Abstract
Significance: Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), can persist in a latent state for decades without causing overt disease. Since latent Mtb is refractory to current antimycobacterial drugs, the discovery and characterization of the biological mechanisms controlling the entry, maintenance, and emergence from latent infection is critical to the development of novel clinical therapies. Recent Advances: Recently, Mtb WhiB3, a member of the family of intracellular iron–sulfur (Fe-S) cluster proteins has emerged as a redox sensor and effector molecule controlling several aspects of Mtb virulence. WhiB3 was shown to contain a 4Fe–4S cluster that specifically reacts with important host gases (O2 and NO), and exogenous and endogenous metabolic signals to maintain redox balance. Notably, the concept of reductive stress emerged from studies on WhiB3. Critical Issues: The detailed mechanism of how WhiB3 functions as an intracellular redox sensor is unknown. Sustaining Mtb redox balance is particularly important since the bacilli encounter a large number of redox stressors during infection, and because several antimycobacterial prodrugs are effective only upon bioreductive activation in the mycobacterial cytoplasm. Future Directions: How Mtb WhiB3 monitors its internal and external surroundings and modulates endogenous oxido-reductive pathways which in turn alter Mtb signal transduction, nucleic acid and protein synthesis, and enzymatic activation, is mostly unexplored. Modern expression, metabolomic and proteomic technologies should provide fresh insights into these yet unanswered questions. Antioxid. Redox Signal. 16, 687–697.
Introduction
The ability of Mtb to maintain a state of latent TB infection is responsible for its remarkable success as a pathogen. Approximately 2.3 million people die each year of this disease and the devastation caused by this pathogen is exacerbated by the coexistent HIV epidemic and the emergence of multidrug resistant, extensively drug-resistant, and totally drug-resistant Mtb strains (19).
A major obstacle to the development of successful therapeutic strategies is the lack of a mechanistic understanding of how Mtb maintains a persistent, nonreplicating state in human tissues for decades, unresponsive to antimycobacterial drugs, to then abruptly resume growth and cause disease (24). This requires a detailed understanding of the metabolic flexibility of the bacilli that survive in the varied environments within the human host, ranging from high oxygen tension in the lung alveolus to hypoxic conditions within the tuberculous granuloma (6, 10). An important challenge is to understand how in vivo gases [e.g., oxygen (O2), nitric oxide (NO), carbon monoxide (CO), carbon dioxide (CO2), hydrogen (H2), and hydrogen sulfide (H2S)] and nutrients (e.g., host fatty acids), several of which are thought to play a crucial role in persistence, affect Mtb physiology and consequently redox homeostasis (Fig. 1). Furthermore, frontline anti-Mtb drugs such as isoniazid (34), ethionamide (7), and PA-824 (32) require bioreductive activation to exert antimycobacterial effects, which magnifies the importance of understanding how Mtb maintains redox balance during treatment. It is this lack of drug efficacy against dormant Mtb that represents a fundamental challenge to investigators worldwide. A better understanding of how environmental host factors affect Mtb redox homeostasis (Fig. 1) may shed light on the biology of Mtb persistence and lead to the development of new and improved antimycobacterial drugs.
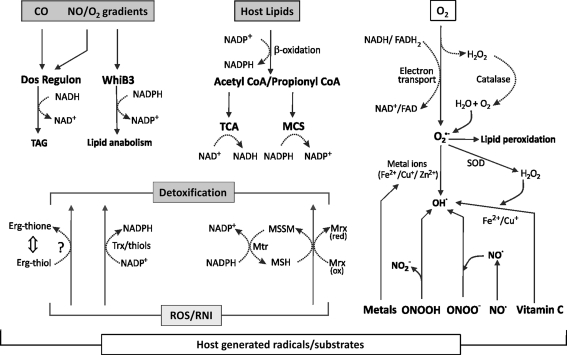
FIG. 1.
Redox machinery of Mtb. During infection, Mtb encounters redox active compounds that have the capacity to alter or skew intracellular redox balance. Mtb has evolved several mechanisms to maintain redox homeostasis. Mtb utilizes host lipids as a source of carbon in vivo, which are metabolized via the (-oxidation pathway. This leads to the generation of high concentrations of NAD(P)H during the conversion of fatty acids to acetate and propionate. The accumulation of high levels of NAD(P)H causes reductive stress in Mtb (18, 40). Furthermore, host-generated free radicals such as reactive oxygen species (ROS) and reactive nitrogen species (RNS) are also capable of disturbing Mtb redox balance. However, Mtb counterbalances the stress generated by these radicals primarily via the abundant intracellular redox buffer, mycothiol (MSH). In addition, protein thiols and mycobacterial thioredoxins (Trx) are also involved in detoxifying the adverse effect of free radicals. Another yet uncharacterized redox active molecule, ergothioneine (Erg), is also expected to play a role in detoxification of free radicals and maintaining redox balance. Endogenous byproducts of mycobacterial respiration such as H2O2 and O2•− may react with mycobacterial lipids to generate lipid peroxides. In the presence of iron or copper, these ROS can generate the highly redox active OH•. ROS or RNS such as peroxide, peroxynitrous acid (ONOOH), and peroxynitrite (ONOO-), and compounds such as vitamin C (in the presence of metal ions) may also lead to the production of intramycobacterial OH• that can cause the oxidation of intracellular substrates. Host-generated gases also play a role in inducing mycobacterial redox changes. Mtb has well-defined sensor systems such as the DosS/T/R and WhiB3 signaling pathways, which specifically sense NO, CO, and changes in pO2. CO, NO, and O2 interact with the Dos signaling pathway, leading to the induction of the 47-member Dos regulon. It also leads to the production of the storage lipid TAG, which requires large quantities of NAD(P)H for synthesis. Similarly, WhiB3 also senses and responds to alterations in cytoplasmic NO and O2. Mrx; mycoredoxin, Mtr; mycothiol disulfide reductase.
Exciting progress has been made in understanding how members of the WhiB family sense and respond to host gases NO and O2 through their iron–sulfur (Fe-S) clusters, and how they may modulate Mtb virulence. Here we review the current knowledge of the Mtb WhiB-like family of proteins, focusing on the best characterized member, WhiB3.
Phylogenetic Distribution of the Wbl Proteins
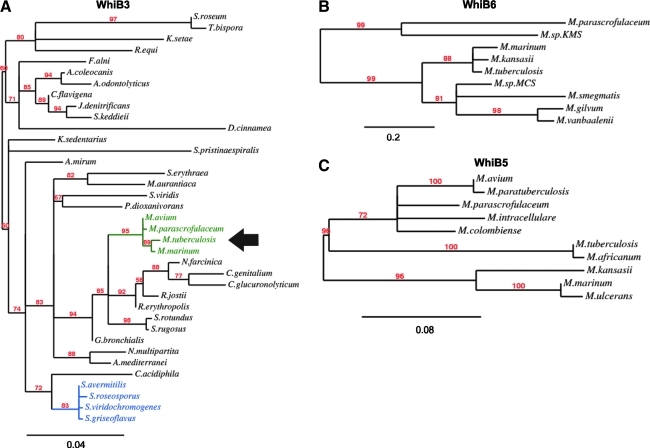
Comprehensive phylogenetic analysis showed that Mtb WhiB1, WhiB2, WhiB3, WhiB4, and WhiB7 are evolutionary closer to Corynebacterium and Nocardia WhiB-like (Wbl) proteins (Fig. 2A). Furthermore, using Interpro and Pfam databases, protein bioinformatic analyses have identified more than 1600 Wbl homologues (database ID: IPR003482/PF02467), which are exclusive to actinobacteria and the double-stranded DNA (dsDNA) containing siphoviridae family of phages. Notably, WhiB5 and WhiB6 are restricted to mycobacteria (Figs. 2B and 2C) with WhiB5 only present in virulent strains (Fig. 2C). All Wbl proteins with the exception of WhiB5 contain four invariant Cys residues arranged in a Cys-X14-22-Cys-X2-Cys-X5-Cys motif (Fig. 3A), which is typically associated with metal co-coordinating DNA-binding proteins. Secondary structure prediction of Wbl proteins indicates an acidic amphiphatic α-helical region near the N-terminus and a basic α-helical region at its C-terminus with a central β-sheet region (43).
FIG. 2.
Phylogenetic analysis of Mtb (A) WhiB3, (B) WhiB6, and (C) WhiB5. BLAST analyses were performed against the NCBI ‘Nr’ database. All the hits (except candidates from the same species) with e ≥1 e−10 were considered significant and further used in Clustal W alignment. A maximum likelihood method was used for tree construction, and boot strap values are indicated along the tree nodes. This analysis demonstrates that Mtb WhiB3 constitute a separate node (arrow) (A) from Streptomyces spp. and is closer related to Nocardia and Clostridium spp. A similar distribution pattern and tree architecture also hold true for Mtb WhiB1, WhiB2, WhiB4, and WhiB7 (not shown), but not for WhiB6 (B) and WhiB5 (C), which are restricted to mycobacteria. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
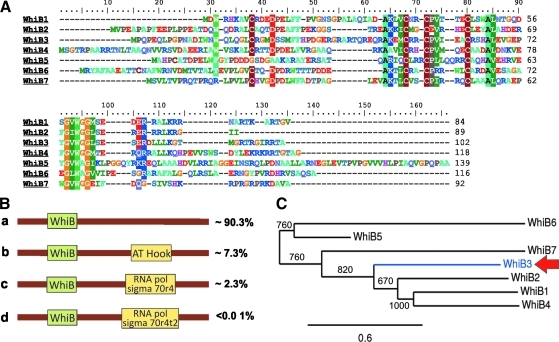
FIG. 3.
Phylogenetic relationship among Mtb Wbl proteins and domain organization across species. (A) Mtb Wbl protein sequences were extracted from NCBI and aligned using Clustal W. In addition to four invariant Cys residues that are a characteristic signature of the Wbl family, the small G-X-W motif, typical of fatty acid binding proteins, is also present. Note that WhiB5 lacks a Cys residue. (B) Domain architecture of the Wbl family indicates that the “WhiB” domain is the most widely represented since it is present in ∼90% of Wbl members. All Streptomyces, mycobacterial phages, and mycobacterial WhiB1, WhiB2, WhiB4, WhiB5, and WhiB6 homologues share this architecture. (C) Phylogenetic tree was constructed using the maximum likelihood method and revealed that Mtb Wbl's are clustered in two nodes comprising of WhiB5 and WhiB6 in one node, and WhiB1, WhiB2, WhiB3 (red arrow), WhiB4, and WhiB7 in another. This indicates that WhiB5 and WhiB6 are functionally distinct from the other Mtb Wbl's. The numbers along the nodes denote bootstrap values, and the phylogenetic scale represents the number of differences between different sequences.
Most Wbl proteins (∼90%) contain a “WhiB” domain (including all Wbl homologues in phages, and Mtb WhiB1, WhiB2, WhiB4, WhiB5, and WhiB6), whereas ∼7% possess an additional AT-hook like domain at their C-terminus (e.g., Mtb WhiB3 and WhiB7). The AT-hook like domain is a DNA-binding motif with a preference for A/T-rich regions (4) (Fig. 3B). Approximately 2% of Wbl members contain an additional RNA polymerase (RNAP) sigma70r4 binding region and a clearly distinguished DNA-binding 'helix-turn-helix' (HTH) motif that binds to the −35 promoter region. A small percentage of Wbl members contain a RNAP sigma70r4t2 signature (Fig. 3B). Although a systematic functional analysis of Wbl protein domains is lacking, the diversity in protein architecture may be responsible for the varied functions including redox homeostasis(40), cAMP regulation (1), oxidative stress (28), cell division (23), sporulation (16), quorum sensing (5), drug resistance (33), and virulence (44).
Mtb harbors seven Wbl proteins, WhiB1 (Rv3219), WhiB2 (Rv3260c), WhiB3 (Rv3416), WhiB4 (Rv3681c), WhiB5 (Rv0022c), WhiB6 (Rv3862c), and WhiB7 (Rv3197A) (Fig. 3C). Only moderate aa sequence similarity exists between members of the Mtb Wbl family (<70%). BLAST analysis of Mtb Wbl proteins against A CLAssification of Mobile genetic Elements (ACLAME), a database of mobile genetic elements such as plasmids, phages, and viruses, identified several Wbl homologues that are present on plasmids. For example, WhiB1 and WhiB4 share significant similarity (E-value=≤ e−15) with gene products from Streptomyces plasmids. Also, WhiB2 and WhiB3 show similarity with Rhodococcus plasmid-encoded gene products (Table 1). Thus, it is unlikely that Mtb Wbl members evolved through gene duplication events. Rather, it is tempting to hypothesize that the Mtb Wbl proteins have been independently acquired from mycobacterial ancestors via lateral gene transfer from mobile elements such as plasmids or phages. The selective distribution of Mtb Wbl proteins in different mycobacterial species (e.g., the absence of WhiB5 in nonpathogenic species) coupled with considerable sequence heterogeneity, suggest a discrete functional role for each Mtb Wbl protein.
Table 1.
Similarity of Mtb Wbl Proteins with Mobile Genetic Elements
| Protein name | Query length | Hit length | Plasmids | Host organism | E value* | Identity (%) |
|---|---|---|---|---|---|---|
| WhiB1 | 84 | 105 | SAP1 | Streptomyces avermitilis MA-4680 | 4.00E-21 | 63 |
| WhiB2 | 89 | 89 | pREL1 | Rhodococcus erythropolis PR4 | 9.00E-20 | 66 |
| WhiB3 | 102 | 119 | pRHL1 | Rhodococcus sp. RHA1 | 8.00E-19 | 50 |
| WhiB4 | 118 | 268 | SCP1 | Streptomyces coelicolor | 5.00E-15 | 50 |
| WhiB6 | 116 | 117 | pMKMS01 | Mycobacterium sp. KMS | 3.00E-19 | 53 |
| Phages | ||||||
| WhiB1 | 84 | 86 | gp94 | Mycobacterium smegmatis | 3.00E-12 | 61 |
| WhiB2 | 89 | 215 | gp65 | Mycobacterium smegmatis | 5.00E-20 | 44 |
| WhiB4 | 118 | 215 | gp68 | Mycobacterium smegmatis | 5.00E-10 | 41 |
Mtb Wbl proteins were used in a BLAST analysis against the ACLAME database that contains mobile genetic elements such as plasmids and phages. Only hits with e−10 were considered as significant. The presence of Wbl proteins on plasmids and phages suggest that Mtb wbl genes were laterally acquired. WhiB5 and WhiB7 did not yield any significant hits.
Genetic Regulation of Mtb wbl Genes
To better understand the genetic regulation of Mtb wbl genes, Geiman et al. (22) performed an elegant real-time PCR expression study that examined the differential expression of all seven Mtb wbl genes following exposure to a variety of environmental stresses, including detergent exposure, oxidative stress, acid exposure, nutrient starvation, ethanol, antibiotic, heat, and low iron stress. WhiB3 expression was highly increased upon acid stress, but weakly upon exposure to oxidants. WhiB7 was highly upregulated under low iron conditions, whereas whiB7 and whiB2 expression was shown to be significantly increased upon exposure to antibiotics (22). The whiB7 finding is consistent with a previous study implicating members of the WhiB family in antimicrobial stress (33). Overall, whiB6 appeared to be responsive to the widest variety of stress conditions (22).
Exploiting an Mtb transposon library screen, whiB1 and whiB6 were both implicated in the switchover from a slow to fast growth rate (8). In another genome-wide expression study, whiB6 expression was consistently upregulated under prolonged hypoxia, a condition associated with dormant infection, whereas whiB4 and whiB2 were moderately upregulated (35, 36). Last, whiB2 expression was shown to be significantly increased upon re-aeration following hypoxia, suggesting a possible role for whiB2 in the activation of Mtb from the latent state (39).
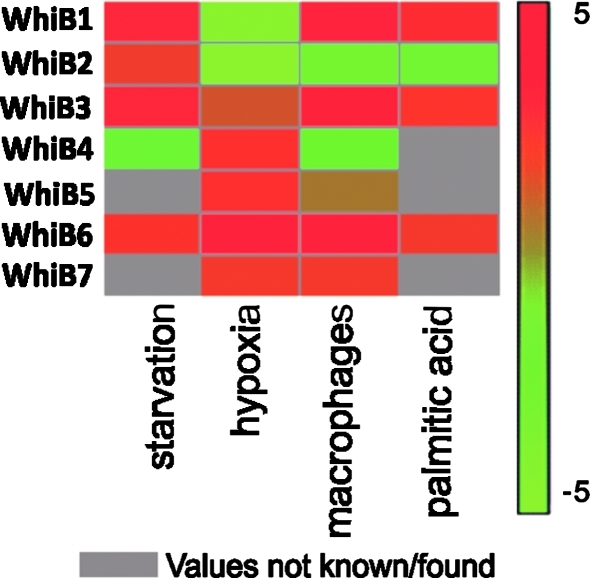
When comparing the microarray expression profiles of several clinical Mtb strains within macrophages (27), it became clear that the wbl expression profiles are highly variable. Figure 4 depicts a summary of the relative wbl expression profiles obtained from the literature. Noticeably, whiB6 appears to be prominently upregulated under several conditions, consistent with the report by Geiman et al., (22). WhiB4 expression was upregulated under hypoxia, but downregulated during starvation and within macrophages. Surprisingly, starvation in the presence of oxygen leads to increased whiB4 expression (9), suggesting a possible role in Mtb reactivation. Mtb whiB3 appeared to be upregulated under a range of conditions.
FIG. 4.
Relative transcription profiles of Mtb wbl genes. Graphic representation of the independent Mtb wbl transcription profiles extracted from published studies (9, 35, 45). WhiB3 and especially whiB6 appear to be induced under a wide range of conditions including starvation, hypoxia, and within macrophages. Expression of whiB4 was repressed under nutrient-starved conditions and upon infection of macrophages, whereas whiB4 expression was slightly induced during hypoxia. Notably, whiB1 and whiB2 appears to be strongly repressed under hypoxic conditions.
Metabolic Regulation of Mtb WhiB3
Metabolism
Mtb WhiB3 senses fluctuations in the intracellular redox environment associated with O2 depletion, including the metabolic switchover to the favored in vivo carbon source, fatty acids (40). This has important consequences for understanding how Mtb persists within the host, as it is widely believed that fatty acids serve as a main source of carbon and energy during long-term infection
Several lines of evidence implicate WhiB3 as a metabolic regulator. First, WhiB3 is essential for maintaining bacterial cell shape and size (40, 41). Second, WhiB3 controls the anabolism of complex virulence lipids such as polyacyltrehaloses (PAT), diacyltrehaloses (DAT), sulfolipids (SL-1), and phthiocerol dimycocerosates (PDIM), which requires large quantities of reducing equivalents (NAD[P]H). Third, WhiB3 was shown to maintain redox homeostasis during infection of macrophages (40). Fourth, the MtbΔwhiB3 mutant showed substantially decreased production of the methyl-branched polar lipids PAT, DAT, and SL-1, but increased production of PDIM, and to a lesser extent triacylglycerol (TAG) (40). Mtb ΔwhiB3 grows on much higher concentrations of propionate compared to wild-type (wt) Mtb, and accumulates PDIM and TAG during intra-macrophage growth, suggesting that the increased resistance to propionate toxicity is because this intermediate is channeled into PDIM via the methyl-malonyl CoA (MMCoA) pathway and into TAG (40). These metabolic analyses implicate WhiB3 in core intermediary metabolism and are reminiscent of studies executed on FNR and ArcA, which regulate key enzymes in the tricarboxylic acid cycle (TCA) cycle under carbon source starvation conditions (31, 38).
Protein networks and Wbl function
Originally, Mtb WhiB3 was discovered through a yeast two-hybrid screen using SigA as bait (44). Recently, an E. coli two-hybrid system was exploited and showed that 113 proteins interacted with WhiB3 and 52 with WhiB7 (average number of interacting neighbors ∼6). As is typically the case with microbial protein linkage maps, many interactions are difficult to explain in the context of what is biologically known about WhiB3 (or WhiB7) function, and await future verification. Also, the frequency of false positives and negatives, often due to “sticky” interacting proteins (a common observation in yeast two-hybrid screens) is unknown. Nonetheless, the potentially large number of proteins interacting with WhiB3 and WhiB7 is difficult to ignore and may point to biological properties that are distinct among Mtb Wbl proteins. Lastly, it is uncertain to what extent post-translational modification (e.g., assembly of the WhiB3 and WhiB7 Fe–S clusters) of Mtb proteins expressed in E. coli influences protein–protein interaction. Thus, although an admirable resource, these data await further verification and should be interpreted with caution in order to generate accurate knowledge on the biological function of Mtb Wbl proteins.
A provocative finding by one group was that several Wbl proteins in their apo- forms demonstrated nonspecific protein disulfide reductase activity (2, 3, 20, 21). The authors proposed that the Fe–S cluster blocks the four active Cys residues until endogenous oxidants destroys the Fe–S cluster, which then leads to the formation of an intramolecular disulfide bond. Subsequent reduction of the disulfide bond (through a yet unknown mechanism) stimulates disulfide reductase activity. This debate was fueled further when independent groups were unable to detect such activity with WhiB1 or the S. coelicolor orthologue of WhiB3 (13, 42), and also when it was reported that Mtb WhiB1 interacts with GlgB (an α-1,4-glucan branching enzyme) to reduce the intramolecular GlgB disulfide bond (20). Although these enzymatic studies are in contrast with increasing evidence suggesting that the Wbl proteins are regulatory DNA-binding proteins (37, 40, 42), or transcription factors as was recently demonstrated for Mtb WhiB1 (42), it cannot be discounted that WhiB1 may function as a highly selective reductase.
In sum, in the past few years novel genome-wide technologies (46) and comprehensive biochemical analyses (3) have been employed to characterize the Mtb Wbl proteins. However, the methodology and exact experimental conditions should be critically evaluated in order to yield accurate information on the biological functions of Wbl proteins.
The WhiB3 Fe–S Cluster
To date, biochemical evidence suggests that all Wbl proteins contain Fe–S clusters, and it is implicitly assumed that this prosthetic group plays a crucial role in Wbl protein function. However, evidence suggests that the Fe–S clusters from different Mtb Wbl proteins vary in their behavior towards O2 exposure (3) (Fig. 5). Furthermore, the fact that Fe–S cluster proteins are stable under anaerobic conditions and unstable under aerobic conditions directly implicates the Fe–S clusters in Wbl function. Host factors may also react with Fe–S clusters. For example, Fe–S cluster enzymes in the TCA cycle are known to be inhibited by O2•− and by high pO2 (26). In addition, NO can react with the Fe–S clusters of TCA cycle enzymes, leading to the formation of a protein-bound dinitrosyl-iron complex (DNIC), which can alter enzymatic activity (17). Also, in elegant biochemical studies on Streptomyces WhiD (the homologue of Mtb WhiB3) and Mtb WhiB1, protein stability in the presence of H2O2 and O2•− (13) and NO (14) was examined. The reaction of S. coelicolor WhiD and Mtb WhiB1 with NO was particularly interesting as each reacted 104-fold faster with NO than O2 in a multiphasic reaction containing eight NO molecules per [4Fe–4S] cluster (14). Using electron paramagnetic resonance spectroscopy, Singh et al. (41) demonstrated that O2 exposure can cause cluster degradation, and that Mtb WhiB3 can function as a sensor of the two dormancy gases O2 and NO.
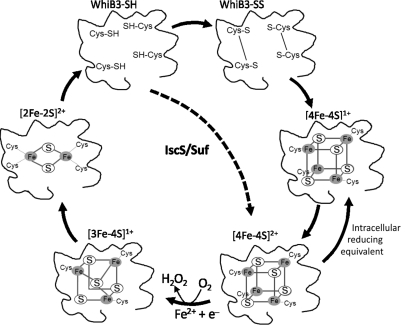
FIG. 5.
Diagrammatic representation of WhiB3 Fe-S cluster loss and assembly. In the presence of high concentrations of oxygen, WhiB3 [4Fe-4S]1+ cluster is oxidized to [4Fe-4S]2+ and is subsequently converted into a [3Fe-4S]1+ with the simultaneous generation of H2O2. The [3Fe-4S]1+ further yields [2Fe-2S]2+ intermediates, followed by a complete loss of the cluster wherein WhiB3 either exists in a reduced WhiB3-SH state, or oxidized WhiB3-SS state. Mtb IscS converts apo-WhiB3 to WhiB3 [4Fe-4S]2+. As suggested for Mtb WhiB1 (42), it is tempting to speculate that thioredoxin and mycothiol may be involved in stabilizing the WhiB3 [4Fe-4S] cluster.
What protects the WhiB3 Fe–S cluster against oxidation and eventually complete loss of the cluster during aerobic growth? One possibility is that a mycobacterial redox buffer in the cytoplasm protects cellular Fe–S clusters. Using methyl-mycothiol, it was shown that the extent of cluster loss was substantially less compared to untreated Streptomyces WhiD samples (13). Methyl-mycothiol also reduced the sensitivity of native WhiD against low pH. Lastly, Cys and thioredoxin were also shown to be protective against cluster loss due to O2 exposure (13). This provides theoretical evidence for how mycobacteria may maintain cytoplasmic Fe–S cluster integrity during aerobic growth.
An important physicochemical factor that may influence Fe–S cluster assembly (and thus WhiB3 function) is the acid dissociation constant (pKa) of residues adjacent to the conserved Cys residues and factors that may modify their properties. For example, WhiB3 Cys residues are flanked by multiple arginine residues (Cys23-Arg24-Cys53-Arg54-Arg55-Cys56-Cys62-Arg63). At neutral pH, Arg residues located close to Cys residues can significantly lower the pKa of Cys, resulting in Cys thiolates, which are highly susceptible to oxidation. Not surprisingly, Mtb whiB3 was highly upregulated when exposed to acidic stress (22). In sum, the inherent biochemical properties of the 4Fe–4S cluster allow WhiB3 to function as a sensitive redox switch that responds to a range of environmental signals implicated in virulence.
Mtb WhiB3 DNA-Binding and Transcriptional Regulation
Since the identification of a Wbl protein in Streptomyces spp. in 1992, it has been hypothesized that Wbl proteins bind DNA due of the presence of putative DNA-binding motifs (16). Initial evidence became stronger when it was demonstrated that Mtb WhiB3 physically interacts with the principle sigma factor, SigA (RpoV) (44). However, it took nearly two decades to provide experimental proof that a Wbl protein, WhiB3, binds DNA (40). The evidence came from characterizing the DNA-binding properties of apo- and holo- forms of WhiB3 to the upstream sequence of lipid biosynthesis genes pks2 and pks3 (40). Important findings were: (i) the redox state of reconstituted holo-WhiB3 did not affect DNA binding, (ii) oxidized apo-WhiB3 bound DNA much stronger than either holo-WhiB3 or reduced apo-WhiB3, and (iii) the oxidized apo-WhiB3 form demonstrated the strongest DNA binding. It was further demonstrated that WhiB3 regulates Mtb pks2 and pks3 genes in a redox-dependent manner, which provides further proof of a role for WhiB3 in the redox-dependent regulation of virulence factors (40). In another study, an E. coli bacterial one-hybrid system was used to screen for Mtb regulatory proteins that bind to promoter DNA (25). Mtb WhiB3 was shown to bind to the promoter regions of several in vivo induced genes. Although an important resource, the use of E. coli as surrogate host to examine Mtb DNA-binding proteins has the same drawbacks as the E. coli protein–protein interaction technology (see Protein Networks and Wbl Function) and awaits independent verification.
Since the WhiB3 4Fe–4S cluster reacts with NO to form a DNIC complex, any structural or redox-mediated changes associated with complex formation could potentially affect DNA binding and/or transcriptional regulation of WhiB3 target genes (Fig. 6). Notably, while the redox sensing capacity of WhiB3 is based on the presence of 4Fe–4S cluster and Cys residues, there is no clearly defined DNA-binding domain. Nonetheless, the C-terminus of WhiB3 contains a weak AT-hook motif TMGRTRGIRRTA, which is prevalent in eukaryotic nuclear proteins that nonspecifically interact with DNA (4). AT-hook motifs are known to interact with the minor groove of AT-rich DNA sequences, which in Mtb are typically restricted to the promoter regions. Similar to other AT-hook-containing proteins (4), the AT-hook like domain may enable WhiB3 to nonspecifically bind, or “coat” DNA as a protection against free radical-mediated damage.
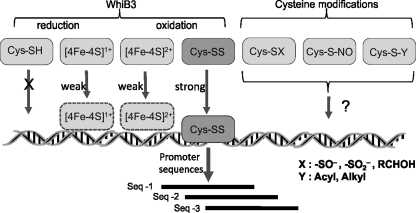
FIG. 6.
Model depicting the mechanisms of Mtb WhiB3 DNA binding. WhiB3 DNA binding is influenced by the redox state of the Fe–S cluster (in the case of holo-WhiB3), or alternatively by the oxidation states of the Cys residues (in the case of apo-WhiB3). As previously demonstrated (40), oxidized apo-WhiB3 binds DNA strongly, whereas reduced apo-WhiB3 does not bind DNA at all, and holo-WhiB3 ([4Fe-4S]1+ or [4Fe-4S]2+) binds DNA weakly. It is reasonable to assume that the redox state of the mycobacterial cytoplasm would influence protonation and deprotonation of the WhiB3 Cys residues to affect DNA binding. Since Mtb experiences gradients of in vivo signals, an alteration in the concentration of effector molecules may lead to differential responses. It is therefore proposed that WhiB3 may sense and differentiate between effector molecules and their concentrations Fe–S cluster intermediates (3Fe-4S, 2Fe-2S), oxidation states of the Cys thiols, or via post-translational modifications of the Cys residues. These redox modifications likely dictate WhiB3 binding and modulation of its target genes. Since Cys residues within a protein can be modified to -SH, disulfide (-SS-), sulfenate (-SO–), sulfinate (-SO2–), or s-nitrosylated states, it is likely that these modifications could also modulate DNA-binding activity.
Collectively, these WhiB3 studies suggested that the Wbl proteins interact strongly with DNA in their oxidized apo-forms and weakly in the holo-forms. It was subsequently shown that Mtb apo-WhiB1 (42), and Mtb apo-WhiB2 and its mycobacteriophage homologue apo-WhiBTM4 bind DNA strongly, as opposed to the holo-form of these proteins that bind DNA weakly or not at all (37). Interestingly, Mtb WhiB2 and WhiBTM4 bind to a conserved promoter sequence upstream of whiB2 to regulate its transcription (37).
Recent studies have shown that oxidized Mtb apo-WhiB1 as well as nitrosylated WhiB1 bind DNA, and that DNase I footprinting of apo-WhiB1 bound to whiB1 promoter DNA revealed a 37-bp protected region (42). Importantly, transcriptional analyses demonstrated that apo-WhiB1 repressed whiB1 transcription in vitro, and provided evidence for the first time that a Wbl member functions as a transcription factor (42).
In sum, it is clear that the oxidation states of holo- or apo-Wbl proteins alter their DNA binding and likely also transcriptional activity. Physiological modifications to either WhiB3 Cys residues (e.g., oxidized, reduced) or the Fe–S cluster (e.g., the apo- or holo-states, the Fe–S cluster oxidation state, nitrosylation, etc) represent a flexible switch that allows the microbial cell to rapidly respond to a wide range of environmental conditions (Fig. 6). Since virtually all DNA-binding proteins influence transcription to some extent, this suggests that the Wbl proteins are transcription regulators with DNA binding capability.
Role of WhiB3 in Mtb Disease
The first in vivo genetic complementation studies identified the principle sigma factor SigA (RpoV) as the first mycobacterial virulence factor (12). A caveat of this finding is that sigA is an essential gene, thereby making it exceedingly difficult to study. In this regard, WhiB3 was identified as a putative regulatory protein that interacts with SigA (44). This was determined by using a yeast two-hybrid screen to examine the independent interactions of wt SigA or the mutated allele of SigA (Arg515-His) with WhiB3. Noticeably, the WhiB3:SigA interaction was strong, whereas the SigA(Arg515-His):WhiB3 interaction was significantly decreased. This finding suggests that the SigA:WhiB3 interaction is necessary for full virulence and that the Arg515-His mutation contributes to the attenuation observed in the avirulent M. bovis strain. It was hypothesized that the disruption of this interaction by the SigA Arg515-His mutation caused alterations in expression of virulence genes under WhiB3 control (44).
Further studies demonstrated that deletion of whiB3 in Mtb and M. bovis caused no phenotype in vitro, but led to the loss of virulence in mouse and guinea pig models of TB (44). The authors proposed that virulence lipids were responsible for triggering the severe pathology. Indeed, subsequent lipid analyses by the same authors (40) have shown that the lack of WhiB3 was strongly associated with decreased production of the virulence lipids SL-1, PAT, and DAT, albeit with increased synthesis of PDIM (see section on Metabolic Regulation of Mtb WhiB3). The fact that the levels of some lipids are upregulated and others downregulated makes it difficult to accurately predict the exact contribution of each lipid species towards virulence. The identification of gene products under WhiB3 control that are responsible for the differences in host lung pathology is eagerly awaited.
Mtb WhiB3 Modulates Oxido-Reductive Stress
In vitro metabolic labeling studies performed under defined thiol-specific oxidizing (diamide) or reducing (dithiothreitol) conditions demonstrated that Mtb WhiB3 regulates the production of the lipids PAT, PDIM, and TAG in a redox-dependent manner. Furthermore, MtbΔwhiB3 is able to survive in the presence of toxic concentrations of propionate in vitro and within macrophages (40). A related finding was that reductive stress (dithiothreitol) exposure upregulated TAG anabolism. Earlier studies of Mtb showed that TAG production is highly upregulated under hypoxic conditions and exposure to NO and CO (29, 30). Oxygen, CO, and NO are modulatory ligands of the heme sensor kinases DosT and DosS, which modulate DosR, the regulator of the 47-member Dos dormancy regulon, which includes the tgs1 gene that controls TAG biosynthesis (45). It was recently suggested that TAG is the prime carbon source utilized upon reactivation from a nonreplicating persistent state (15). Thus, these findings provide a link between the intracellular WhiB3 pathway and the two-component DosR/S/T signaling pathway.
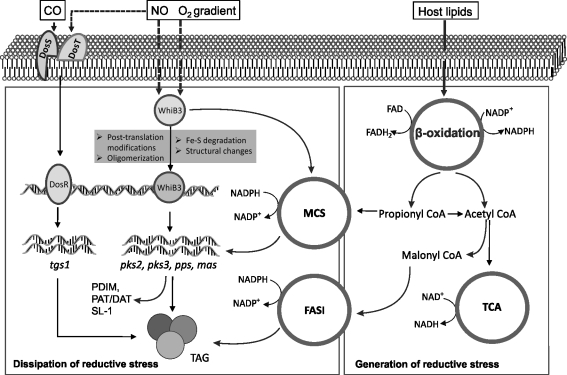
During intracellular growth, Mtb utilizes host fatty acids as a primary energy source via β-oxidation, wherein a large quantity of toxic propionate is generated. WhiB3 appears to play a central role in dissipating propionate toxicity and regulates the metabolic switchover between the in vitro carbon source (e.g., glucose) and the in vivo carbon source (e.g., acetate) (40, 41). Furthermore, β-oxidation is accompanied with the generation of high concentrations of reducing equivalents such as NADH and NADPH (Fig. 7). Extraordinarily high levels of NAD(P)H, but not NAD(P)+ were identified in Mtb isolated from infected mouse lungs, demonstrating that Mtb experiences reductive stress in vivo (11). In macrophages, MtbΔwhiB3 was shown to experience more severe reductive stress upon infection compared to Mtb. These observations point to a major role for WhiB3 in dissipating excessive reductive stress (40) [for a review on reductive stress see (18)]. While the mechanism of how WhiB3 maintains redox balance is largely unknown, a likely explanation is that excess reducing equivalents are assimilated via lipid anabolism (Fig. 7).
FIG. 7.
Diagram illustrating the role of Mtb WhiB3 in redox regulation. Mtb WhiB3 is a DNA-binding protein that senses changes in the environmental gases such as NO and gradients of O2. WhiB3 also senses or responds to fluctuating levels of reducing equivalents (NADH) that are generated by the TCA cycle (respiration), and NADPH that accumulates during (β-oxidation of host fatty acids. If not properly balanced, the bacillus will experience reductive stress (18). As a result, WhiB3 channels these excess reducing equivalents into virulence lipid anabolic pathways, which requires NADPH as cofactor. Thus, virulence lipid anabolism (e.g., SL-1, PDIM, PAT, DAT, and trehalose dimycolate) functions as an electron sink or reserve storage material, which is available for future use. This is also, in part, a mechanism to maintain intracellular redox balance. WhiB3 also overlaps with the DosR/S/T signaling pathway since both regulates TAG synthesis.
Conclusion and Future Directions
Understanding the mechanisms of how Mtb is able to persist in host tissue for decades without causing disease, and the development of new therapeutic strategies against latent TB depends on an advanced understanding of the role of redox homeostasis in Mtb pathogenesis. Novel paradigms are urgently needed, and the Wbl protein family represents an unexplored avenue of research. This poorly characterized group of Fe–S cluster proteins influences a diverse range of biological events including redox homeostasis (40), cAMP regulation (1), oxidative stress (28), cell division (23), drug resistance (33), and virulence (44), and makes them ideal candidates for further study.
Notably, the existence of more than 1600 Wbl homologues across numerous bacterial species and the fact that the ds DNA siphoviridae bacteriophage family contains Wbl homologues may shed light on the mechanism of horizontal and vertical gene transfer of wbl gene products. Subsequent to the original WhiB3 virulence study (44), a rapid surge in Wbl studies followed, particularly in the area of biochemistry (3, 13, 14, 41, 42), which has established a broad mechanistic framework for understanding the mode of action of Wbl members. Combining these mechanistic findings with in vivo Mtb virulence studies should lead to a better understanding of the biological function of these proteins. Demonstrating that Mtb WhiB3 binds DNA (40) was an important finding and has paved the way for studying how other Wbl proteins may bind DNA. Although technically challenging, obtaining the crystal structure of any Wbl protein would significantly contribute to the field. The identification of Mtb WhiB3 as a metabolic redox sensor (41) is particularly interesting as the concept of reductive stress emerged from these findings (18).
In sum, the Wbl family represents an important group of Mtb proteins of which WhiB3 represents the prevailing paradigm. Employing genome-wide technologies geared towards identifying new pathways and metabolites should lead to increased understanding of the biological function of Wbl proteins.
Abbreviations Used
- Aa
amino acid
- DAT
diacyltrehalose
- DNIC
dinitrosyl-iron complex
- Fe–S
iron–sulfur
- MMCo
methyl-malonyl CoA
- Mtb
Mycobacterium tuberculosis
- PAT
polyacyltrehalose
- PDIM
phthiocerol dimycocerosates
- pO2
partial pressure of O2
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SL
sulfolipids
- TAG
triacylglycerol
- TB
tuberculosis
- TCA
tricarboxylic acid cycle
- Wbl
WhiB-like
Acknowledgments
We thank Joel N. Glasgow for critical reading of this manuscript. Research in the laboratory is supported in whole or in part by the National Institutes of Health Grants AI058131, AI076389 (to AJCS). This work is also supported by the University of Alabama at Birmingham (UAB) Center for AIDS Research, and UAB Center for Free Radical Biology (AJCS). AJCS is a Burroughs Welcome Investigator in the Pathogenesis of Infectious Diseases.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Agarwal N. Raghunand TR. Bishai WR. Regulation of the expression of whiB1 in Mycobacterium tuberculosis: Role of cAMP receptor protein. Microbiology. 2006;152:2749–2756. doi: 10.1099/mic.0.28924-0. [DOI] [PubMed] [Google Scholar]
- 2.Alam MS. Garg SK. Agrawal P. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: A [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol Microbiol. 2007;63:1414–1431. doi: 10.1111/j.1365-2958.2007.05589.x. [DOI] [PubMed] [Google Scholar]
- 3.Alam MS. Garg SK. Agrawal P. Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. FEBS J. 2009;276:76–93. doi: 10.1111/j.1742-4658.2008.06755.x. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L. Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banaiee N. Jacobs WR., Jr. Ernst JD. Regulation of Mycobacterium tuberculosis whiB3 in the mouse lung and macrophages. Infect Immun. 2006;74:6449–6457. doi: 10.1128/IAI.00190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry CE. Boshoff HI. Dartois V. Dick T. Ehrt S. Flynn JA. Schnappinger D. Wilkinson RJ. Young D. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baulard AR. Betts JC. Engohang-Ndong J. Quan S. McAdam RA. Brennan PJ. Locht C. Besra GS. Activation of the pro-drug ethionamide is regulated in mycobacteria. J Biol Chem. 2000;275:28326. doi: 10.1074/jbc.M003744200. [DOI] [PubMed] [Google Scholar]
- 8.Beste DJV. Espasa M. Bonde B. Kierzek AM. Stewart GR. McFadden J. The genetic requirements for fast and slow growth in mycobacteria. PLoS One. 2009;4:e5349. doi: 10.1371/journal.pone.0005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betts JC. Lukey PT. Robb LC. McAdam RA. Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 10.Boshoff HI. Barry CE., 3rd. Tuberculosis. Metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 11.Boshoff HI. Xu X. Tahlan K. Dowd CS. Pethe K. Camacho LR. Park TH. Yun CS. Schnappinger D. Ehrt S, et al. Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J Biol Chem. 2008;283:19329–19341. doi: 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins DM. Kawakami RP. de Lisle GW. Pascopella L. Bloom BR. Jacobs WR., Jr Mutation of the principal sigma factor causes loss of virulence in a strain of the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 1995;92:8036–8040. doi: 10.1073/pnas.92.17.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crack JC. den Hengst CD. Jakimowicz P. Subramanian S. Johnson MK. Buttner MJ. Thomson AJ. Le Brun NE. Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD. Biochemistry. 2009;48:12252–12264. doi: 10.1021/bi901498v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crack JC. Smith LJ. Stapleton MR. Peck J. Watmough NJ. Buttner MJ. Buxton RS. Green J. Oganesyan VS. Thomson AJ. Mechanistic insight into the nitrosylation of the [4Fe- 4S] cluster of WhiB-like proteins. J Am Chem Soc. 2011;133:1112–1121. doi: 10.1021/ja109581t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel J. Deb C. Dubey VS. Sirakova TD. Abomoelak B. Morbidoni HR. Kolattukudy PE. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis NK. Chater KF. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol Gen Genet. 1992;232:351–358. doi: 10.1007/BF00266237. [DOI] [PubMed] [Google Scholar]
- 17.Duan X. Yang J. Ren B. Tan G. Ding H. Reactivity of nitric oxide with the [4Fe-4S] cluster of dihydroxyacid dehydratase from Escherichia coli. Biochem J. 2009;417:783. doi: 10.1042/BJ20081423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farhana A. Guidry L. Srivastava A. Singh A. Hondalus MK. Steyn AJC. Reductive stress in microbes: Implications for understanding Mycobacterium tuberculosis disease and persistence. Adv Microb Physiol. 2010;57:43–117. doi: 10.1016/B978-0-12-381045-8.00002-3. [DOI] [PubMed] [Google Scholar]
- 19.Gandhi NR. Moll A. Sturm AW. Pawinski R. Govender T. Lalloo U. Zeller K. Andrews J. Friedland G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. The Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 20.Garg S. Alam MS. Bajpai R. Kishan KV. Agrawal P. Redox biology of Mycobacterium tuberculosis H 37 Rv: Protein–protein interaction between GlgB and WhiB 1 involves exchange of thiol-disulfide. BMC Biochem. 2009;10:1. doi: 10.1186/1471-2091-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg SK. Suhail Alam M. Soni V. Radha Kishan KV. Agrawal P. Characterization of Mycobacterium tuberculosis WhiB1/Rv3219 as a protein disulfide reductase. Protein Expr Purif. 2007;52:422–432. doi: 10.1016/j.pep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Geiman DE. Raghunand TR. Agarwal N. Bishai WR. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob Agents Chemother. 2006;50:2836. doi: 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez JE. Bishai WR. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc Natl Acad Sci USA. 2000;97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez JE. McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis. 2004;84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Guo M. Feng H. Zhang J. Wang W. Wang Y. Li Y. Gao C. Chen H. Feng Y. He ZG. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009;19:1301. doi: 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halliwell BaG J.M.C. Chemistry of Free Radicals and Related 'Reactive species'. In: Halliwell BaG J.M.C., editor. Free Radicals in Biology and Medicine. Oxford University Press; 2008. pp. 1–29. [Google Scholar]
- 27.Homolka S. Niemann S. Russell DG. Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: Delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathogens. 2010;6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim TH. Park JS. Kim HJ. Kim Y. Kim P. Lee HS. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem Biophys Res Commun. 2005;337:757–764. doi: 10.1016/j.bbrc.2005.09.115. [DOI] [PubMed] [Google Scholar]
- 29.Kumar A. Deshane JS. Crossman DK. Bolisetty S. Yan BS. Kramnik I. Agarwal A. Steyn AJ. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J Biol Chem. 2008;283:18032–18039. doi: 10.1074/jbc.M802274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A. Toledo JC. Patel RP. Lancaster JR., Jr. Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci USA. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levanon SS. San KY. Bennett GN. Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol Bioeng. 2005;89:556–564. doi: 10.1002/bit.20381. [DOI] [PubMed] [Google Scholar]
- 32.Manjunatha UH. Boshoff H. Dowd CS. Zhang L. Albert TJ. Norton JE. Daniels L. Dick T. Pang SS. Barry CE. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:431. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris RP. Nguyen L. Gatfield J. Visconti K. Nguyen K. Schnappinger D. Ehrt S. Liu Y. Heifets L. Pieters J, et al. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quemard A. Sacchettini JC. Dessen A. Vilcheze C. Bittman R. Jacobs WR., Jr Blanchard JS. Enzymic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 35.Rohde KH. Abramovitch RB. Russell DG. Mycobacterium tuberculosis invasion of macrophages: Linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Rustad TR. Harrell MI. Liao R. Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rybniker J. Nowag A. van Gumpel E. Nissen N. Robinson N. Plum G. Hartmann P. Insights into the function of the WhiB-like protein of mycobacteriophage TM4–a transcriptional inhibitor of WhiB2. Mol Microbiol. 2010;77:642–657. doi: 10.1111/j.1365-2958.2010.07235.x. [DOI] [PubMed] [Google Scholar]
- 38.Shalel-Levanon S. San KY. Bennett GN. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metab Eng. 2005;7:364–374. doi: 10.1016/j.ymben.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Sherrid AM. Rustad TR. Cangelosi GA. Sherman DR. Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis. PLoS One. 2010;5:e11622. doi: 10.1371/journal.pone.0011622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh A. Crossman DK. Mai D. Guidry L. Voskuil MI. Renfrow MB. Steyn AJC. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathogens. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh A. Guidry L. Narasimhulu KV. Mai D. Trombley J. Redding KE. Giles GI. Lancaster JR., Jr. Steyn AJ. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci USA. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith LJ. Stapleton MR. Fullstone GJM. Crack JC. Thomson AJ. Le Brun NE. Hunt DM. Harvey E. Adinolfi S. Buxton RS. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide sensitive iron-sulphur cluster. Biochem J. 2010;432:417. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soliveri JA. Gomez J. Bishai WR. Chater KF. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000;146:333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 44.Steyn AJ. Collins DM. Hondalus MK. Jacobs WR., Jr. Kawakami RP. Bloom BR. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voskuil MI. Schnappinger D. Visconti KC. Harrell MI. Dolganov GM. Sherman DR. Schoolnik GK. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y. Cui T. Zhang C. Yang M. Huang Y. Li W. Zhang L. Gao C. He Y. Li Y, et al. Global protein-protein interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J Proteome Res. 2010;9:6665–6677. doi: 10.1021/pr100808n. [DOI] [PubMed] [Google Scholar]