Abstract
With the advent of novel sequencing technologies, interest in the identification of rare variants that influence common traits has increased rapidly. Standard statistical methods, such as the Cochrane-Armitage trend test or logistic regression, fail in this setting for the analysis of unrelated subjects because of the rareness of the variants. Recently, various alternative approaches have been proposed that circumvent the rareness problem by collapsing rare variants in a defined genetic region or sets of regions. We provide an overview of these collapsing methods for association analysis and discuss the use of permutation approaches for significance testing of the data-adaptive methods.
Keywords: association, collapsing methods, collection of rare variants, common disease/rare variant hypothesis, contingency table, generalized linear model, next-generation sequencing, pooling methods
Introduction
Genome-wide association studies have been successful in identifying common single-nucleotide polymorphisms (SNPs) that contribute to complex genetic disease [Manolio, 2010]. They are thus supportive of the common disease/common variant hypothesis, which states that frequent SNPs contribute to widespread disease. However, only a portion of the heritability as estimated from twin, adoption, and family studies can be explained by loci identified in genome-wide association studies. Although both the validity and the accuracy of the heritability estimates are questionable [Ziegler and König, 2010, ch. 6], it is widely believed that there is missing heritability [Maher, 2008; Eichler et al., 2010]. This hypothesis, if true, suggests that other genetic mechanisms, such as gene-gene interaction, epigenetics, and rare variants, contribute to disease susceptibility. Indeed, although the exploration of these factors has just begun, there already is increasing evidence that gene-gene interaction and epigenetics play a role [Cordell, 2009; Petronis, 2010].
The investigation of rare variants (rare generally corresponds to a minor allele frequency [MAF] < 1%) is complicated because relatively few rare variants are well represented in current genome-wide association arrays; in addition, the methods used for genome-wide association analysis have low power for low-MAF SNPs unless the effect size is large, and untyped rare variants are poorly tagged by common SNPs. The combination of low MAFs and poor tagging properties makes rare variants unsuitable for analysis with the microarrays used in genome-wide association studies [Asimit and Zeggini, 2010]. However, with the development of novel technologies for high-throughput next-generation sequencing [Metzker, 2009; Meyerson et al., 2010], it is now possible to sequence regions of interest, the exome, or even the entire genome. Unfortunately, the costs are still high, there is a trade-off between cost and accuracy, and the post-processing and analysis of sequence data are challenging.
The reasons that multiple rare variants might play a role in complex genetic disease have been summarized by Bansal et al. [2010]: With the recent expansion of the human population, a large number of segregating, functionally relevant rare variants have emerged that mediate phenotypic variation. Furthermore, multiple rare variants within the same gene contribute to largely monogenic disease [Fitze et al., 2002; Easton et al., 2007]. It is therefore reasonable to assume that the same genetic mechanisms that operate for complex disease also apply to common disease. Finally, and most important, sequencing studies that focus on specific genes have shown that collections of rare variants can indeed associate with particular phenotypes [Bansal et al., 2010, Table 1].
Several strategies for identifying rare variants that contribute to disease susceptibility have been proposed and include the study of families and studies that place increasing emphasis on other structural variants, such as insertions, deletions, inversions, or translocations [Manolio et al., 2009, Box 1]. The study of large families is a promising approach in this context. Specifically, the study of extended pedigrees has several advantages over unrelated subjects. First, some rare variants can be observed at higher frequencies in extended pedigrees compared to the general population. Second, rare variants that segregate with reasonably high penetrance in extended pedigrees can provide a linkage signal so that deep sequencing in extended pedigrees is not required for the entire genome but only in those chromosomal regions that show a linkage signal. Thus the sequencing effort is substantially reduced. Third, one can expect larger effect sizes of the rare variants. Fourth, the results are simpler to interpret because the rare variants, together with the disease, run within the families and therefore provide a proof of the genetic basis. Finally, families are generally simpler to follow up than individuals.
As an alternative to the study of families, the investigation of subjects with unusual phenotypes or from the extreme ends of the phenotype distribution can be reasonable, as can studies in isolated or founder populations or of subjects of recent African ancestry. These study designs generally lead to analyses that are analogous to those of standard case-control studies. In addition, population-based cohort studies might be of interest to obtain unbiased estimates of population parameters. In all scenarios involving unrelated subjects, the statistical analysis of rare variants remains challenging because of low power. One approach to overcome the problem of low power is to pool rare variants for analysis, and these methods are generally called collapsing methods.
Several novel collapsing approaches were proposed during Genetic Analysis Workshop 17 (GAW17), and these were often compared with already published collapsing approaches. Furthermore, GAW17 contributors compared various approaches discussed in the literature using the simulated data provided for the workshop. Our aim in this paper is to provide a comprehensive overview of published collapsing methods and the permutation approaches most studies require to assess statistical significance. For simplicity, we restrict the description of the statistical approaches to dichotomous phenotypes.
Indicator and Proportion Coding for Collapsing Methods
Let na and nu denote the number of affected and unaffected subjects, respectively, with n = na + nu. We consider a region of interest (ROI), and this term will be discussed in greater detail in the next section. Two fundamentally different ideas are generally used for collapsing. The first, termed indicator coding, results in a dichotomous variable that indicates presence or absence of any rare variant within the ROI. Formally, the expression
| (1) |
denotes whether a variant is present at chromosomal position i in the ROI in subject j. Then, with K denoting the number of sites with variants in the ROI,
| (2) |
denotes whether subject j carries any rare variant in the ROI.
In proportion coding we count the number of variants of subject j over all the K sites; that is, denotes the number of variants at position i of subject j so that the proportion of all variants is given by
| (3) |
Both Eqs. (2) and (3) indicate that all rare variants are weighted equally, independent of their frequency. Because the effect size of a variant may depend on its frequency, several approaches to up- or down-weight variants in a ROI have been proposed. For example, Madsen and Browning [2009] suggested weighting variants according to their frequency in unaffected subjects. Specifically, they weighted site i inversely proportional to its variance:
| (4) |
where
| (5) |
is an estimate of the MAF in unaffected subjects and denotes the number of variants in the unaffected subject j. The correction factors 1 and 2 in the numerator and denominator, respectively, of Eq. (5) are included because a rare variant might be present only in case subjects. Price et al. [2010] used a similar weighting approach with weights
| (6) |
where is the estimated MAF at site i. However, Price and colleagues did not consider the critical case that a rare variant is present only in affected subjects.
Another weighting alternative is to jointly consider affected and unaffected subjects so that
| (7) |
where p̂i is the estimated MAF at site i estimated over all subjects.
Finally, we note that the coding of Eq. (3) for each site corresponds to the coding of an additive genetic model, and generalizations to other genetic models are straightforward.
Options in Coding and Analyzing Rare Variants
Both the indicator and proportion coding approaches allow flexibility with respect to the units that are considered for collapsing. The ROI can be a gene, a gene cluster, a combination of different genes (e.g., several genes from the same pathway), or even an arbitrary chromosomal region defined by base-pair positions on a chromosome.
In addition, different thresholds can be used for collapsing. More specifically, rare variants with MAF ≤ p are considered collapsed, and the standard choices for collapsing are p1 (i.e., p = 0.01) and p5 (i.e., p = 0.05). The threshold may be either fixed or variable, and this has been discussed in detail by Price et al. [2010]. Furthermore, the analysis in the ROI can be restricted to rare variants only. Alternatively, frequent SNPs and rare variants can be analyzed simultaneously.
Finally, rare variants can be grouped according to their functionality. More specifically, one can restrict counting to synonymous or nonsynonymous rare variants only. Another option is to group rare variants according to their predicted effect, which may be beneficial, neutral, or deleterious. For these groupings, standard packages and databases, such as PolyPhen2 [Ramensky et al., 2002], SIFT [Ng and Henikoff, 2003], SNAP [Bromberg et al., 2008], or SNPs3D [Yue et al., 2006], can be used.
Implementation of these three approaches—unit definition, threshold type, and rare variant grouping—can be accomplished by integrating indicator variables that flag variants for inclusion in Eqs. (2) and (3).
Statistical Tests for Rare Variants
Now that we have described the collapsing approaches and the flexibilities in the up- or down-weighting of rare variants, we can now outline the different statistical approaches for analysis.
Standard Methods for Analysis: Trend Test and Regression
If the MAF is sufficiently high at a specific site, standard approaches, such as the Cochran-Armitage trend test or a logistic regression, can be conducted (see, e.g., Morris and Zeggini [2010]); for a detailed description of these methods, see, for example, the textbook of Ziegler and König [2010]. Single-site analysis is similar to indicator coding in the sense that only a specific site is filtered. However, the power of this approach is extremely low if the MAF is low. For example, with an odds ratio of 2 and a type I error level of 5 × 10−8, in order to detect the association at a site with a MAF of 0.005 in control subjects under the assumption of Hardy-Weinberg equilibrium, the required sample size would have to exceed 20,000.
It may therefore be more reasonable to use indicator or proportion coding over all sites in a logistic regression or any other model from the class of generalized linear models. In this case, one models
| (8a) |
or
| (8b) |
where yj denotes the phenotype of subject j, zj is a vector of covariates, and is the parameter vector to be estimated. The effects of β1 or can be tested using likelihood ratio, score, or Wald-type tests. Morris and Zeggini [2010] developed the likelihood ratio indicator coding test (rare variant test 2, or RVT2) and the likelihood ratio proportion coding test (RVT1). A limitation of both RVT1 and RVT2 is that they can be applied only to rare variants (for a discussion, see the next subsection). The inclusion of frequent variants is not intended, and a multivariate model to include both rare variants and frequent SNPs from different sites has not been formulated so far.
Collapsing and Summation Test and Combined Multivariate and Collapsing Method
The collapsing and summation test (CAST) is a simple approach for comparing case subjects with control subjects. More precisely, it compares the number of case subjects with a variant with the number of control subjects with a variant in a 2 × 2 frequency table. CAST has been applied in different studies (see, e.g., Fitze et al. [2002]) and has been described in detail by Morgenthaler and Thilly [2007]. A restriction of CAST is that it is limited to rare variants. Rare and frequent variants cannot be investigated jointly in a multivariate test. Therefore the approach of Li and Leal [2008] is a straightforward generalization. For the joint analysis of rare variants and frequent SNPs, they proposed a three-step approach. In the first step, a test statistic is calculated for the collapsed rare variants using CAST. Next, univariate test statistics are calculated for frequent SNPs. Finally, a standard combination rule, such as the inverse normal method or Fisher’s combination rule, can be applied to all rare variants and frequent SNPs in the ROI.
More elegant is a multivariate test statistic, termed the combined multivariate and collapsing (CMC) method, such as Hotelling’s T2 for both rare variants and frequent SNPs [Li and Leal, 2008].
Weighted-Sum Collapsing
The weighted-sum (WS) collapsing approach [Madsen and Browning, 2009] gives more weight to rare variants because stronger effects are expected for rare variants than for more frequent ones. The original WS approach of Madsen and Browning is performed in five steps:
-
Step 1.
Site-specific weights wi are estimated using Eq. (4).
-
Step 2.Subject-specific genetic scores are estimated using
(9) where denotes the number of variants of subject j at site i.
-
Step 3.The sum of the rank of the score is determined over all affected subjects; that is,
(10) -
Step 4.
The affection status is permuted B times. For each permutation, steps 1 to 3 are repeated, yielding in permutation b.
-
Step 5.
The p-value is not estimated as the number of permutations exceeding r. Instead, the mean (μ̂B) and the standard deviation (σ̂B) of the permutation distribution are estimated. This permutation distribution should reflect the distribution of the test statistic under the null hypothesis of no association. The p-value is estimated from the standard normal distribution using (r − μ̂B)/σ̂B.
Data-Adaptive Summation
None of the approaches discussed so far differentiates between beneficial, neutral, or deleterious variants. One approach in this direction has been proposed by Han and Pan [2010]. They introduced data-adaptive weights and summed over all variants, and their method is therefore termed adaptive summation (aSUM). Specifically, they considered the regression model with proportion coding of Eq. (8b), and embedded this regression approach in the following four-step procedure:
-
Step 1.
A logistic regression is fitted using the original data for every site i.
-
Step 2.If the p-value pi of the statistical test at site i is less than 0.1 and if the estimated regression coefficient β̂i is negative (i.e., the variant has a beneficial effect in the data), then the coding of all subjects j is altered at site i. Otherwise, the coding is left unchanged. That is,
(11) -
Step 3.
With the recoded data , the RVT1 model of Eq. (8b) is estimated and a test statistic is formed.
-
Step 4.
Because the coding at the sites is changed data adaptively, the p-value from this procedure is determined using permutations. Specifically, the affection status is permuted B times, and steps 1 to 3 are repeated for each permutation sample b. The p-value is determined in the classical way as the proportion of permutations for which the permuted test statistic exceeds the test statistic of the original data.
Variable Threshold Collapsing
The aSUM method is one approach to differentiating between beneficial and strongly deleterious variants. The variable threshold (VT) approach of Price et al. [2010] permits another flexibility. All statistical methods discussed so far assume a fixed threshold for collapsing rare variants. However, the optimal threshold τ for which rare variants with MAF < τ are more likely to be functional than SNPs with a MAF ≥ τ is unknown. For this reason, Price et al. [2010] proposed computing the maximum test statistic over all reasonable thresholds τ, and they used the following three-step procedure:
-
Step 1.Determine the proportion coding of subject j when all rare variants with MAF < τ are included in
(12) The MAF is estimated in the entire sample.
-
Step 2.The test statistic is
(13) with(14) This means that the maximum of threshold-specific test statistics T(τ) is calculated, and T(τ) is obtained by summing the proportion of coded variants at threshold τ over all affected subjects.
-
Step 3.
The p-value from this procedure is determined using permutations. Specifically, the affection status is permuted B times, and steps 1 and 2 are repeated for each permutation sample b. The p-value is determined in the classical way as the proportion of permutations for which the permuted test statistic exceeds the test statistic of the original data.
A disadvantage of the VT approach is that the variability at a site increases with the MAF. Therefore there might be a trade-off between summation over all rare variants with a large effect and the increased variability, and this needs to be investigated further.
Permutation Approaches in Significance Testing
All the approaches described in the “Statistical Tests for Rare Variants” section use estimates from the observed data (e.g., MAF in control subjects, direction of effect at a variant) in a data-adaptive manner where both model selection and model testing steps occur. These approaches depend on permutations to estimate significance. Determining an estimate of statistical significance using permutations requires permuting enough times to reach the desired level of significance. Because the data from high-throughput sequencing studies can be quite large, the calculations require a substantial amount of computer-processing time.
Classical permutation tests have been advised in most of the papers involving permutation procedures [Han and Pan, 2010; Price et al., 2010]. A sufficient condition for a permutation test to be exact is exchangeability under the null hypothesis [Good, 1994]. However, it is unclear whether the test statistics of interest remain exchangeable in next-generation sequencing studies.
Because the number of required permutations can be quite large for a reliable estimation of the p-value, Madsen and Browning [2009] proposed a different approach. They estimated the mean and standard deviation from the permutation distribution using a relatively small number of permutations (e.g., 1,000), assuming that these represent the moments of the distribution under the null hypothesis of no association; they then used the standard normal distribution to derive a p-value. However, the permutation distribution need not adequately reflect the distribution under the null hypothesis (Fig. 1). Furthermore, it is by no means clear that the permutation distribution will be close to a normal distribution in the tail of the permutation distribution, and the estimation of small p-values using a normal distribution might be misleading. Alternative approaches in the same direction have been proposed by, for example, Qian [2004] and Knijnenburg et al. [2009], who also estimated the tails of the permutation distribution. Whereas Qian linearly extrapolated the tail of the normal distribution, Knijnenburg and colleagues considered the generalized Pareto distribution (GPD) for p-value approximation in the extreme tails of the distribution. Further work is required to determine the optimal approximation and the validity of the permutation approaches.
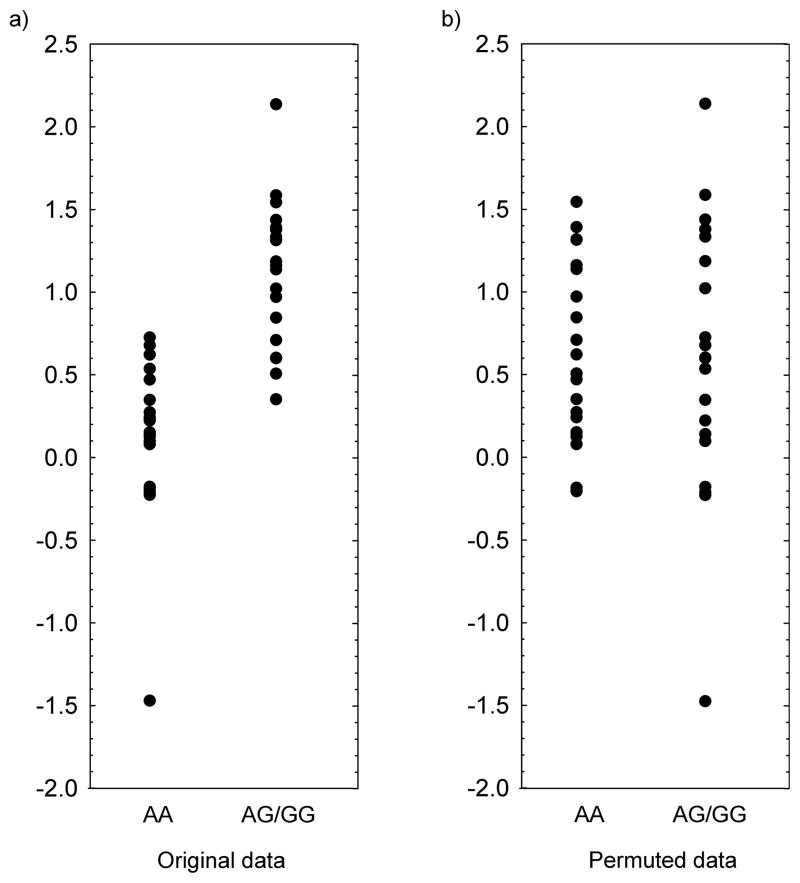
Fig. 1. Failure of standard permutation methods.
Consider an autosomal SNP with an allele frequency of 0.7 and a recessive genetic model for the A allele. The phenotype is assumed to be normally distributed. (a) Two genotype groups in the original sample differ in their means but the variances are identical. The assumptions of the standard t-test are met. (b) One permutation sample of the same data as in panel (a) is shown. Note that the permuted data are not unimodal and that, although the variances in the permuted sample are similar for both genotype groups, the group-specific permutation variance is substantially larger than the group-specific variance in the original sample. In fact, the standard two-sided t-test yields p = 0.03, whereas the Madsen and Browning permutation approach with 1,000 permutations gives p = 0.23 and the standard permutation approach, also with 1,000 permutations, results in p = 0 because none of the permuted samples reveal a t statistic larger than the original data. As a result, the Madsen and Browning permutation approach leads to a different conclusion compared with the standard t test. Furthermore, the classical permutation method most likely yields p = 0, even for a high number of permutations, and thus underestimates the p-value.
Final Remarks
We have provided an overview of the different collapsing methods that were published in the literature until October 2010. Since then, several other approaches have been proposed [Hoffmann et al., 2010; Li et al., 2010; Liu and Leal, 2010], and two review papers on association studies involving rare variants have become available [Asimit and Zeggini, 2010; Bansal et al., 2010]. Novel approaches for the analysis of rare variants were also proposed during GAW17, and the new collapsing methods are summarized by Sun et al. [2011] and Tintle et al. [2010].
Acknowledgments
The work of CD was supported by an intramural grant from the Medical Faculty of the University at Lübeck. The Genetic Analysis Workshops are supported by National Institutes of Health grant R01 GM031575 from the National Institute of General Medical Sciences.
References
- Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annu Rev Genet. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet. 2010;11(11):773–85. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24(20):2397–8. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ. Genome-wide association studies: Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81(5):873–83. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitze G, Cramer J, Ziegler A, Schierz M, Schreiber M, Kuhlisch E, Roesner D, Schackert HK. Association between c135G/A genotype and RET proto-oncogene germline mutations and phenotype of Hirschsprung’s disease. Lancet. 2002;359(9313):1200–5. doi: 10.1016/S0140-6736(02)08218-1. [DOI] [PubMed] [Google Scholar]
- Good P. Permutation Tests: A Practical Guide to Resampling Methods for Testing Hypotheses. New York: Springer; 1994. [Google Scholar]
- Han F, Pan W. A data-adaptive sum test for disease association with multiple common or rare variants. Hum Hered. 2010;70(1):42–54. doi: 10.1159/000288704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, Marini NJ, Witte JS. Comprehensive approach to analyzing rare genetic variants. PLoS One. 2010;5(11):e13584. doi: 10.1371/journal.pone.0013584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knijnenburg TA, Wessels LF, Reinders MJ, Shmulevich I. Fewer permutations, more accurate P-values. Bioinformatics. 2009;25(12):161–8. doi: 10.1093/bioinformatics/btp211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Byrnes AE, Li M. To identify associations with rare variants, just WHaIT: weighted haplotype and imputation-based tests. Am J Hum Genet. 2010;87(5):728–35. doi: 10.1016/j.ajhg.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6(10):e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456(7218):18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Manolio TA. Genomewide association studies and assessment of the risk of disease. New Engl J Med. 2010;363(2):166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies: the next generation. Nat Rev Genet. 2009;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat Res. 2007;615(1–2):28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34(2):188–93. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465(7299):721–7. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86(6):832–8. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D. Haplotype sharing correlation analysis using family data: a comparison with family-based association test in the presence of allelic heterogeneity. Genet Epidemiol. 2004;27(1):43–52. doi: 10.1002/gepi.20005. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV, Sung YJ, Tintle N, Ziegler A. Identification of genetic association of multiple rare variants using collapsing methods. Genet Epidemiol. 2011;X(suppl X):X–X. doi: 10.1002/gepi.20658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintle N, Aschard H, Hu I, Nock N, Wang H, Pugh E. Inflated type I error rates when using aggregation methods to analyze rare variants in the 1000 Genomes Project exon sequencing data in unrelated individuals: summary results from Group 7 at Genetic Analysis Workshop 17. Genet Epidemiol. 2011;X(suppl X):X–X. doi: 10.1002/gepi.20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinform. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Konig IR. A Statistical Approach to Genetic Epidemiology: Concepts and Applications. 2. Weinheim, Germany: Wiley-VCH; 2010. [Google Scholar]