and tryptophan mutants were formed at 8°C and 11°C, respectively.
The detailed procedures are described in Supplementary Methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Y. Sakamaki and T. Akagi for help with the EM experiments and S. Akiyama for discussion on the SAXS measurements. Y.O. is supported by JSPS postdoctoral fellowships. Funding was provided by JST PRESTO, Grants in MEXT and MHLW, The Uehara Memorial Foundation (M.T.), Howard Hughes Medical Institute and the National Institutes of Health (J.S.W.).
Footnotes
AUTHOR CONTRIBUTIONS
M.T. and Y.O. designed the experiments. Y.O. performed most experiments, K. I. contributed to setup of the SAXS experiments and B.H.T. helped the NMR experiments. B.H.T. and J.S.W. provided new reagents. Y.O., J.S.W. and M.T. analyzed, discussed the data and wrote the paper.
COMPETING FINANCIAL INTEREST
I declare that the authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
References
- 1.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu. Rev. Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr. Opin. Struct. Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 5.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 6.Tuite MF, Koloteva-Levin N. Propagating prions in fungi and mammals. Mol Cell. 2004;14:541–552. doi: 10.1016/j.molcel.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Tessier P, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat. Struct. Mol. Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kochneva-Pervukhova NV, et al. [PSI+] prion generation in yeast: characterization of the ‘strain’ difference. Yeast. 2001;18:489–497. doi: 10.1002/yea.700. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 11.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 12.Johnson SM, et al. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses. Acc. Chem. Res. 2005;38:911–921. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 13.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell. Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 14.Balch WE, Morimoto RI, Dillin A, Kelly JW. Science. 2009;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 15.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 16.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 17.Scheibel T, Lindquist SL. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat. Struct. Biol. 2001;11:958–962. doi: 10.1038/nsb1101-958. [DOI] [PubMed] [Google Scholar]
- 18.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka M, et al. Expansion of polyglutamine induces the formation of quasi-aggregate in the early stage of protein fibrillization. J. Biol. Chem. 2003;278:34717–34724. doi: 10.1074/jbc.M209852200. [DOI] [PubMed] [Google Scholar]
- 20.Glatter O, Kratky O. Small Angle X-ray Scattering. Academic Press; New York: 1982. [Google Scholar]
- 21.Walsh DM, et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 22.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 23.Lee S, Eisenberg D. Seeded conversion of recombinant prion protein to a disulfide-bonded oligomer by a reduction-oxidation process. Nat. Struct. Biol. 2003;10:725–730. doi: 10.1038/nsb961. [DOI] [PubMed] [Google Scholar]
- 24.Plakoutsi G, et al. Evidence for a mechanism of amyloid formation involving molecular reorganisation within native-like precursor aggregates. J. Mol. Biol. 2005;351:910–922. doi: 10.1016/j.jmb.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 25.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat. Struct. Mol. Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 26.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 27.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 28.Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature. 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- 29.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447:556–561. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, Ferrone FA, Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue WF, Homans SW, Radford SE. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8926–8931. doi: 10.1073/pnas.0711664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 38.Inoue Y, Taguchi H, Kishimoto A, Yoshida M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J. Biol. Chem. 2004;279:52319–52323. doi: 10.1074/jbc.M408159200. [DOI] [PubMed] [Google Scholar]
- 39.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 41.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thakur AK, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujisawa T, et al. Small-angle X-ray scattering station at the SPring-8 RIKEN beamline. J. Appl. Crystallogr. 2000;33:797–800. [Google Scholar]
- 44.Ameyama Y, et al. Large-aperture TV detector with a beryllium-windowed image intensifier for x-ray diffraction. Rev. Sci. Instrum. 1995;66:2290–2294. [Google Scholar]
- 45.Guinier A, Fournet G. Small-Angle Scattering of. X-rays. John Wiley & Sons; New York: 1955. [Google Scholar]
- 46.Svergun DI, Semenyuk AV, Feigin LA. Small-angle-scattering-data treatment by the regularization method. Acta. Cryst. 1988;44:244–250. [Google Scholar]
- 47.Lehrer SS. Intramolecular Pyrene Excimer Fluorescence: A Probe of Proximity and Protein Conformational Change. Methods Enzymol. 1997;278:286–295. doi: 10.1016/s0076-6879(97)78015-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.