Abstract
Aims: To establish conditions to study the oligomeric assembly of heme-binding protein (HbpS) in solution by applying the tools of site-directed spin labeling combined with pulse electron paramagnetic resonance (SDSL EPR) spectroscopy, as well as to analyze redox stress-based conformational changes in HbpS subunits within the oligomer in solution. In vivo elucidation of molecular mechanisms that control the downregulation of the novel redox-system HbpS-SenS-SenR. Results: Using a set of specifically generated HbpS mutants, and SDSL EPR spectroscopy, we show the octomeric assembly of HbpS in solution, and demonstrate that iron-mediated stress induces conformational changes in HbpS subunits within the octamer. We further demonstrate that the catalase-peroxidase CpeB protects HbpS from hydrogen peroxide (H2O2)-mediated oxidative attack in vivo. Moreover, chromosomal inactivation of cpeB results in an enhanced sensitivity of the mutant to redox-cycling compounds. Innovation: SDSL EPR has been used in this work for the first time to monitor redox-mediated conformational changes in a redox-sensing protein in solution. This work substantially explains redox-dependent dynamics in HbpS at the atomic level, and presents novel molecular mechanisms supporting downregulation of a signaling cascade. Conclusion: Iron-mediated stress induces movements of subunits within the HbpS octomeric assembly. We suggest a motion of the C-terminal α-helix toward the preceding helical segment. These events upregulate the activity of the HbpS-SenS-SenR system, in which HbpS acts as an accessory element. The mycelia-associated CpeB, under the control of HbpS-SenS-SenR, protects the extracellular HbpS from oxidation in vivo. Thus, de novo synthesized HbpS proteins downregulate the HbpS-SenS-SenR signaling cascade. Antioxid. Redox Signal. 16, 639–648.
Introduction
As soil-dwelling organisms, streptomycetes should be well equipped to coexist with their natural neighbors, including bacteria, fungi, plants, and insects (3, 7, 17). They should also respond to highly variable conditions, including changes in environmental temperature, pH, humidity, and redox state. The redox state in soil is strongly influenced by iron, which can form a range of oxidation states, including ferrous (II) and ferric (III). In the ferrous form, it plays a crucial role in many biological pathways, such as respiration, photosynthesis, nitrogen fixation, and gene regulation (2, 27). In the presence of oxygen, ferrous iron ions frequently result in oxidative stress via the Fenton reaction-mediated generation of highly reactive hydroxyl radicals (8), which damage nucleic acids, proteins, and lipids. Moreover, iron has also evolved as a sensor of oxygen and reactive oxygen species (ROS). Thus, iron or an iron cofactor that is coordinated in the metal center of a polypeptide chain fine tunes the protein's reactivity. The protein subsequently translates the iron-mediated redox reaction into a biochemical effect in form of a change in activity (27).
Recently, a novel type of redox-sensing system has been discovered: the three-component signaling system HbpS-SenS-SenR from the cellulose degrader Streptomyces reticuli (24). This system provides S. reticuli with an efficient defense mechanism against the toxicity of redox-cycling compounds (9). Since homologs of HbpS-SenS-SenR have been identified in a number of ecologically and medically relevant bacteria, it has been proposed as a bacterial model of a redox-sensing pathway (24).The membrane protein SenS is a sensor kinase that, after autophosphorylation, transfers the phosphate group to its cognate response regulator SenR. In dependence of its phosphorylation state, SenR regulates transcription of the senS-senR operon, the hbpS gene encoding a secreted heme-binding protein (HbpS), and the furS-cpeB operon encoding the redox regulator (FurS) and the mycelia-associated catalase-peroxidase (CpeB) (4). It is noteworthy that CpeB is highly homologous to the catalase-peroxidase KatG from Mycobacterium tuberculosis (36). KatG plays an essential role in the survival of the organism in the environment of the phagocyte oxidative burst. Interestingly, the front-line anti-tuberculosis drug isoniazid requires KatG activation before exerting a lethal effect (1, 35). Further, CpeB has been shown to degrade H2O2 in vitro (37). However, the in vivo role of CpeB during oxidative stress has not yet been explored.
Innovation.
Despite the fact that the novel bacterial redox-sensing system HbpS-SenS-SenR has been well characterized, the following questions remained to be addressed: (1) which structural events at the atomic level lead to upregulation, and occur on iron-mediated redox-stress within the octomeric heme-binding protein (HbpS), that is, the sensory and modulatory element of the HbpS-SenS-SenR system; (2) what kind of molecular mechanisms are responsible for the downregulation of the HbpS-SenS-SenR-mediated signaling cacade. In this study, using a set of HbpS mutants and spin labeling combined with pulse electron paramagnetic resonance (SDSL EPR), we show that iron-mediated stress induces conformational changes in HbpS subunits within the octamer. We suggest that a motion of the C-terminal α-helix toward the preceding helical segment ultimately results in an upregulation of HbpS-SenS-SenR. Remarkably, SDSL EPR has been used in this work for the first time to monitor redox-mediated conformational changes in a sensory protein in solution. This report can, therefore, serve as a guide for related studies. Further, we demonstrate that the catalase-peroxidase CpeB, being controlled by HbpS-SenS-SenR, protects HbpS in vivo from H2O2-mediated oxidative attack. Moreover, CpeB provides Streptomyces reticuli with a nonstressed environment, in which freshly synthesized HbpS proteins downregulate the HbpS-SenS-SenR signaling cascade. Since homologs to HbpS-SenS-SenR are widespread in different bacteria with ecological and medical relevance, the data presented here will be useful in investigating the biological role of these homologs.
HbpS is an extracellular octomeric protein that not only binds heme but also degrades it via a nonenzymatic pathway known as coupled oxidation (20), leading to free iron ions that are captured and displayed at the protein surface (25). It specifically interacts with SenS and modulates its autophosphorylation. Although HbpS inhibits the autophosphorylation of SenS under nonstressed conditions, in the additional presence of iron ions or heme combined with redox stress, HbpS activates SenS activity (5). Further fluorescence resonance energy transfer (FRET) and circular dichroism (CD) spectroscopy analyses revealed that iron-mediated oxidative stress induces both secondary structure and overall intrinsic conformational changes within HbpS (26). However, detailed structural analyses that reveal molecular movement of HbpS subunits within the octomeric assembly on stress are lacking.
Here, we used SDSL EPR (12) to analyze iron-based conformational changes within the octomeric HbpS in solution. Additionally, we report the generation of a S. reticuli cpeB disruption mutant and comparative in vivo studies that together allow the elucidation of the role of CpeB in the anti-oxidative stress response as well as in the turnover of HbpS.
Results
Design and spin-labeling of cysteine-containing HbpS mutants
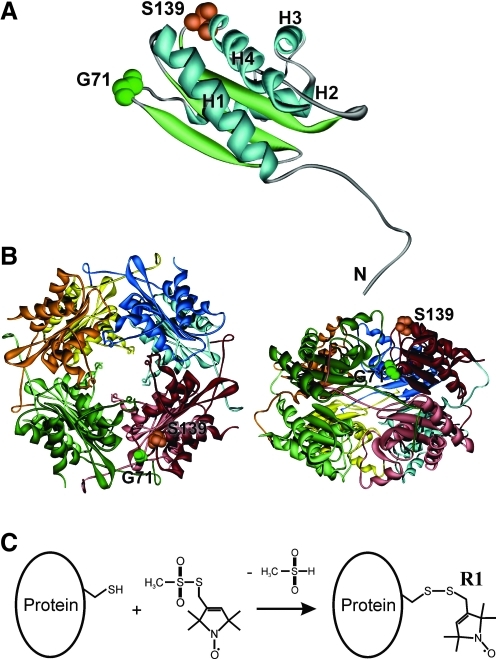
To label the proteins with the [(1-Oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate] spin label (MTSSL), cysteine-containing HbpS proteins were generated. Substitution of single amino acids for cysteine (HbpS does not contain native cysteine residues) was done by site-directed mutagenesis (see Supplementary Materials; Supplementary Data are available online at www.liebertonline.com/ars). Using the available crystal structure of native HbpS (protein data bank [PDB]: 3FPV), amino acids Ser-139 and Gly-71 were selected, as they display high solvent accessibility and are located within HbpS loops (Fig. 1A, B), thereby minimizing possible influences on the protein structure. In addition, Gly-71 is located near the postulated iron-center, which includes Tyr-77, Glu-78, and Glu-81 (26). After isolation of cysteine-containing HbpS by a combination of affinity, anion-exchange, and size-exclusion chromatographies (for details see Supplementary Materials), the proteins were labeled with MTSSL (Fig. 1C). The resulting spin label side chain is abbreviated as R1 hereafter, and the proteins are, therefore, named HbpS-Gly71R1, HbpS-Ser139R1, and HbpS-Gly71R1-Ser139R1.
FIG. 1.
Positions of selected amino acids and site-directed spin labeling. (A) An image showing the amino acid residues Ser-139 (S139) and Gly-71 (G71) on the crystal structure of a single heme-binding protein (HbpS) unit (PDB: 3FPV). α-Helices are numbered H1–H4. The N-terminus is indicated. (B) Octomeric HbpS assembly viewed down the fourfold axis (left) and from the “side” (right) (PDB: 3FPV). A single HbpS monomer is highlighted. (C) Site-directed spin labeling. Reaction of the [(1-Oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate] spin label (MTSSL) with the sulfhydryl group of a cysteine side chain, generating the spin label side chain R1. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Assembly and functionality of spin labeled HbpS
CD spectroscopic analysis of unlabeled and labeled cysteine-containing HbpS proteins in their native states revealed that all proteins contained an almost identical distribution of secondary structures: α-helices, 42%±0.5%; β-sheet; 15%±0.2%; turn 19%±0.6%; random coil 24%±0.6%. This is in line with the calculated amounts for the native wild-type (WT) HbpS (26), thus indicating that neither the insertion of a cysteine residue nor the spin labeling has an effect on the protein structure. Additionally, native-polyacrylamide gel electrophoresis (PAGE) indicated that all proteins retained the oligomeric assembly, as they show almost identical migration behavior to the WT on the gel (see Supplementary Fig. S1).
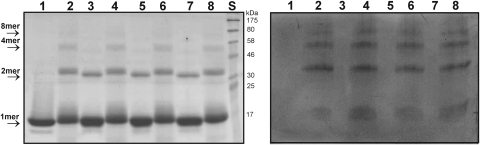
After optimization, including use of different reducing agents, reaction times, and order of addition of the redox-sensitive spin labels, we could successfully label the stressed HbpS samples. The functionality of the labeled samples, in terms of response to iron-mediated oxidative stress, was tested by sodium dodecyl sulfate (SDS)-PAGE combined with immunoblotting using the OxyBlot Kit (Chemicon International, Temecula, CA) that contains anti-DNP antibody. This kit allows the identification of protein carbonyl groups that are introduced into amino acid side chains during protein oxidation (26). As for the WT, the formation of SDS-insensitive crosslinked forms could be detected in all mutants on iron-mediated stress (Fig. 2, left). Each untreated sample showed a protein band with a molecular weight of ∼16 kDa, representing the monomeric form of the protein (15.5 kDa). Formation of a dimeric form (∼32 kDa) was observed with all untreated cysteine-containing mutants. This might be due to the presence of intermolecular disulfide bridges. Using the OxyBlot kit, it could be observed that, in contrast to the untreated samples, those subjected to stress immuno reacted with the anti-DNP antibody (Fig. 2, right), thus indicating that iron-based stress led to the introduction of carbonyl groups in these proteins. These data demonstrate that spin labeling does not interfere with the ability of the proteins to sense and react to iron-based stress.
FIG. 2.
Iron-mediated crosslinking and carbonylation in HbpS proteins. Left: An aliquot of unstressed (wild-type [WT], lane 1; Ser139R, lane 3; Gly71R, lane 5; Ser139R-Gly71R, lane 7) or iron-stressed (WT, lane 2; Ser139R, lane 4; Gly71R, lane 6; Ser139R-Gly71R, lane 8) HbpS samples were subjected to 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Coomassie staining. Arrows indicate positions for the monomeric (1mer), dimeric (2mer), tetrameric (4mer), or octomeric (8mer) HbpS forms. The approximate sizes of prestained protein markers are also indicated (lane S). Right: A second aliquot of unstressed and iron-stressed samples were subjected to DNP derivatization as described by (26). Reaction products were loaded onto a 12% SDS-PAA gel (in the same order as in the gel showed in the left part of this figure). After electrophoresis, proteins were transferred on to a fluorotrans membrane and treated with anti-DNP antibodies as described earlier (26).
Continuous wave EPR measurements
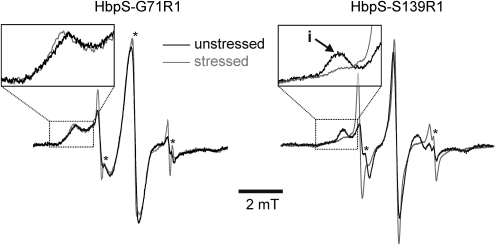
Figure 3 shows the room temperature continuous wave (cw) EPR spectra for the spin labeled HbpS single mutants Gly71R1 and Ser139R1 in the native (unstressed) form and after oxidative stress. For Gly71R1, the spectra for stressed and unstressed HbpS are almost identical, thus indicating that no significant structural changes take place in the vicinity of Gly71. An exception is the significantly increased amount of unbound spin label (*) in the stressed sample due to the protocol used to stress and label the sample (see Materials and Methods section). Nevertheless, some immobilization of the spin label side chain on stress is indicated by a slight shift of the low-field peak to lower field values. In contrast, for Ser139R1, significant spectral changes can be recorded that indicate a structural rearrangement of either Ser139 itself or structural elements/side chains in the direct vicinity of this residue. The low-field peak indicative of R1 side chains with significantly restricted mobility (marked with “i” in the inset showing the low field area) is strongly reduced. Consequently, oxidative stress leads to structural rearrangements in HbpS, which lead to an increased motional freedom of Ser139R1.
FIG. 3.
Room temperature continuous wave electron paramagnetic resonance spectra (cw EPR) for HbpS-G71R1 (left panel) and HbpS-S139R1 (right panel) in native (unstressed) form (black) and after iron-mediated oxidative stress (gray). Original spectra contained a significant amount of contributions of unbound spin label due to the corresponding sample preparation/labeling procedure (described in Materials and Methods section) that have been largely removed by subtraction of correspondingly scaled spectra of free SL in buffer solution. Artifacts caused by this procedure are marked with an asterisk. The inset shows the low field area of the spectra for HbpS-G71R1.
Inter spin distance measurements
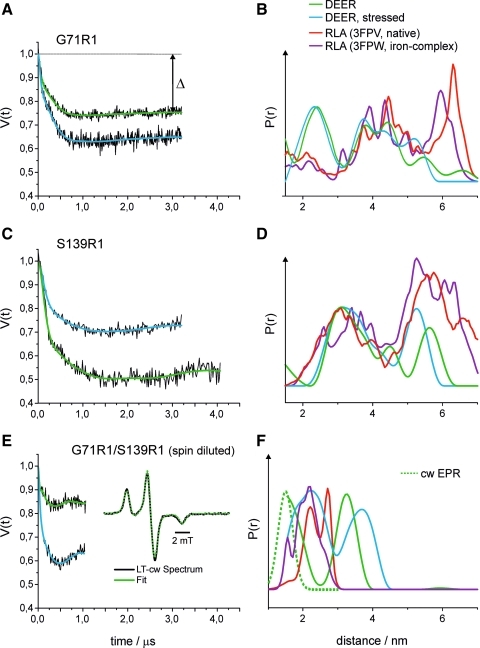
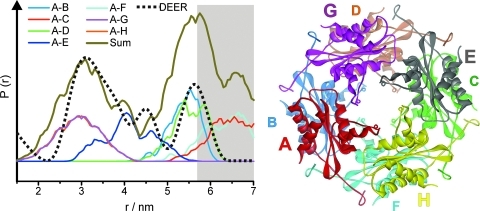
We applied four-pulse DEER spectroscopy (13, 16, 19, 28) and low-temperature cw EPR spectroscopy (34) to determine inter spin distances within the HbpS octamer and, using the double mutant, within the single protomers in the octamer in native and stressed HbpS. The results of the EPR experiments are shown in Figure 4. The obtained distance distributions for native (green) and stressed (blue) HbpS (Fig. 4A, C, E) are compared with the results of a rotamer library analysis (RLA) performed with the published crystal structures of native (red) and iron-bound (purple) HbpS (Fig. 4B, D, F).
FIG. 4.
Influence of oxidative stress on inter spin distances in HbpS. (A, C, E) Background corrected dipolar evolution data (the raw DEER traces together with the background fits are shown in Supplementary Fig. S2 in Supplementary Materials) and corresponding fits obtained by Tikhonov regularization for unstressed (green) and stressed (blue) samples of the respective HbpS mutant. For Gly71R1/Ser139R1 (E), in addition, the low-temperature cw spectrum with the respective fit obtained with DipFit (33) is shown. (B, D, F) Distance distributions obtained by Tikhonov regularization: unstressed–green, stressed–blue. The distance distribution obtained from the low-temperature cw experiment for Gly71R1/Ser139R1 is shown as a broken line (F). The results of the rotamer library analyses are shown in red (PDB: 3FPV, native HbpS) and purple (PDB: 3FPW, HbpS-iron complex). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
For HbpS-Gly71R1 (Fig. 4A, B), the distance distributions for native and stressed HbpS differ only slightly, thus indicating that the relative arrangement of the protein region where Gly-71 is located does not change significantly under the influence of oxidative stress. This observation is in line with the results from the cw experiments, where only minor differences in the spectra are found (Fig. 3). The observed differences in the DEER modulation amplitudes (Fig. 4A, modulation depth Δ) are caused by different labeling efficiencies due to the different protocols used to prepare the samples (see Materials and Methods section) and the resulting variation in unbound spin label in the samples, which contribute to the fraction of noninteracting spins. The comparison with the result from the RLA shows reasonable agreement between experimental and calculated distances in both cases, as the two RLA predicted distance distributions (red: native, purple: stressed) also show only minor differences. This agreement can also be seen in a comparison of the primary DEER data with the corresponding data predicted from the RLA (see Supplementary Fig. S3). Differences visible in the short (1.5–3 nm) and long (>5 nm) distance regions mainly concern the relative probabilities of finding certain inter-spin distances. This could be attributed to the limited signal-to-noise-ratio and incomplete excitation caused by the limited excitation bandwidth of the 12 ns pump pulse used for the short distance range, and the limited dipolar evolution time and uncertainties concerning the background correction (that also result from the limited dipolar evolution time) in the case of distances>5 nm. Hence, the EPR data are in line with the octomeric arrangement of HbpS found in the crystal structure, and further, this arrangement does not seem to be dramatically influenced by oxidative stress.
Oxidative stress also does not significantly influence the inter-spin distance distribution for HbpS-Ser139R1 (Fig. 4C, D), although in this case a significant change of the spin label mobility was deduced from the cw spectra (Fig. 3). The observed shift of the long distance peak from ∼5.7 nm to ∼5.2 for the stressed protein (Fig. 4D) most likely results from the shorter dipolar evolution time in the latter experiment, as distances close to the limits dictated by the length of the dipolar evolution data can be analyzed only qualitatively rather than quantitatively. Nevertheless, good agreement is found between the experimental data and the calculated distances, again confirming the octomeric arrangement of HbpS and the structure of the single subunits. Also here, as for Gly71R1, the RLA yields very similar distance distributions for the native and the stressed form (Fig. 4D). The comparison between the experimental and predicted primary DEER data also confirms these findings (see Supplementary Fig. S3). Figure 5 shows the detailed rotamer analysis for HbpS-Ser139R1 in the native state, relating contributions from single spin label/subunit pairs to the octomeric structure of HbpS.
FIG. 5.
Detailed rotamer analysis for HbpS-S139R1. Left: Distance distribution showing the overall calculated distribution from the rotamer library analysis (olive) and the corresponding contributions from specific spin label pairs as indicated in the legend compared with the experimental result (dotted black line). Right: Octomeric assembly of HbpS (PDB: 3FPV). The subunits are named with letters according to the protomer pairs indicated in the distance distribution. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The DEER experiment with the double mutant HbpS-Gly71R1-Ser139R1 was carried out with a spin diluted sample (Fig. 4E). In this case, the distance distribution should mainly reflect the distance between the two spin labels within the subunits, and intermolecular interactions are strongly suppressed. In contrast to the results for the HbpS single mutants, in this case, stress causes drastic changes in the inter-spin distance distribution (Fig. 4F). Native HbpS shows one distance peak centered at ∼3.3 nm and contributions <2 nm, which have been analyzed by low temperature cw EPR (see inset, Fig. 4E). The cw data revealed an inter spin distance of 1.5 nm with a distribution width of ∼0.4 nm (dotted line, Fig. 4F). For the stressed sample, a similar distance distribution is obtained, which is only slightly shifted by ∼0.5–0.7 nm to longer distances (maxima at 2.2 and 3.8 nm). Surprisingly, the comparison with the RLA shows the opposite tendency. The mean distances of the calculated distributions are located at ∼2.5 and 2.1 nm for native HbpS and the HbpS-iron complex, respectively, and are not bimodal as in the experiments, where the distance peaks are separated by ∼1.5–1.8 nm. It should be noted that we had carried out spin dilutions in a 1:4 stoichiometry (higher dilutions lead to echo amplitudes not sufficient for the DEER experiment) that were not sufficient to completely suppress intermolecular interactions. Therefore, the long-distance peaks in both cases could also arise from intermolecular interactions within the octomer. Nevertheless, even with this assumption, both the experimental distance distributions and the direction of the observed distance change do not correlate with the RLA results. The strong disagreement between the experimental results and the predictions from the RLA for the double mutant can also clearly be seen in the comparison of the experimental and predicted primary DEER data (see Supplementary Fig. S3).
Taken together, the EPR data show significant conformational changes within single HbpS units induced by iron-mediated oxidative stress. The observed distance increase between G71R1 and S139R1 suggests that at least one of the residues is involved in a conformational change, and according to the observed stress-induced mobilization, this is most likely S139R1. G71R1 and S139R1 are located on neighboring edges of the β-sheet structure, and, therefore, a separation of these residues by several Angstroms could be achieved by slight disturbance of this structure. Indeed, changes in the secondary structure distribution of the WT HbpS protein on oxidative stress have been earlier reported (26).
CpeB and response to oxidative stress in vivo
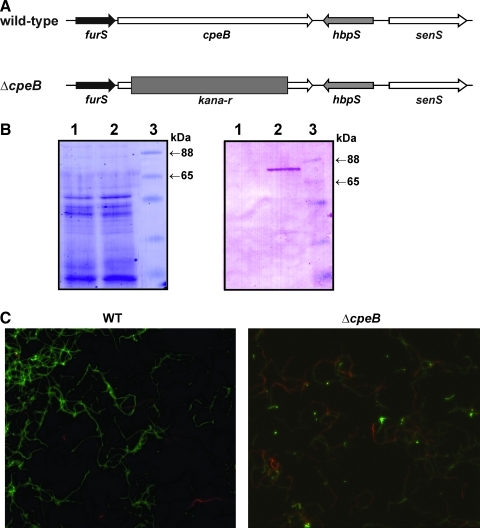
Our previously published data showed that conformational changes in HbpS are accompanied by oxidative modifications. Additionally, we have proposed that the catalase-peroxidase CpeB is an important player in the in vivo turnover of HbpS during oxidative stress (26). To further test this hypothesis and investigate the in vivo role of CpeB during the anti-oxidative stress response in S. reticuli, the chromosomal cpeB gene was disrupted by insertional inactivation (Fig. 6A) (see Supplementary Materials). Western blot analysis revealed that within the mycelia-associated proteins of the strain lacking cpeB (named S. reticuli ΔcpeB), the CpeB protein is missing (Fig. 6B). Further growth inhibition assays showed that in contrast to the WT, S. reticuli ΔcpeB is more sensitive to the following stressing agents: H2O2, cumene hydrogen peroxide (CHP), and plumbagin, because in the presence of increasing concentrations of these compounds, zones of growth inhibition appeared or increased (Table 1). Interestingly, CpeB seems to provide S. reticuli with specifically high resistance against CHP. Moreover, viability tests revealed that on the treatment of Streptomyces mycelia with plumbagin, the proportion of damaged mycelia is considerably enhanced in S. reticuli ΔcpeB (Fig. 6C). Due to instability of plasmids in S. reticuli strains (31), S. reticuli ΔcpeB complemented with a plasmid to over-express CpeB could not be obtained. Therefore, to investigate the in vivo role of CpeB in greater detail, we instead used Streptomyces lividans transformants that carry either the plasmid pWKS13 (cpeB+) or pWHM3 (cpeB-) (22). In contrast to Streptomyces lividans pWHM3, which does not produce any CpeB, S. lividans pKWS13 produces a high quantity of the mycelia-associated CpeB (see Supplementary Fig. S4). Inhibition of growth assays demonstrated that S. lividans transformants over-producing CpeB are clearly more resistant to the redox-cycling compounds tested (Table 1). All these data together imply that CpeB actively participates in the anti-oxidative stress response in vivo.
FIG. 6.
The Streptomyces reticuli ΔcpeB mutant. (A) Relative location of the genes furS, cpeB, hbpS, and senS within a DNA segment of the S. reticuli WT chromosome (WT, top). In the ΔcpeB mutant, the cpeB gene within the chromosomal DNA has been disrupted by the insertion of the kanamycin resistance (kana-r) cassette (ΔcpeB, bottom). (B) S. reticuli ΔcpeB (lane 1) and S. reticuli WT (lane 2) were cultivated in minimal medium containing 1% crystalline cellulose, as previously described (21). Proteins were released from the mycelium, subjected to 10% SDS-PAGE, and either stained with Coomassie (left) or transferred to a fluorotrans membrane for Western analysis by using anti-CpeB antibodies (right). The approximate sizes of two prestained protein markers are also indicated (lane 3). (C) S. reticuli WT and S. reticuli ΔcpeB (ΔcpeB) were cultivated in the presence of the redox-cycling compound plumbagin. Samples were subjected to a viability assay and subsequently examined by fluorescence microscopy. Living mycelia exhibit green fluorescence, whereas dead mycelia are seen in red. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Table 1.
Sensitivity of Streptomyces Strains Against Plumbagin, Hydrogen Peroxide, and Cumene Hydroperoxide
| |
Zone of inhibition (mm)a |
|||
|---|---|---|---|---|
| Streptomyces reticuli | Streptomyces lividans transformants | |||
| Redox-cycling compound | Wild type | cpeB mutant | pWKS13 (cpeB+) | pWHM3 (cpeB−) |
| Plumbagin (50 mM) | 18 | 27 | 12 | 22 |
| Plumbagin (100 mM) | 24 | 34 | 18 | 34 |
| H2O2 (0.25%) | 0 | 30 | 0 | 20 |
| H2O2 (0. 5%) | 20 | 50 | 5 | 36 |
| CHP (0.25%) | 0 | 10 | 0 | 8 |
| CHP (0.5%) | 0 | 20 | 0 | 14 |
Assays were repeated four times.
CHP, cumene hydrogen peroxide; H2O2, hydrogen peroxide.
CpeB and turnover of HbpS
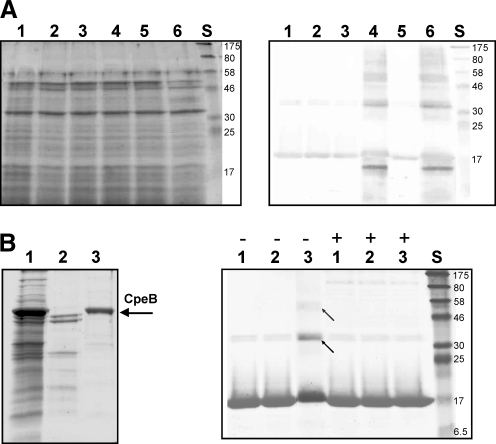
Oxidative modifications accompanied by formation of crosslinkages in HbpS upregulate the HbpS-SenS-SenR-mediated expression of cpeB and hbpS (26). Therefore, oxidized HbpS induces SenS autophosphorylation, which, in turn, phosphorylates SenR, leading to SenR∼P that activates hbpS and cpeB transcription. Since oxidative modifications in HbpS are not reversible and lead to the degradation of the protein, a mechanism should exist which ensures that freshly synthesized HbpS remains unmodified. Unmodified HbpS forms can inhibit SenS autophosphorylation, and, hence, block the signaling cascade. Comparative analyses using extracellular proteins of S. reticuli strains grown under iron-based stress demonstrated that the presence of CpeB blocked the formation of SDS-insensitive crosslinked forms of HbpS (Fig. 7A right; lanes 1, 3 and 5). However, in the absence of CpeB and with increasing cultivation time, a higher production of HbpS accompanied by the formation of crosslinked forms was observed (Fig. 7A right; lanes 2, 4 and 6). After 2 h of incubation, degradation of HbpS was detected in the mutant strain (Fig. 7A right; compare lane 4 with 3 and 6 with 5). In contrast, at the same time point, the WT strain produced lower amounts of HbpS that remained apparently unmodified (Fig. 7A right; lane 5).
FIG. 7.
CpeB and the turnover of HbpS. (A) S. reticuli WT (lanes 1, 3 and 5) and S. reticuli ΔcpeB (lanes 2, 4 and 6) strains were cultivated in the presence of 1 mM FeCl2 previously treated with 10 mM DTT (see Material and Methods section). After 1 h (lanes 1 and 2), 2 h (lanes 3 and 4), and 3 h (lanes 5 and 6) of cultivation, the precipitated extracellular protein extracts were subjected to 12% SDS-PAGE, and either stained with Coomassie (left) or transferred to a fluorotrans membrane for Western analysis by using anti-HbpS antibodies to specifically identify HbpS within the protein extracts (right). The approximate sizes (in kDa) of prestained protein markers are also shown (lane S). (B, left) Mycelia-associated proteins were released from a Streptomyces strain over-producing CpeB (lane 1) by using 0.01% Triton X-100 (see Material and Methods section). After the third anion-exchange chromatography over a DEAE-Sepharose column (lane 2), CpeB was eluted from the DEAE-Sepharose column by using 100 mM NaCl (lane 3). The arrow indicates the position of CpeB on the 10% SDS PAA gel. (B, right) HbpS native (lane 1) or treated with FeCl2 and DTT in the presence (lane 2) or absence (lane 3) of EDTA were subjected to 12% SDS-PAGE and subsequently stained with Coomassie. The absence (−) or presence (+) of the isolated CpeB (see the left part of this figure) in the reactions is indicated. Crosslinked HbpS forms (lane 3,−) are indicated with arrows. The approximate sizes (in kDa) of prestained protein markers are also shown (lane S).
To test the influence of the catalase-peroxidase CpeB on oxidation of HbpS, the active enzyme was isolated from Streptomyces (Fig. 7B left). In the absence of CpeB, the treatment of HbpS with FeCl2 (500 μM) and DTT (5 mM) led to the formation of SDS-resistant crosslinked forms (Fig. 7B right, lane 3,−). When purified CpeB was simultaneously added to the reaction mixture, crosslinking in HbpS was blocked (Fig. 7B right, lane 3,+). The control reaction (Fig. 7B right, lane 2) containing EDTA (10 mM) that chelates iron ions also did not produce crosslinked HbpS forms. All these data indicate that CpeB protects HbpS in vivo from iron-dependent oxidation.
Discussion
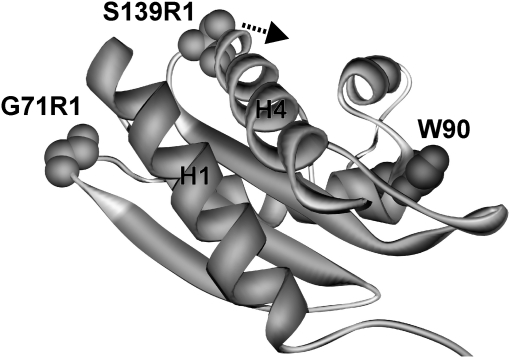
In this article, SDSL EPR was used to investigate redox-dependent conformational changes in the sensory protein HbpS in solution. Utilizing an optimized stress treatment/labeling protocol, we were able to circumvent the principle obstacle in using nitroxide spin labels under strongly reducing or oxidizing conditions, namely destruction of the radical function of the nitroxide. To our knowledge, this is the first time that SDSL EPR has been successfully used to monitor iron stress-mediated conformational changes in a protein in solution. Generally, comparisons of X-ray structure and SDSL EPR data allow a more detailed analysis of the molecular dynamics that regulate the function of proteins, as described, for example, for the proteins NpSRII/NpHtrII (6, 19) or MnmE (18). For HbpS, comparison of the iron-bound X-ray structure with the “native” crystal form shows residues 71 and 139 approaching each other (25). In contrast, the DEER data indicate that the distance between these two side chains actually increases by about 0.6 nm. Earlier, FRET analysis of HbpS revealed a decrease in the distance between an IAEDANS fluorophore attached to Ser139Cys and Trp90 from 2.0 to 1.6 nm (26). Figure 8 depicts the relative positions of Gly71R1, Ser139R1, and Trp90. A distance increase between Gly71R1 and Ser139R1, if caused by a movement of Ser139R1 (arrow in Fig. 8) as deduced from our EPR data, could lead to a concomitant distance decrease between Ser139 and Trp90, as observed in the FRET experiment. Consequently, the SDSL EPR and FRET data corroborate each other, thus indicating an iron-mediated movement of Ser139. This can also be related to the observed mobility increase for Ser139R1, as a motion of the N-terminal end of H4 toward Trp90 would separate Ser139 from H1, and consequently lead to a larger motional freedom for the spin label. Nevertheless, although both SDSL EPR and FRET indicate a movement of helix H4 in HbpS on iron-based stress, modeling of the conformational changes is not currently possible. For this purpose, further studies employing other selected spin label positions in HbpS are necessary.
FIG. 8.
Iron-mediated conformational changes in HbpS. The DEER experiments in this study and previously obtained fluorescence resonance energy transfer data indicated a motion of Ser-139 away from Gly-71 toward Trp-90, as indicated by the arrow.
Previously published in vitro and in vivo data have demonstrated that iron-mediated oxidative modifications (i.e., carbonylation and dityrosine formation) in extracellular HbpS enhance autophosphorylation of the membrane-embedded sensor kinase SenS. SenS, in turn, phosphorylates the response regulator SenR, thus leading to SenR∼P that subsequently activates the transcription of target genes, including hbpS and cpeB (4, 5, 26). Oxidatively modified HbpS tends to degrade autonomously (26), or enzymatically through action of naturally occurring extracellular Streptomyces proteases (7). Here, we provide evidence that the mycelia-associated catalase-peroxidase CpeB protects HbpS in vivo from iron-mediated oxidative modifications, thus indicating that freshly synthesized HbpS proteins remain unmodified. In this state and under nonstressing conditions, HbpS inhibits SenS autophosphorylation (5), in turn preventing phosphorylation of SenR. Dephosphorylated SenR can subsequently repress the expression of hbpS as well as of cpeB (4, 23). Taken together with our previous work, the data presented here suggest that under conditions of redox-stress, the HbpS-SenS-SenR system is upregulated. On the removal of hazardous ROS, leading to a nonstressed environment, this system is capable of self-downregulation.
It is noteworthy that the presence of CpeB considerably enhances the resistance of S. reticuli and S. lividans transformants (carrying cpeB on the plasmid) against different redox-cycling compounds, including H2O2, plumbagin, and CHP. The resistance against CHP seems to be especially strong (Table 1). The organic peroxide CHP may enter the soil environment from industrial discharges and spills, and also as a by-product of fuel oil slicks exposed to ultraviolet radiation. In some eucaryotic cells, CHP promotes lipid peroxidation that can provoke cell death (11, 14). Since CpeB associates with the mycelia of streptomycetes (36), it is expected that this enzyme protects lipid-rich cellular compartments, namely the cell wall and cytoplasmic membrane. Despite the fact that different studies have been performed to monitor ROS-mediated damage of proteins and DNA in bacteria, the influence of such events on lipids remains poorly investigated. Thus, it will be challenging to investigate lipid peroxidation in the cpeB disruption mutant.
Materials and Methods
Growth and viability assays
The sensitivity of S. reticuli WT and S. reticuli cpeB mutant against the redox stressors H2O2, CHP, and plumbagin was determined by using a disk inhibition assay (for details see Supplementary Materials). Viability tests were performed by using the DEAD/LIFE®-BacLight Kit™ (Invitrogen, Darmstadt, Germany) (for details see Supplementary Materials).
Purification of native CpeB
The S. lividans strain carrying the plasmid construct pWKS12 was used for the purification of CpeB, as it produces high levels of the functional enzyme (22). Purification was performed as described earlier (37) with slight modifications. Briefly, Streptomyces mycelia were cultivated in minimal medium (MM) containing yeast extract (final concentration of 0.25%). After overnight incubation, mycelia were treated with acetate buffer (pH 7.0) containing Triton X-100 (final concentration of 0.01%) for 30 min to achieve the release of mycelia-associated proteins, which were subsequently chromatographed by anion-exchange chromatography over a DEAE-Sepharose column. This chromatography was repeated thrice. CpeB was eluted with 100 mM NaCl.
Detection of HbpS in streptomyces
To obtain well-grown mycelia, S. reticuli strains were grown in complete medium. The cultures were subsequently washed four times with MM and cultivated in MM containing crystalline cellulose (1% final concentration) in the presence of 1 mM FeCl2 previously treated with 10 mM DTT (26). After different times (1, 2 and 3 h) of incubation, mycelia were centrifugated. The extracellular proteins from the culture filtrate were precipitated by adding (NH4)2SO4 (90% w/v saturation). The precipitated proteins were analyzed by PAGE and Western blotting (see Supplementary Materials) using anti-HbpS antibodies.
Spin labeling of HbpS
Native (unstressed) HbpS proteins were incubated with 10 mM of freshly prepared DTT for 5 h at 25 °C. DTT was removed by buffer exchange with 10 mM potassium phosphate buffer (pH 7.0) using a PD10 (Sephadex G-25 medium) column (GE Healthcare, Heidelberg, Germany). MTSSL spin label (100 mM in DMSO) was added to a final concentration of 1 mM to the concentrated (50 μM) protein solutions and incubated overnight at 30 °C under N2. Unbound spin label was removed by dialysis against phosphate buffer (pH 7.0). Subsequently, the solutions were buffer exchanged with deuterated 10 mM potassium phosphate buffer (pH 7.0) containing 10% glycerol-d8 and concentrated to 100–300 μM with the use of PEG (polyethylene glycol) 20000 and a Microcon YM-3 Centrifugal Filter Units (Millipore, Schwalbach, Germany).
Nitroxide spin labels are redox sensitive, whereas the HbpS stressed samples were obtained before labeling, as previously described (26), using FeCl2 (500 μM) and DTT (5 mM). Subsequently, stressed samples were incubated with an excess of EDTA (10 mM) for 1 h to remove iron. Before labeling, the reaction mixtures were purified by using a PD10 gel filtration column previously equilibrated and eluted with potassium phosphate buffer. Thereafter, the entire labeling procedure (see above) beginning with the treatment by DTT was performed.
The labeling efficiencies for both procedures were determined to be 40%–70%. Spin diluted samples have been prepared by mixing the spin-labeled protein with unlabeled HbpS in a 1:4 molar ratio.
Cw EPR measurements
Room temperature cw EPR spectra were recorded on a Miniscope X-band EPR spectrometer (MS200; Magnetech GmbH, Berlin, Germany; microwave power: 10 mW; B-field modulation amplitude: 0.125 mT). Cw EPR spectra for inter-spin distance determination in the range from ∼0.8 to 2.0 nm were obtained on a homemade cw X-band EPR spectrometer (for details see Supplementary Materials) at 160 K (microwave power: 0.2 mW; B-field modulation: 0.25 mT). Fitting of simulated dipolar broadened EPR powder spectra to the experimental ones was carried out by using the program DipFit (33).
Pulse EPR measurements
Pulse EPR (DEER) experiments to determine distances >1.5 nm were carried out at X-band frequencies (9.3–9.5 GHz) with a Bruker Elexsys 580 spectrometer. All measurements were performed by using the four-pulse DEER sequence (16, 28, 30). Analysis of the data was performed with DeerAnalysis 2010 (10). For further details, see Supplementary Materials.
Rotamer library analysis
The rotamer library analyses have been carried out by using the program MMM Version 2010 (29) on the crystal structures of HbpS (PDB: 3FPV, native HbpS octomeric; 3FPW, HbpS-iron complex, monomer). For more details, see Supplementary Materials.
Supplementary Material
Abbreviations Used
- CD
circular dichroism
- cw EPR
continuous wave EPR
- DEER
double electron-electron resonance spectroscopy
- DTT
dithiothreitol
- EPR
electron paramagnetic resonance
- FRET
fluorescence resonance energy transfer
- H2O2
hydrogen peroxide
- HbpS
hem-binding protein
- MM
minimal medium
- MTSSL
[(1-Oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate] spin label
- PAGE
polyacrylamide gel electrophoresis
- PDB
protein data bank
- RLA
rotamer library analysis
- ROS
reactive oxygen species
- SDS
sodium dodecyl sulfate
- SDSL
site-directed spin labeling
- WT
wild-type
Acknowledgments
The authors are very grateful to Prof. Dr. H. Schrempf (Applied Genetics of Microorganisms, University of Osnabrueck) and Prof. Dr. H.-J. Steinhoff (Physics, University of Osnabrueck) for their helpful comments. They also thank C. Gloger (Applied Genetics of the Microorganisms, University of Osnabrueck) for fluorescence microscopy. This work was supported by Grant OR 224/1-3 of the Deutsche Forschungsgemeinschaft (DFG).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ando H. Kitao T. Miyoshi-Akiyama T. Kato S. Mori T. Kirikae T. Downregulation of katG expression is associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 2011;79:1615–1628. doi: 10.1111/j.1365-2958.2011.07547.x. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC. Robinson AK. Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 3.Baltz RH. Molecular engineering approaches to peptide, polyketide and other antibiotics. Nat Biotechnol. 2006;24:1533–1540. doi: 10.1038/nbt1265. [DOI] [PubMed] [Google Scholar]
- 4.Bogel G. Schrempf H. Ortiz de Orué Lucana D. DNA-binding characteristics of the regulator SenR in response to phosphorylation by the sensor histidine autokinase SenS from Streptomyces reticuli. FEBS J. 2007;274:3900–3913. doi: 10.1111/j.1742-4658.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- 5.Bogel G. Schrempf H. Ortiz de Orué Lucana D. The heme-binding protein HbpS regulates the activity of the Streptomyces reticuli iron-sensing histidine kinase SenS in a redox-dependent manner. Amino Acids. 2009;37:681–691. doi: 10.1007/s00726-008-0188-5. [DOI] [PubMed] [Google Scholar]
- 6.Bordignon E. Klare JP. Holterhues J. Martell S. Krasnaberski A. Engelhard M. Steinhoff HJ. Analysis of light-induced conformational changes of natronomonas pharaonis sensory rhodopsin II by time resolved electron paramagnetic resonance spectroscopy. Photochem Photobiol. 2007;83:263–272. doi: 10.1562/2006-07-05-RA-960. [DOI] [PubMed] [Google Scholar]
- 7.Chater KF. Biró S. Lee KJ. Palmer T. Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010;34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- 8.Fenton HJH. Oxidation of tartaric acid in the presence of iron. J Chem Soc. 1986;65:899–910. [Google Scholar]
- 9.Groves MR. Ortiz de Orué Lucana D. Adaptation to oxidative stress by Gram-positive bacteria: the redox sensing system HbpS-SenS-SenR from Streptomyces reticuli. In: Mendez-Vilas A, editor. Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology. Badajoz, Spain: Formatex Research Centre; 2010. pp. 33–42. [Google Scholar]
- 10.Jeschke G. Chechik V. Ionita P. Godt A. Zimmermann H. Banham JE. Timmel CR. Hilger D. Jung H. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 11.Johansson K. Järvliden J. Gogvadze V. Morgenstern R. Multiple roles of microsomal glutathione transferase 1 in cellular protection: a mechanistic study. Free Radic Biol Med. 2010;49:1638–1645. doi: 10.1016/j.freeradbiomed.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Klare JP. Steinhoff HJ. Spin Labelling EPR. Photosynth Res. 2009;102:377–390. doi: 10.1007/s11120-009-9490-7. [DOI] [PubMed] [Google Scholar]
- 13.Klare JP. Steinhoff HJ. Site-directed spin labelling and pulse dipolar electron paramagnetic resonance. Encyclop Anal Chem. 2010;a9024 [Google Scholar]
- 14.Linden A. Gülden M. Martin HJ. Maser E. Seibert H. Peroxide-induced cell death and lipid peroxidation in C6 glioma cells. Toxicol In Vitro. 2008;22:1371–1376. doi: 10.1016/j.tiv.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Mackerell AD., Jr. Feig M. Brooks CL., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 16.Martin RE. Pannier M. Diederich F. Gramlich V. Hubrich M. Spiess HW. Determination of end-to-end distances in a series of TEMPO diradicals of up to 2.8 nm length with a new four-pulse double electron electron resonance experiment. Angew Chem Int Ed. 1998;37:2833–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Meschke H. Schrempf H. Streptomyces lividans inhibits the proliferation of the fungus Verticillium dahliae on seeds and roots of Arabidopsis thaliana. Microb Biotechnol. 2010;3:428–443. doi: 10.1111/j.1751-7915.2010.00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer S. Böhme S. Krüger A. Steinhoff HJ. Klare JP. Wittinghofer A. Kissing G domains of MnmE monitored by X-ray crystallography and pulse EPR Spectroscopy. PloS Biol. 2009;7:e1000212. doi: 10.1371/journal.pbio.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moukhametzianov R. Klare JP. Efremov R. Baeken C. Göppner A. Labahn J. Engelhard M. Büldt G. Gordeliy VI. Development of the signal in sensory rhodopsin and its transfer to the cognate transducer. Nature. 2006;440:115–119. doi: 10.1038/nature04520. [DOI] [PubMed] [Google Scholar]
- 20.Nagababu E. Rifkind JM. Heme degradation by reactive oxygen species. Antioxid Redox Signal. 2004;6:967–978. doi: 10.1089/ars.2004.6.967. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz de Orué Lucana D. Schrempf H. The DNA-binding characteristics of the Streptomyces reticuli regulator FurS depend on the redox state of its cysteine residues. Mol Gen Genet. 2000;264:341–353. doi: 10.1007/s004380000328. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz de Orué Lucana D. Tröller M. Schrempf H. Amino acid residues involved in reversible thiol formation and zinc ion binding in the Streptomyces reticuli redox regulator FurS. Mol Genet Genomics. 2003;268:618–627. doi: 10.1007/s00438-002-0776-0. [DOI] [PubMed] [Google Scholar]
- 23.Ortiz de Orué Lucana D. Zou P. Nierhaus M. Schrempf H. Identification of a novel two-component system SenS/SenR modulating the production of the catalase-peroxidase CpeB and the haem-binding protein HbpS in Streptomyces reticuli. Microbiology. 2005;151:3603–3614. doi: 10.1099/mic.0.28298-0. [DOI] [PubMed] [Google Scholar]
- 24.Ortiz de Orué Lucana D. Groves MR. The three-component signaling system HbpS-SenS-SenR as an example of a redox sensing pathway in bacteria. Amino Acids. 2009;37:479–486. doi: 10.1007/s00726-009-0260-9. [DOI] [PubMed] [Google Scholar]
- 25.Ortiz de Orué Lucana D. Bogel G. Zou P. Groves MR. The oligomeric assembly of the novel haem degrading protein HbpS is essential for interaction with its cognate two-component sensor kinase. J Mol Biol. 2009;386:1108–1122. doi: 10.1016/j.jmb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Ortiz de Orué Lucana D. Roscher M. Honigmann A. Schwarz J. Iron-mediated oxidation induces conformational changes within the redox-sensing protein HbpS. J Biol Chem. 2010;285:28086–28096. doi: 10.1074/jbc.M110.127506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Outten FW. Theil EC. Iron-based redox switches in biology. Antioxid Redox Signal. 2009;11:1029–1046. doi: 10.1089/ars.2008.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pannier M. Veit S. Godt A. Jeschke G. Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Res. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 29.Polyhach Y. Bordignon E. Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys Chem Chem Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 30.Reginsson GW. Schiemann O. Pulsed electron-electron double resonance: beyond nanometre distance measurements on biomacromolecules. Biochem J. 2011;434:353–363. doi: 10.1042/BJ20101871. [DOI] [PubMed] [Google Scholar]
- 31.Schrempf H. Plasmid loss and changes within the chromosomal DNA of Streptomyces reticuli. J Bacteriol. 1982;151:701–707. doi: 10.1128/jb.151.2.701-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siemieniewicz KW. Schrempf H. Concerted responses between the chitin-binding protein secreting Streptomyces olivaceoviridis and Aspergillus proliferans. Microbiology. 2007;153:593–600. doi: 10.1099/mic.0.2006/001073-0. [DOI] [PubMed] [Google Scholar]
- 33.Steinhoff HJ. Radzwill N. Thevis W. Lenz V. Brandenburg D. Antson A. Dodson GG. Wollmer A. Determination of interspin distances between spin labels attached to insulin: comparison of electron paramagnetic resonance data with the x-ray structure. Biophys J. 1997;73:3287–3298. doi: 10.1016/S0006-3495(97)78353-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinhoff HJ. Pfeiffer M. Rink T. Burlon O. Kurz M. Riesle J. Heuberger E. Gerwert K. Oesterhelt D. Azide reduces the hydrophobic barrier of the bacteriorhodopsin proton channel. Biophys J. 1999;76:2702–2710. doi: 10.1016/S0006-3495(99)77422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y. Heym B. Allen B. Young D. Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 36.Zou P. Borovok I. Ortiz de Orué Lucana D. Müller D. Schrempf H. The mycelium-associated Streptomyces reticuli catalase-peroxidase, its gene and regulation by FurS. Microbiology. 1999;145:549–559. doi: 10.1099/13500872-145-3-549. [DOI] [PubMed] [Google Scholar]
- 37.Zou P. Schrempf H. The heme-independent manganese-peroxidase activity depends on the presence of the C-terminal domain within the Streptomyces reticuli catalase-peroxidase CpeB. Eur J Biochem. 2000;267:2840–2849. doi: 10.1046/j.1432-1327.2000.01259.x. [DOI] [PubMed] [Google Scholar]
- 38.Zou P. Groves MR. Viale-Bouroncle SD. Ortiz de Orué Lucana D. Cristallization and preliminary characterization of a novel haem-binding protein of Streptomyces reticuli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:386–390. doi: 10.1107/S1744309108008348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.