Summary
Atg7 is a noncanonical, homodimeric E1 enzyme that interacts with the noncanonical E2 enzyme, Atg3, to mediate conjugation of the ubiquitin-like protein (UBL) Atg8 during autophagy. Here we report that the unique N-terminal domain of Atg7 (Atg7NTD) recruits a unique “flexible region” from Atg3 (Atg3FR). The structure of an Atg7NTD-Atg3FR complex reveals hydrophobic residues from Atg3 engaging a conserved groove in Atg7, important for Atg8 conjugation. We also report the structure of the homodimeric Atg7 C-terminal domain, which is homologous to canonical E1s and bacterial antecedents. The structures, SAXS, and cross-linking data allow modeling of a full-length, dimeric (Atg7∼Atg8-Atg3)2 complex. The model and biochemical data provide a rationale for Atg7 dimerization: Atg8 is transferred in trans from the catalytic Cys of one Atg7 protomer to Atg3 bound to the N-terminal domain of the opposite Atg7 protomer within the homodimer. The studies reveal a distinctive E1∼UBL-E2 architecture for enzymes mediating autophagy.
Introduction
In response to cues such as low cellular energy, eukaryotic cells initiate the degradative process of macroautophagy (hereafter referred to as autophagy) (McEwan and Dikic, 2011; Mizushima and Levine, 2010; Nakatogawa et al., 2009; Yang and Klionsky, 2009; Yorimitsu and Klionsky, 2005). During autophagy, a double-membrane vesicle, termed autophagosome, engulfs part of the cytoplasm and fuses with a lysosome or vacuole, and its cytosolic contents are ultimately degraded and recycled. At least 18 distinct so-called “Atg” proteins are key core components for autophagosomal membrane formation (Nakatogawa et al., 2009; Suzuki et al., 2001). These include the ubiquitin-like protein (UBL) Atg8 in yeast, which has six orthologs in mammals including LC3 (Ichimura et al., 2000; Schulman and Harper, 2009). Atg8 is unique among UBLs in being ligated to a lipid, phosphatidylethanolamine (PE) (Ichimura et al., 2004; Ichimura et al., 2000). Atg8∼PE (∼ = covalent bond) adducts are incorporated into the growing autophagosomal membrane (Nakatogawa et al., 2007).
As with other UBLs, Atg8 becomes ligated via a multistep process (Geng and Klionsky, 2008; Nakatogawa et al., 2009). First, the E1 enzyme Atg7 binds MgATP and Atg8, and catalyzes adenylation of Atg8's C-terminus. Next, the resulting Atg8-acyl adenylate is attacked by Atg7's catalytic Cys to generate a thiolester-bonded Atg7∼Atg8 intermediate. A transthiolation reaction occurs whereby Atg8's C-terminus is transferred from Atg7's catalytic Cys to that of the E2 enzyme, Atg3, generating a thiolester-linked Atg3∼Atg8 conjugate. Finally, Atg8 is transferred from Atg3's catalytic Cys to PE in a reaction stimulated by an unusual multiprotein E3 containing the Atg12∼Atg5 complex (Hanada et al., 2007; Ichimura et al., 2000). This is partly composed of Atg12, another UBL involved in autophagy. Like Atg8, Atg12 is activated by Atg7, although Atg12 has its own E2 (Atg10) and target (Atg5) (Mizushima et al., 1998).
Models for the initial steps of Atg8 activation may be extrapolated from 1) structural similarity of Atg8 to canonical UBLs such as ubiquitin, NEDD8, and SUMO, and bacterial antecedents MoaD and ThiS (Noda et al., 2009; Sugawara et al., 2004), and 2) sequence homology between roughly half of Atg7 to the adenylation domain found in canonical E1s and homodimeric bacterial E1 antecedents MoeB and ThiF (Mizushima et al., 1998; Noda et al., 2009; Streich and Haas, 2010; Schulman and Harper, 2009). However, detailed mechanisms of Atg8 conjugation for the steps following the initial adenylation reaction cannot be extrapolated from knowledge of canonical UBL pathways due to numerous fundamental differences. First, as in ThiF, Atg7's catalytic Cys resides in a loop (Duda et al., 2005; Lehmann et al., 2006). By contrast, in canonical E1s such as for ubiquitin, NEDD8, and SUMO, the catalytic Cys is in a distinct domain, which undergoes a remarkable conformational change involving structural remodeling and moving close to the adenylation active site for covalent linkage to the UBL C-terminus (Olsen et al., 2010). Another difference is that Atg7 is a homodimer, with two potential adenylation and thiolester active sites per Atg7 homodimer (Komatsu et al., 2001). By contrast, canonical E1s are pseudosymmetric and share the same MoeB/ThiF-like adenylation domain fold, but contain only a single adenylation active site, a single catalytic cysteine, and a single E2-binding site, all within the same subunit (reviewed in Schulman and Harper, 2009). Furthermore, Atg7-Atg3 interactions differ from canonical E1-E2 interactions: Atg7 binds a unique Atg3 “flexible region” (referred to hereafter as Atg3FR) not found in canonical E2s (Yamada et al., 2007). The Atg3FR has many acidic residues (theoretical pI of yeast Atg3FR = 3.54) interspersed with some hydrophobic residues and is disordered in solution (Yamada et al., 2007). Atg7 also has unique features, most notably a ∼300-residue N-terminal region that lacks any known function, or sequence homology to any other proteins. In this study, we identify Atg7's N-terminal domain (Atg7NTD) as the binding site for the Atg3FR. We find that Atg7 utilizes a unique mechanism that relies on its homodimerization to mediate E1-to-E2 UBL transfer in trans. Biophysical, X-ray crystallographic and biochemical experiments reveal the structural basis and functional importance of distinctive E1-E2 interactions required for optimal Atg8 conjugation in the autophagy pathway.
Results
Dimerization and structure of Atg7CTD – overall similarity to MoeB, ThiF, and canonical E1s
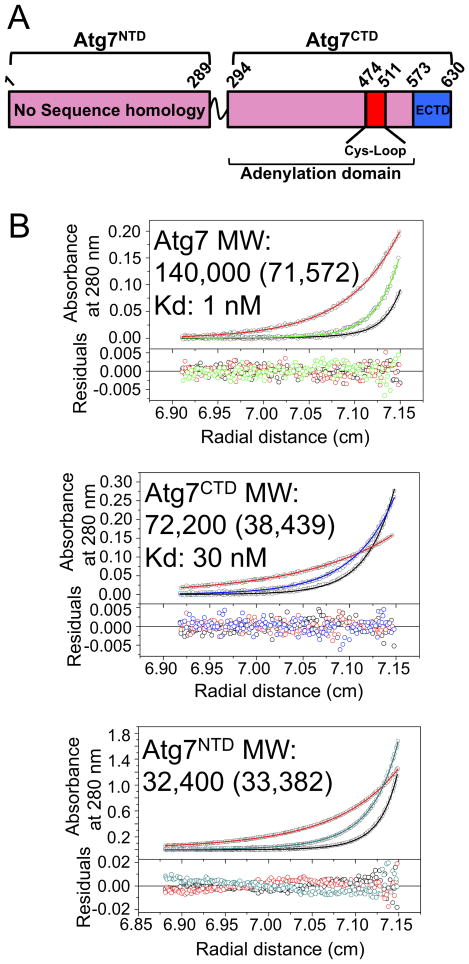
To gain insight into the overall architecture of Atg7, we mapped two structural domains in the yeast protein (Fig. 1A) based on prior studies (Ichimura et al., 2000; Komatsu et al., 2001), sequence analysis, and deletion mutants (data not shown). The Atg7NTD spans residues 1 through 289 and does not display sequence homology to indicate structure or function. The Atg7 C-terminal domain (Atg7CTD) sequence has three regions. First, residues 294-473 and 512-572 are homologous to the adenylation domains of canonical E1s for ubiquitin, NEDD8, and SUMO, as well as to the bacterial enzymes MoeB and ThiF, which catalyze C-terminal acyl-adenylation of MoaD and ThiS respectively; both MoaD and ThiS have a UBL fold (Lake et al., 2001; Lehmann et al., 2006). Second, residues 474-511 contain the catalytic Cys507 and correspond to a “Cys-loop” or “crossover loop” crossing over the adenylation domain in MoeB/ThiF-like enzymes. Finally, Atg7's ∼60 extreme C-terminal residues lack homology to other E1s, but are required for expression in E. coli (data not shown) and presumably folding, and for homooligomerization and UBL-binding in yeast (Komatsu et al., 2001). A short linker connects the Atg7NTD and Atg7CTD.
Figure 1. Homodimerization of Atg7 via Atg7CTD.
A) Schematic of Atg7 showing positions of NTD and CTD [adenylation domain, Cys-loop, extreme C-terminal domain (ECTD)].
B) Sedimentation equilibrium AUC data for Atg7, Atg7CTD, and Atg7NTD. Molecular weight (MW) was determined using sedimentation velocity AUC, with theoretical values in parenthesis.
Previous studies indicated that Atg7 forms a homodimer in vivo (Komatsu et al., 2001). To understand the roles of our identified domains in assembly of exogenously expressed Atg7, we performed analytical ultracentrifugation experiments on full-length yeast Atg7, Atg7NTD and Atg7CTD. The experimental molecular weights reveal that Atg7 and Atg7CTD are dimeric in solution, whereas the Atg7NTD is monomeric (Fig. 1B). Sedimentation equilibrium data indicate high-affinity dimer equilibrium dissociation constants for full-length Atg7 and Atg7CTD of ∼1 nM and ∼30 nM respectively (Fig. 1B).
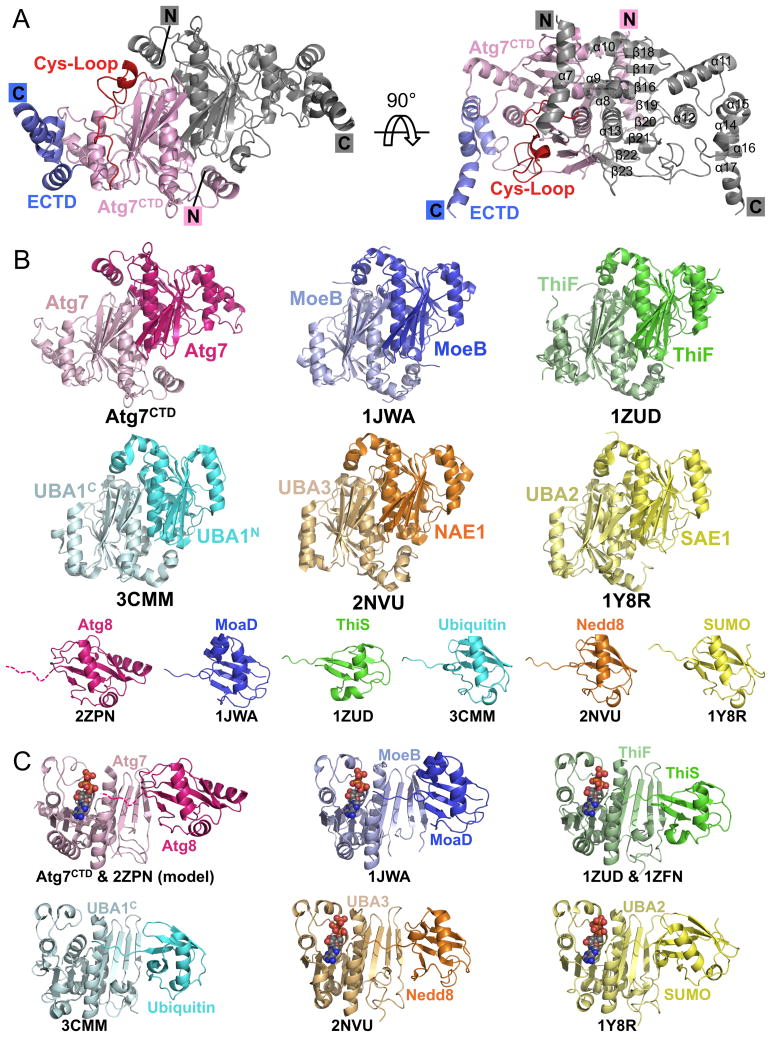
A 2.25 Å resolution crystal structure of yeast Atg7CTD confirmed a homodimeric structure closely resembling MoeB and ThiF (2.2 and 2.1 Å root-mean-square deviation (rmsd), respectively) and the pseudosymmetric adenylation domains of canonical E1s (Figs. 2A, B, C, S1; Table 1). Similarities include conservation of several MgATP binding residues and the β-sheet structure MoeB/ThiF/E1s use for UBL-binding during catalysis. Indeed, mutation of these regions confirm their importance in forming the Atg7∼Atg8 thiolester intermediate (Fig. S1).
Figure 2. Structure of Atg7CTD: structural similarity among E1 adenylation domains.
A) Structure of crystallographic Atg7CTD homodimer, with one protomer colored as in Figure 1A, and the other in gray.
B) Structural comparison of Atg7CTD adenylation domain with those of MoeB, ThiF, and E1s for ubiquitin (UBA1), NEDD8 (NAE1-UBA3), and SUMO (SAE1-UBA2), depicted without Cys-loops/domains for clarity, with cognate UBLs in corresponding colors below (Huang et al., 2007; Lake et al., 2001; Lee and Schindelin, 2008; Lehmann et al., 2006; Lois and Lima, 2005; Noda et al., 2008). UBA1 (ubiquitin E1) is one polypeptide, but represented according to N- and C-terminal halves. Atg8's C-terminal tail is missing in the structure and is therefore depicted as a dashed line. PDB codes are shown below each structure.
C) Model of Atg7CTD (adenylation domain)-Atg8-ATP based on superposition with MoeB-MoaD-ATP (Lake et al., 2001) compared with other E1-UBL structures +/- ATP. PDB codes are shown below each structure.
Table 1. Crystallographic data collection and refinement statistics.
| Structure | Atg7CTD | Atg7NTD | Atg7NTD-Atg3FRpep | Atg7NTD (P283D) |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | NECAT ID24-C | NECAT ID24-C | ALS 822 | SERCAT ID22 |
| Wavelength (Å) | 1.28255 | 0.97924 | 1 | 1 |
| Space group | P43212 | P21212 | C2 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | a=66.56, b=66.56, c=222.27 | a=56.86, b=74.68, c=76.05 | a=152.14, b=99.43, c=71.50 | a=56.18, b=74.44, c=153.37 |
| α, β, γ (°) | α= β= γ=90 | α= β= γ=90 | α= γ=90, β=113.13 | α= β= γ=90 |
| Resolution (Å) | 50-2.25 (2.33-2.25) | 40.0-1.9 (1.97-1.90) | 50.0-2.1 (2.18-2.10) | 50-1.6 (1.66-1.60) |
| Total reflections | 218208 | 343047 | 217998 | 596964 |
| Unique reflections | 24766 | 26234 | 58224 | 85691 |
| Rmerge (%) | 7.6 (72.6) | 8.1 (25.0) | 6.7 (51.2) | 5.8 (45.0) |
| Average I/σ | 302.9 (3.0) | 43.3 (10.5) | 22.4 (2.6) | 31.5 (3.0) |
| Completeness (%) | 99.7 (99.5) | 97.3 (84.7) | 99.9 (100.0) | 99.8 (98.4) |
| Redundancy | 8.8 (7.1) | 13.4 (9.5) | 3.7 (3.7) | 7.0 (5.7) |
| Refinement | ||||
| Resolution range (Å) | 50-2.25 | 33.8-1.9 | 33.2-2.1 | 45.3-1.6 |
| No. of reflections (σ≥0) | 24572 | 25621 | 58121 | 81152 |
| Rwork (%) | 19.3 | 17.5 | 18.2 | 18.6 |
| Rfree (%) | 23.3 | 19.7 | 20.6 | 20.6 |
| Protein atoms (Average B-factor) | 2477 (57.4) | 2343 (16.1) | 4576 (62.3) | 4636 (20.2) |
| Waters (Average B-factor) | 86 (52.5) | 323 (27.8) | 190 (50.9) | 689 (34.1) |
| Zinc (Average B-factor) | 1 (61.5) | |||
| RMSD | ||||
| Bond lengths (Å) | 0.008 | 0.007 | 0.008 | 0.007 |
| Bond angles (°) | 1.205 | 1.025 | 1.033 | 1.036 |
| Ramachandran plot statistics (PHENIX) | ||||
| Preferred (%) | 97.8 | 99.7 | 97.2 | 98.9 |
| Additionally allowed (%) | 2.2 | 0.0 | 2.8 | 1.1 |
| Disallowed (%) | 0.0 | 0.3 | 0.0 | 0.0 |
Data for highest resolution shell are shown in parentheses. Rwork=Σ|Fo-Fc|/ΣFo. Rfree is the cross-validation of R-factor, with 5% or more of the reflections omitted in refinement.
Consistent with sequence analysis, the Atg7CTD structure also has some distinctive features. The first helix is less kinked and extended, perhaps for stability for connection to the Atg7NTD. Second, the Cys-loop is slightly longer than that in MoeB/ThiF and in our structure packs against the adenylation domain. The crystallized conformation of the Cys-loop would block access of Atg8's C-terminal tail to the adenylation and Cys active sites. Furthermore, although the Cα of the catalytic Cys507 is only ∼8 Å from both a modeled Atg8 C-terminus and ATP α-phosphate, the Cys side-chain faces the opposite direction from that necessary for forming an Atg7∼Atg8 intermediate. Thus, despite its proximity to the adenylation active site, the Cys-loop must adopt different conformations during catalysis. Consistent with conformational flexibility, the Cys-loop is only partially ordered and has high B-factors in our structure. Notably, the corresponding regions of MoeB, ThiF, and E1s also fluctuate upon UBL binding and/or formation of an E1∼UBL thiolester intermediate (Duda et al., 2005; Lake et al., 2001; Lehmann et al., 2006; Lois and Lima, 2005; Olsen et al., 2010; Walden et al., 2003). Finally, although the 16 C-terminal amino acids are not visible in the structure, residues 573-614 form a 4-helix platform extreme C-terminal domain (ECTD) that makes extensive hydrophobic interactions with the remainder of the CTD, explaining their requirement for Atg7 folding. The most C-terminally observed residue is poised near Atg8 modeled based on other E1-UBL structures. Future studies will be required to understand detailed functions of the extreme C-terminal portion of Atg7.
Atg7 N-terminal domain recruits the Atg3 “flexible region”
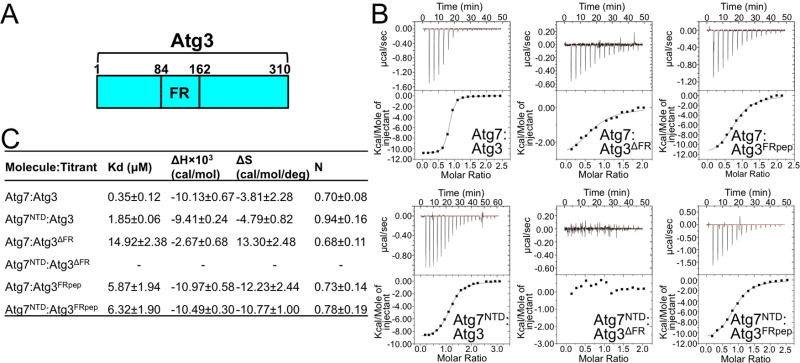
We next mapped high-affinity binding between Atg7 and Atg3, which interact with a Kd of 0.35 μM as measured by isothermal titration calorimetry (ITC) (Fig. 3). Atg7NTD retains much affinity for Atg3, with a Kd of 1.85 μM (Fig. 3C). A previous study showed that Atg3FR (residues 84-162) binds Atg7, and an Atg3 mutant lacking the FR region was defective for autophagy (Yamada et al., 2007). Indeed, we found that deleting the FR region (residues 86-159, hereafter referred to as Atg3ΔFR) reduces the Kd for full-length Atg7 by more than 40-fold, and eliminates binding to the Atg7NTD under our assay conditions (Fig. 3C).
Figure 3. Roles of Atg7NTD and Atg3FR in Atg7-Atg3 interactions.
A) Schematic of Atg3, highlighting location of flexible-region (FR).
B) ITC data for binding between indicated Atg7 and Atg3 constructs. Upper panels - raw power data from titrations. Lower panels - fits of standard binding equations using MicroCal software.
C) Summary of thermodynamic parameters determined by ITC. N value is based on total concentration, not of homodimers. Error ranges refer to standard deviation.
To identify the region within the >80-residue Atg3FR essential for binding Atg7NTD, we examined co-pulldowns of His6-MBP-tagged Atg3FR deletion mutants and GST-tagged Atg7NTD upon their coexpression in E. coli. Atg3 residues 130-142 appear to be both necessary and sufficient for binding to Atg7NTD in this assay (Fig. S2). Furthermore, a synthetic peptide encompassing Atg3 residues 128-144 (Atg3FRpep) binds similarly to both full-length Atg7 and Atg7NTD, with 5.9 and 6.3 μM Kd values, respectively (Figs. 3B, 3C).
To understand details of Atg7-Atg3 interactions, we determined, using yeast proteins, crystal structures of Atg7NTD by single wavelength anomalous diffraction using selenomethionine substituted protein, and of an Atg7NTD-Atg3FRpep complex by molecular replacement using the Atg7NTD structure as a searchmodel (Figs. S2, S3, 4A Table 1). The Atg7NTD fold is comprised of two structurally similar subdomains each with a 5-stranded mixed β-sheet sandwiched between two α-helices, with distinct flanking helices and loops (Fig. S3). The subdomains are interconnected by three extended loops spanning the entire length of the Atg7NTD such that the N- and C-termini are at opposite ends (Fig. S3).
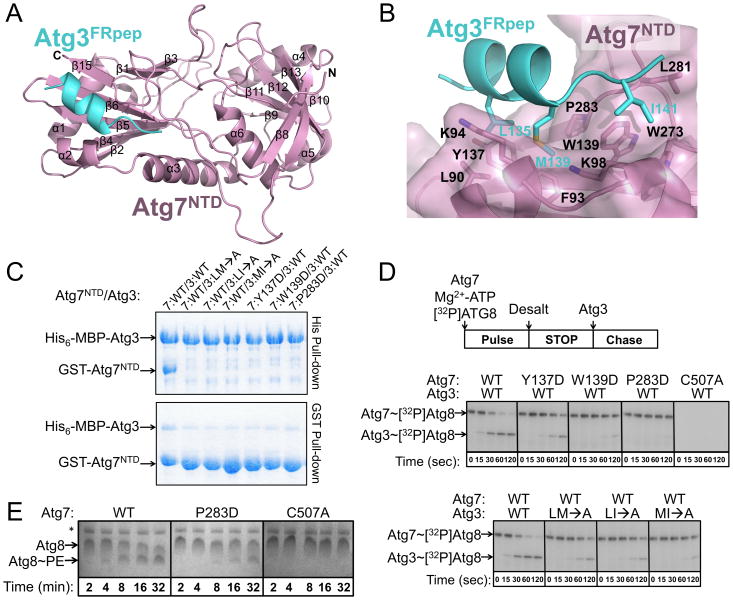
Figure 4. Structural basis for Atg7NTD-Atg3FRpep interaction.
A) Structure of Atg7NTD (pink)-Atg3FRpep (cyan).
B) Residues involved in Atg7NTD (pink, semi-transparent surface)-Atg3FRpep (cyan) interaction are shown in sticks. Oxygens are colored red, nitrogens blue, and sulfurs yellow.
C) Pull-downs from E. coli co-expressing wild-type (WT) and mutant versions of Atg7NTD and Atg3FR. LM→A = Atg3(L135A, M139A); LI→A = Atg3(L135A, I141A); and MI→A = Atg3(M139A, I141A).
D) Pulse-chase assays showing time courses of [32P]Atg8 transfer from indicated versions of Atg7 to Atg3.
E) Atg8 lipidation in presence of wild-type (WT), P283D, or catalytic cysteine (C507A) mutant versions of Atg7. * = E. coli nickel-binding Crp and SlyD (Bolanos-Garcia and Davies, 2006) copurifying with Atg12∼Atg5.
A hydrophobic groove in Atg7, which has β-strands-5, -6 and -15 as its floor and α-helix-3 and the β14/β15-loop as its sides, cradles the Atg3FRpep. In complex with Atg7NTD, the Atg3FRpep adopts a 2-turn helix followed by a short extension (Fig. 4A). Interestingly, NMR data previously indicated that in the absence of Atg7, the Atg3FR adopts a disordered structure both on its own and in the context of full-length Atg3 (Yamada et al., 2007). Furthermore, it seems that the residues corresponding to the Atg3FRpep can adopt a range of conformations because the corresponding sequence is the only portion of the FR visible in a prior crystal structure of full-length Atg3 (PDB code: 2DYT), where the Atg3FRpep sequence forms a helix (Yamada et al., 2007). Consistent with its structural flexibility, in the crystal structure of full-length Atg3, this region has substantially higher B-factors than the rest of Atg3 (91 v. 51), half the side-chains including that of Met139 are not observed, and lack of the majority of the FR raises the possibility that the observed Atg3FRpep sequence comes from an adjacent Atg3 in the crystal to mediate packing (Fig. S2). It seems likely that the structural malleability of the Atg3FRpep region facilitates accessibility of this region for binding to Atg7.
The Atg7-Atg3FRpep interaction is mediated by hydrophobic side-chains from Atg3 inserting into the Atg7 hydrophobic groove (Fig. 4B). The interface is centered around Atg3 residue Met139 docking in a hydrophobic pocket formed by Atg7 residues Phe93, Lys94, Trp139, and Pro283. This is buttressed on one side by Atg3 residue Leu135 making hydrophobic contacts with Atg7 residues Leu90, Phe93, Lys94, and Tyr137. On the other side, Atg3 residue Ile141 interacts with Atg7 residues Lys98, Trp139, Trp273, and Leu281. Overall, the interaction buries ∼940 Å2 of surface area.
Notably, the Atg3FRpep–binding groove in Atg7 is continuous through to the junction of the two subdomains, where this portion of Atg7NTD is strikingly basic in character. On this basis, we can approximate the trajectory for more of Atg3's FR: the 10 residues following the structurally observed sequence are almost entirely acidic, and likely traverse through the basic cleft in the Atg7NTD (Fig. S3).
Role for Atg7NTD-Atg3FRpep interface in Atg7-Atg3 interactions and Atg8 transfer
We investigated whether the structurally observed interface is important for the Atg7NTD-Atg3 interaction by pull-down after bacterial coexpression of several versions of GST-Atg7NTD and His6-MBP-Atg3. Because non-bulky hydrophobic residues mediate the interaction, we tested effects of pairs of hydrophobic to Ala mutations in Atg3. From Atg7, we made single hydrophobic to Asp mutations, to diminish the overall hydrophobic character of the groove. Pulling down on the His6-MBP tag shows that wild-type Atg3 is able to copurify similar amounts of wild-type Atg7NTD. Pairwise Ala mutations in Atg3 Leu135, Met139, and Ile141, or individual Asp substitutions in place of Atg7NTD Tyr137, Trp139, or Pro283 diminished the interaction (Fig. 4C). In the reciprocal pull-down for GST-Atg7NTD, even for the wild-type proteins, only a substoichiometric amount of His6-MBP-Atg3 copurifies, as would be the case if GST-Atg7NTD were present in significant excess during the coexpression. Nonetheless, reduced interaction is observed for all the mutants, which are expressed at wild-type levels (Fig. 4C).
We next tested whether the structurally observed Atg7NTD-Atg3FRpep interaction plays a role in Atg8 transfer from Atg7 to Atg3 using a pulse-chase assay. Briefly, in the “pulse” reaction we incubated Atg7 with [32P]Atg8 and MgATP to form the thiolester linked Atg7∼[32P]Atg8 intermediate, and stopped this reaction by desalting into buffer without MgATP. We added Atg3, and monitored transfer of [32P]Atg8 from Atg7 onto Atg3, yielding the thiolester-linked Atg3∼[32P]Atg8 conjugate (Fig. 4D). In agreement with the structure, mutations at the Atg7NTD-Atg3FRpep interface cripple transfer of [32P]Atg8 from Atg7 to Atg3 (Fig. 4D). This effect is mirrored in an in vitro reaction reconstituting the Atg8 pathway: in the presence of MgATP, Atg3, an Atg12∼Atg5 E3, and PE-containing liposomes, Atg8 lipidation is impaired in reactions containing the Pro283Asp mutant version of Atg7 (Fig. 4E). Furthermore, an important role for the hydrophobic Atg7-Atg3 interactions may be inferred from our finding that salt has little effect on Atg8 transfer from Atg7 to Atg3 (Fig. S3).
To provide support for the notion that the deleterious effects of Atg7NTD mutations arose from eliminating the interaction with Atg3FR and not from global misfolding, we determined the crystal structure of the most impaired mutant. Despite subtle differences in crystal packing, the Pro283Asp mutant structure superimposes with wild-type Atg7NTD with 0.55 Å rmsd, confirming correct folding (Figs. S2, S3).
The Atg7NTD binding surface for Atg3FRpep is conserved across species
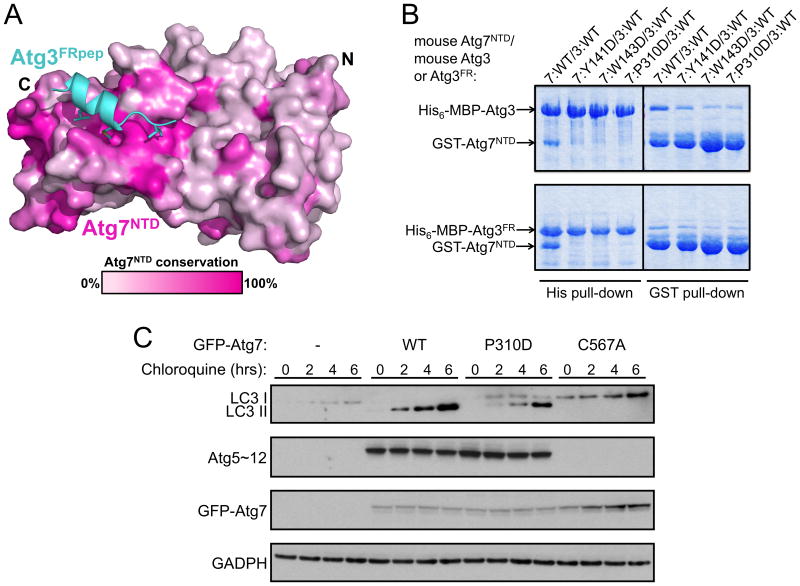
Sequence analysis of the Atg7NTD suggests that its interaction with the Atg3FR is conserved across eukaryotes (Figs. 5A, S2). Indeed, as with the proteins from budding yeast, after coexpression in E. coli we observed interaction between GST-tagged mouse Atg7NTD (spanning residues 1-317 of mouse Atg7) and either His6-MBP-tagged mouse Atg3 or mouse Atg3FR (residues 91-193 of mouse Atg3). Individual Asp mutations in place of mouse Atg7NTD Tyr141, Trp143, and Pro310, which correspond to yeast Tyr137, Trp139, and Pro283, respectively, diminish the interaction (Fig. 5B). To test function in vivo, we stably expressed an N-terminally GFP-tagged mouse Atg7, the Pro310Asp mutant, or the catalytic Cys567Ala mutant, in Atg7 null MEF cell lines. Chloroquine was added to induce accumulation of autophagosomes. Notably, accumulation of LC3-II was impaired by the mutation in the ATG3FRpep binding site (Fig. 5C). The deleterious effects of the mutation are specific for the Atg3 pathway, because no effect is observed for ligation of Atg12 to Atg5 (Fig. 5C), which is mediated by a different E2, Atg10, that is homologous to part of Atg3 but lacks an obvious FR sequence.
Figure 5. Conservation of the Atg7NTD-Atg3FR interaction across species.
A) Conservation among Atg7s displayed on the surface of Atg7NTD in complex with Atg3FRpep (cyan) White - 0 conservation; magenta - 100% conservation across species. Leu135, Met139 and Ile141 on Atg3FRpep shown in sticks.
B) Pull-downs from E. coli co-expressing indicated versions of mouse Atg7NTD based on conservation with yeast Atg3FR-binding site, and Atg3 or Atg3FR.
C) Anti-LC3, Atg12, Atg7, and GADPH (loading control) westerns of lysates of Atg7 null MEFs expressing the indicated versions of GFP-Atg7 treated with chloroquine.
Model for transfer of Atg8 from Atg7 onto Atg3: a unique trans mechanism
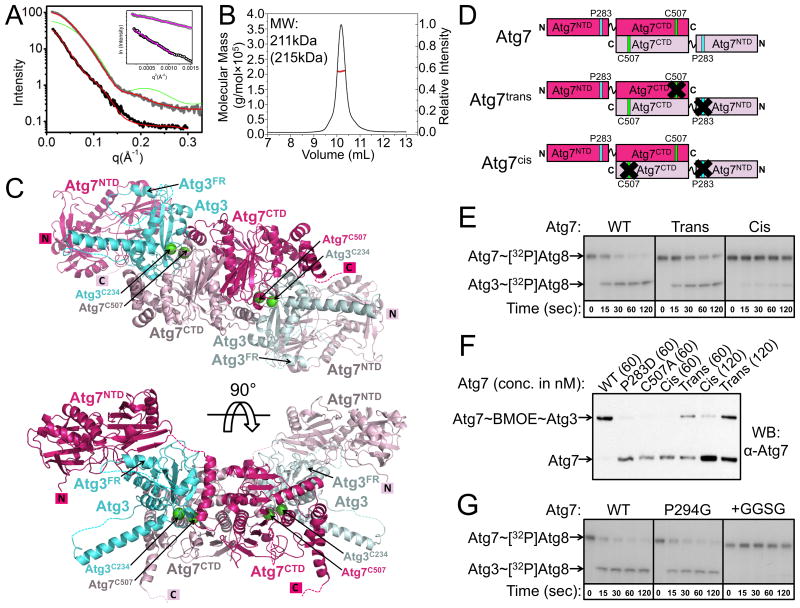
We made a structural model to obtain insights into the role of Atg7NTD-Atg3FRpep interactions in the context of full-length Atg7 and Atg3. First, to understand relative positioning of Atg7's N- and C-terminal domains, we examined Small Angle X-ray Scattering (SAXS) of Atg7 (Figs. 6A, S4) (Hura et al., 2009). It is not possible based on the data to definitively orient the two domains relative to each other, although a model fitting the homodimeric Atg7CTD and two Atg7NTDs generated with the program BILBOMD (Pelikan et al., 2009) yielded a reasonable fit (χ2 = 1.5) (Fig. 6A). Overall, the best-fitting model is consistent with the two Atg7NTDs extending away from a central Atg7CTD homodimeric structure in solution (Fig. S4).
Figure 6. Transthiolation in trans: a working model for transfer of Atg8 from Atg7 to Atg3.
A) Analysis of the interference-free SAXS curve for Atg7CTD (gray) and Atg7 (black), with theoretical scattering calculated with FoXS (Schneidman-Duhovny et al., 2010) from crystal structure and atomistic model, respectively, in red (χ2 = 2.1, Atg7CTD; χ2 = 1.5, Atg7). For reference, a poorly fitting theoretical SAXS curve if Atg7CTD were a monomer is shown in green. Inset – Guinier plots indicating aggregation-free data (magenta).
B) SEC-MALS data plotted as a molar mass distribution (red) superimposed on chromatogram of differential refractive index as a function of elution volume. Shown is molecular weight (MW) determined by SEC-MALS, with theoretical value in parenthesis.
C) Model of homodimeric Atg7 (dark and light pink) bound to two Atg3s (cyan and light blue), with Atg7 and Atg3 catalytic cysteines (green spheres) in close proximity for catalysis. The Cys of the cyan Atg3 bound via its FR to the NTD of the dark pink Atg7 is close to the Cys in the CTD of the light pink Atg7, enabling transthiolation in trans. For clarity, Atg8, which would be transferred from the catalytic Cys of Atg7 to the catalytic Cys of Atg3, is not shown.
D) Schematics of wild-type Atg7, and Atg7trans and Atg7cis dimers. Pro283 that binds Atg3FR is cyan and Atg7's catalytic Cys507 is green. Black Xs indicate mutation of Pro283 to Asp or Cys507 to Ala to impair binding of Atg3 or prevent thiolester formation with Atg8, respectively.
E) Autoradiogram for time course of transfer of [32P]Atg8 from Atg7 to Atg3. WT - wild-type Atg7 homodimer; trans - Atg7 heterodimer composed of Atg7(P283D) and Atg7(C507A); cis - Atg7 heterodimer composed of wild-type Atg7 and Atg7(P283D, C507A).
F) BMOE-crosslinking between Atg3C234only and the indicated variants of Atg7 at concentrations of either 60 or 120 nM, detected by anti-Atg7 western.
G) Autoradiogram showing time course of [32P]Atg8 transfer to Atg3 from the indicated versions of Atg7.
Next, to determine Atg7:Atg3 stoichiometry upon juxtaposition of their catalytic cysteines, we performed Size-Exclusion Chromatography-Multiangle Light Scattering (SEC-MALS) on a crosslinked sample. To attach a crosslinker specifically to the Atg3 active site, we made a mutant in which the only cysteine is the catalytic Cys234 (Atg3C234only). After reacting Atg3C234only with Bis-Maleimidoethane (BMOE, a short-spacer homobifunctional sulfhydryl crosslinker) and desalting to remove any excess unreacted BMOE, Atg3C234only∼BMOE becomes crosslinked to wild-type Atg7 but not a catalytic Cys507Ala mutant, indicating crosslinking between the Atg7 and Atg3 active site cysteines. The SEC-MALS determined molecular weight of the Atg7∼BMOE∼Atg3C234only crosslinked complex closely matches that of a 2:2 dimer (211 kDa vs 215 kDa expected) (Fig. 6B).
Thus, to generate a structural model, we placed the Atg7 and Atg3 catalytic cysteines near each other, and modeled in the Atg7NTD-Atg3FRpep consistent with relative locations of the ATG7CTD and ATG7NTD from SAXS data whilst maintaining reasonable distances for connections to the remainder of Atg3. The model suggests a mechanism of E1-E2 transthiolation that differs from canonical E1s and E2s such as for ubiquitin, NEDD8, and SUMO, for which the same E1 subunit contains both the catalytic Cys and E2-binding regions, for UBL transfer in cis (Schulman and Harper, 2009). Instead, a distinctive “trans” mechanism provides a rationale for Atg7 homodimerization: Atg8 linked to the catalytic Cys of one Atg7CTD would be transferred to Atg3 associated with the Atg7NTD of the opposite Atg7 protomer in the dimer (Fig. 6C, S4).
To test the model, we coexpressed His6- and GST-tagged Atg7 in E. coli, and sequentially performed affinity capture with Ni2+ agarose beads and then with glutathione sepharose beads to obtain Atg7 dimers containing two different protomers. Using this method, we made two mutant Atg7s. Atg7trans contains two mutant versions of Atg7 that together form a dimer: one mutant has an Ala substitution in place of the catalytic Cys507, and the other has the Pro283Asp mutation that hinders the Atg7NTD from binding Atg3. Our sequential affinity capture purification yields a mixed dimer where one protomer is unable to form a thiolester with Atg8 but retains Atg3FR binding, and the other protomer is able to form a thiolester linkage with Atg8, but is defective in recruiting the Atg3FR (Fig. 6D). Therefore, the only way for Atg7trans to mediate Atg8 transfer would be if Atg3 binds one Atg7NTD and accepts Atg8 from the catalytic Cys of the opposite Atg7 in the dimer. By contrast, the Atg7cis dimer contains one wild-type Atg7 and one double Cys507Ala/Pro283Asp mutant that would be defective in both forming a thiolester with Atg8 and binding to Atg3 (Fig. 6D). Thus, Atg7cis would only be active if Atg8 were transferred between the Atg7 catalytic Cys and an Atg3 associated with the Atg7NTD of the same protomer of Atg7. Using our pulse-chase assay, we find that Atg7 preferentially carries out transthiolation in trans (Fig. 6E).
In order to test whether the Atg7NTD-Atg3FRpep interaction helps position the Atg3 active site near that of Atg7 as in our model, we again turned to crosslinking, adding Atg3C234only∼BMOE to either wild-type Atg7, Atg7 with its catalytic Cys507 mutated to Ala, Atg7 Pro283Asp, Atg7trans, or Atg7cis. We observe complete disappearance of wild-type Atg7, and concomitant appearance of a single higher molecular weight band corresponding to Atg7 crosslinked to Atg3 (Fig. 6F). This band does not form with the Atg7 Cys507Ala mutant, in agreement with the notion that BMOE is crosslinking the Atg7 and Atg3 catalytic cysteines (Fig. 6F). Complete crosslinking of wild-type Atg7 is consistent with the Atg7 homodimer binding concurrently to two molecules of Atg3. Significantly less crosslinked product forms with the Pro283Asp mutant (Fig. 6F), supporting the view that the structurally observed interaction helps position Atg3's catalytic cysteine in proximity to that of Atg7. Similar low-level formation of the crosslinked product is observed for Atg7cis, in which one member of each dimer contains both the Cys507Ala and Pro283Asp mutations. Notably, substantially more crosslinked product is formed with Atg7trans, in which one protomer within the dimer contains the Cys507Ala mutation while the other contains the Pro283Asp mutation (Fig. 6F). Crosslinking of the trans complex is also more efficient than the cis complex when a two-fold higher molar concentration of Atg7cis and Atg7trans are used relative to wild-type Atg7, such that the concentrations of Atg7 catalytic cysteines available to form the crosslink are identical in all cases (two catalytic cysteines per wild-type Atg7 homodimer, but only one catalytic cysteine per Atg7cis or Atg7trans dimer) (Fig. 6F). Thus, the crosslinking results agree with our structural model and support an overall “trans” architecture for the Atg7-Atg3 complex.
We wondered whether the flexibility and relative placement of the Atg7NTD, which binds the Atg3FR, and the Atg7CTD, whose catalytic Cys is thiolester-linked to Atg8, influences Atg8 transfer between Atg7 and Atg3. Thus, we made mutations probing flexibility between the domains: a Gly mutation in place of the conserved Pro294 was designed to test effects of increasing flexibility while maintaining linker length, and a mutant with the sequence Gly-Gly-Ser-Gly inserted in between residues Ser290 and Leu291 was designed to test effects of lengthening the linker. The Pro294Gly mutant displayed wild-type ability to mediate transfer of Atg8 to Atg3, whereas the insertion mutant was severely impaired (Fig. 6G). The results are consistent with the notion that the Atg7NTD and Atg7CTD may be flexibly tethered, but that the distance between them is optimized for the transthiolation reaction.
Discussion
Eukaryotic UBLs and E1s appear to have originated from ancient prokaryotic biosynthetic pathways. E. coli MoaD and ThiS represent prokaryotic UBLs, which function in 2:2 complexes with their respective homodimeric E1-like enzymes, MoeB and ThiF, in the biosynthesis of molybdopterin and thiamine, respectively (Hochstrasser, 2000; Schulman and Harper, 2009; Streich and Haas, 2010). The theme represented by MoeB and ThiF has been elaborated upon over the course of evolution, giving rise to eukaryotic E1s that bind both UBLs and E2s, and can be classified as either canonical or non-canonical (Hochstrasser, 2000; Schulman and Harper, 2009; Streich and Haas, 2010). Our structural studies now show that the non-canonical Atg7, like other E1s, possesses at its core a MoeB/ThiF-like adenylation domain. E1s have a variety of additions to this scaffold. Here we found that Atg7's unique N-terminal domain selectively recruits Atg3's FR via a unique type of E1-E2 interaction. First, Atg7NTD forms a roughly stoichiometric complex with Atg3, which is mediated by the Atg3FR. Second, the crystal structure of yeast Atg7NTD reveals, to our knowledge, a previously undescribed fold harboring a conserved hydrophobic groove recruiting Atg3FR. Third, on this basis, we were able to introduce a mutation into yeast Atg7 that leads to defects in Atg3∼Atg8 formation in vitro and into mouse Atg7 that leads to defects in conjugation of LC3, a mammalian Atg8 ortholog, in cells. Finally, we identify a distinctive “trans” E1-to-E2 Atg8 transfer mechanism that provides a rationale for the homodimerization of Atg7: Atg3 bound via its FR to one Atg7NTD accepts Atg8 from the catalytic Cys of the opposite Atg7 protomer in the dimer.
Prior studies on the canonical E1s reveal a trend among these enzymes: their catalytic cysteine domains and E2-binding domains undergo rotations relative to their MoeB/ThiF-like adenylation domain cores (Huang et al., 2007; Olsen et al., 2010). Also, the domain housing a canonical E1's catalytic Cys undergoes striking conformational remodeling during formation of an E1∼UBL intermediate (Olsen et al., 2010). In this study, we find parallel characteristics in Atg7, despite its unique structural features. We observe the potential for flexibility between the Atg7NTD and Atg7CTD due to the presence of a linker between the two domains. Furthermore, catalysis of both Atg8 adenylation and formation of an Atg7∼Atg8 intermediate would require the Atg7 Cys-loop to adopt different conformations from that observed in our crystal structure of Atg7CTD. Future studies will be necessary to understand the extent of structural remodeling that occurs in the Atg7 Cys-loop during catalysis, and how orientation of Atg7 domains might confer functional roles to the enzyme.
In addition to interacting with Atg3 during conjugation of the UBL Atg8, Atg7 has the additional function of activating a second UBL, Atg12, which functions via the distinct E2, Atg10. Notably, we have been able to introduce a mutation in mouse Atg7 that separates these two pathways. The Pro310Asp mutation in the mouse Atg7 NTD, made based on our crystal structure of yeast Atg7NTD-Atg3FR, hinders both binding to Atg3, and LC3 conjugation in cells. This had no observable effect on Atg12 conjugation in cells (Fig. 5C). Consistent with this notion, Atg10 sequences lack an obvious FR. Taken together with our crystallographic data indicating that Atg3FRpep binds in a groove in Atg7, the results raise the possibility that small molecule inhibitors selectively inhibiting Atg8 conjugation may be obtainable.
How might Atg7 interact with its distinct E2s? We speculate that the answer to this question relates to our finding that Atg3 binds full-length Atg7 with a 6-fold greater affinity than it binds Atg7NTD. Thus, in addition to the high-affinity Atg7NTD-Atg3FR interaction investigated here, it seems likely that there are additional weak contacts between the Atg7CTD and Atg3ΔFR. We speculate that these are likely mirrored by Atg10, which is displays some sequence similarity to Atg3ΔFR. Moreover, in our structural model of an (Atg7∼Atg8-Atg3)2 complex, Atg3 is located near Atg8, raising the possibility that the UBLs Atg8 and Atg12 help mediate specificity of transfer to their own distinct E2s, although difficulty in producing purified Atg12 precludes us from currently addressing this. We anticipate many exciting new features of E1s will be revealed from future studies of these and other interactions and mechanisms involved in E1-to-E2 UBL transfer in autophagy.
Experimental Procedures
Details on constructs, protein expression and protein purification are provided in supplemental information.
X-ray crystallography
All crystals were grown in hanging drops. Crystals of selenomethionine-labeled Atg7NTD were grown at 4°C by streak-seeding drops prepared by mixing 1 μL 20-60 mg/mL Atg7NTD in 25 mM Tris pH 7.6, 150 mM NaCl and 10 mM DTT with 1 μL well solution containing 9.2% isopropanol, 0.1 M potassium thiocyanate, and 0.1 M citrate buffer pH 4.0. Crystals were cryoprotected in well solution supplemented with 6% glycerol, 6% ethylene glycol and 6% sucrose. Phasing via identification of the 3 Se sites and initial model building were performed with PHENIX AutoSol (Adams et al., 2010). Crystals of Atg7NTD-Atg3FRpep were grown at room temp by mixing 1 μL Atg7NTD 30 mg/ml premixed with an 8-fold molar excess peptide and 1 μL well solution containing 1.8 M ammonium sulfate, 0.1 M Bis-Tris pH 5.5 and 5 mM DTT. After soaking in well solution supplemented with 6.25% glycerol and 6.25% ethylene glycol, crystals were cryoprotected in well solution supplemented with 12.5% glycerol and 12.5% ethylene glycol. Phases were obtained by molecular replacement with PHENIX AutoMR (Adams et al., 2010) using the Atg7NTD structure as a search model. Atg7NTD(Pro283Asp) at ∼25mg/ml crystallized at 4°C in 10% isopropanol, 0.1 M potassium thiocyanate, and 0.05 M citrate buffer pH 4.4. Crystals were cryoprotected in well solution supplemented with 30% ethylene glycol. Atg7CTD at ∼15mg/ml crystalized at 4°C in 12% PEG 20k, 0.1 M Tris 8.5, 10 mM TCEP, and upon harvesting were soaked in 13% PEG20k, 0.1M Tris 8.5, 10 mM TCEP, 7.5% ethylene glycol, 7.5% glycerol, prior to cryoprotection in 13% PEG20k, 0.1M Tris 8.5, 10 mM TCEP, 15% ethylene glycol, 15% glycerol. Phases were obtained using Molecular Replacement-Single Wavelength Anomalous Diffraction (MR-SAD) at the zinc peak wavelength with ThiF [Chain A, 1ZKM.pdb (Duda et al., 2005)] as the searchmodel, with PHENIX (Adams et al., 2010). All crystals were flash-frozen in liquid nitrogen. Diffraction data were processed with HKL2000 (Otwinowski and Minor, 1997). After phasing, cycles of manual model rebuilding and refinement were performed with Coot (Emsley et al., 2010) and phenix.refine (Adams et al., 2010).
In the crystal, Atg7CTD forms a symmetric homodimer, with one Atg7CTD monomer in the asymmetric unit and the second subunit in the homodimer generated through application of crystallographic symmetry operators.
Biophysical analyses
Isothermal titration calorimetry (ITC) was performed with proteins/peptides in 25 mM HEPES pH 8.0, 150 mM NaCl and 2 mM β-mercaptoethanol (BME) at 30 °C using a MicroCal ITC200. Atg7 or Atg7NTD were placed in the sample cell at concentrations ∼35-90 μM, with titrating Atg3, Atg3ΔFR or AtgFRpep in the syringe. Data were evaluated using Origin (V 7.0) to determine the values of the thermodynamic parameters and are shown as means +/− s.d. (n=3). AUC, SAXS and SEC-MALS analyses are described in detail in supplemental information.
Biochemical analyses
Pulse-chase assay was adapted from (Huang et al., 2009). Atg8 was expressed with a N-terminal Protein Kinase A (PKA) tag for 32P-labeling using PKA (New England Biolabs). Atg7 proteins were charged with [32P]Atg8 in reactions containing 50 mM HEPES pH 7.5, 2 mM ATP, 5 mM MgCl2, 2 mg/ml BSA, 1 μM Atg7 (wild-type or mutants) and 2.5 μM [32P]Atg8 (for assays in Figure S3E, 5 mM ATP, 10 mM MgCl2 and 5 μM [32P]Atg8 was used). For all assays except that in Figure S3E, charging reactions were desalted into 50 mM HEPES pH 7.5, 50 mM EDTA pH 8.0 to prevent reloading of Atg7. For the chase, [32P]Atg8 transfer from Atg7 to Atg3 was assayed in reactions containing 100 mM HEPES pH 7.5, 2 mg/ml BSA, 50 mM EDTA pH 8.0, 500 nM Atg3, with a 1:5 dilution of desalted Atg7∼[32P]Atg8 (for assays in Figure S3E, 5 μM Atg3 was used and chase was performed on ice). Since the Atg7trans and Atg7cis contain half as many active/Atg3-binding sites as wild-type Atg7, twice as much of these enzymes were used relative to wild-type Atg7. Reactions were quenched with 2× SDS loading buffer and run on Nu-PAGE 4-12% Bis-Tris gels in MOPS buffer (Invitrogen).
Cross-linking assays used mutant Atg3 in which all cysteines other than the catalytic Cys234 were mutated to Ala (Atg3C234only). Atg3C234only was reduced with 10 mM DTT then desalted in 20 mM HEPES pH 7.0, 150 mM NaCl. 100 μM Atg3C234only was mixed with five-fold molar excess of Bis-Maleimidoethane (BMOE) (Pierce) and incubated on ice for >1 hour followed by removal of unreacted crosslinker by desalting. Atg7 proteins were reduced with 2 mM TCEP. Crosslinking was initiated by adding Atg3C234only∼BMOE to Atg7 such that the final Atg3 concentration was 120 nM, and the final Atg7 concentration was 60 nM or 120 nM as indicated. After 60 seconds, crosslinking was quenched with 100 mM DTT. Samples were separated by SDS-PAGE, transferred to PVDF membrane and detected by immunoblotting against Atg7 (yN-16, Santa Cruz, #SC8964).
Cell-based assays, pulldowns, Atg8 lipidation, and formation of the Atg7∼Atg8 thiolester intermediate, are described in the supplemental information.
Supplementary Material
Acknowledgments
This work was supported by ALSAC, the St. Jude Cancer Center grant, NIH R01GM077053 to BAS, R01AI40646 to DRG, RO1GM034496 to COR, Hartwell Foundation fellowship to AHW, and the Howard Hughes Medical Institute. BAS is an Investigator of the Howard Hughes Medical Institute. We are grateful to CA Regni for help in early stages of this project, M Frank for providing liposomes, R Cassell and P Rodrigues for expert peptide synthesis and purification, and DW Miller, S Bozeman, DJ Miller, and J Bollinger for administrative/computational support. NECAT is supported by NIH NCRR RR-15301, APS by US DOE W-31-109-ENG-38, ALS by US DOE DE-AC02-05CH11231. SAXS at the SIBYLS beamline BL12.3.1 of ALS is supported in part by the US DOE Integrated Diffraction Analysis Technologies (IDAT).
Footnotes
Accession numbers: Coordinates and structure factors for yeast Atg7CTD, Atg7NTD, Atg7NTD-Atg3FRpep, and Atg7NTD(P283D) have been deposited to the RCSB with accession codes 3T7E, 3T7F, 3T7G, and 3T7H, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia VM, Davies OR. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim Biophys Acta. 2006;1760:1304–1313. doi: 10.1016/j.bbagen.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Duda DM, Walden H, Sfondouris J, Schulman BA. Structural analysis of Escherichia coli ThiF. J Mol Biol. 2005;349:774–786. doi: 10.1016/j.jmb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol. 2000;2:E153–157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Tanida I, Ueno T, Ohsumi M, Ohsumi Y, Kominami E. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1-E2 complex formation. J Biol Chem. 2001;276:9846–9854. doi: 10.1074/jbc.M007737200. [DOI] [PubMed] [Google Scholar]
- Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45:11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Dikic I. The Three Musketeers of Autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 2011 doi: 10.1016/j.tcb.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F. ATG systems from the protein structural point of view. Chem Rev. 2009;109:1587–1598. doi: 10.1021/cr800459r. [DOI] [PubMed] [Google Scholar]
- Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology, Macromolecular Crystallography, part A. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich FC, Jr, Haas AL. Activation of ubiquitin and ubiquitin-like proteins. Subcell Biochem. 2010;54:1–16. doi: 10.1007/978-1-4419-6676-6_1. [DOI] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Suzuki NN, Hanada T, Ichimura Y, Kumeta H, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 2:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.