Abstract
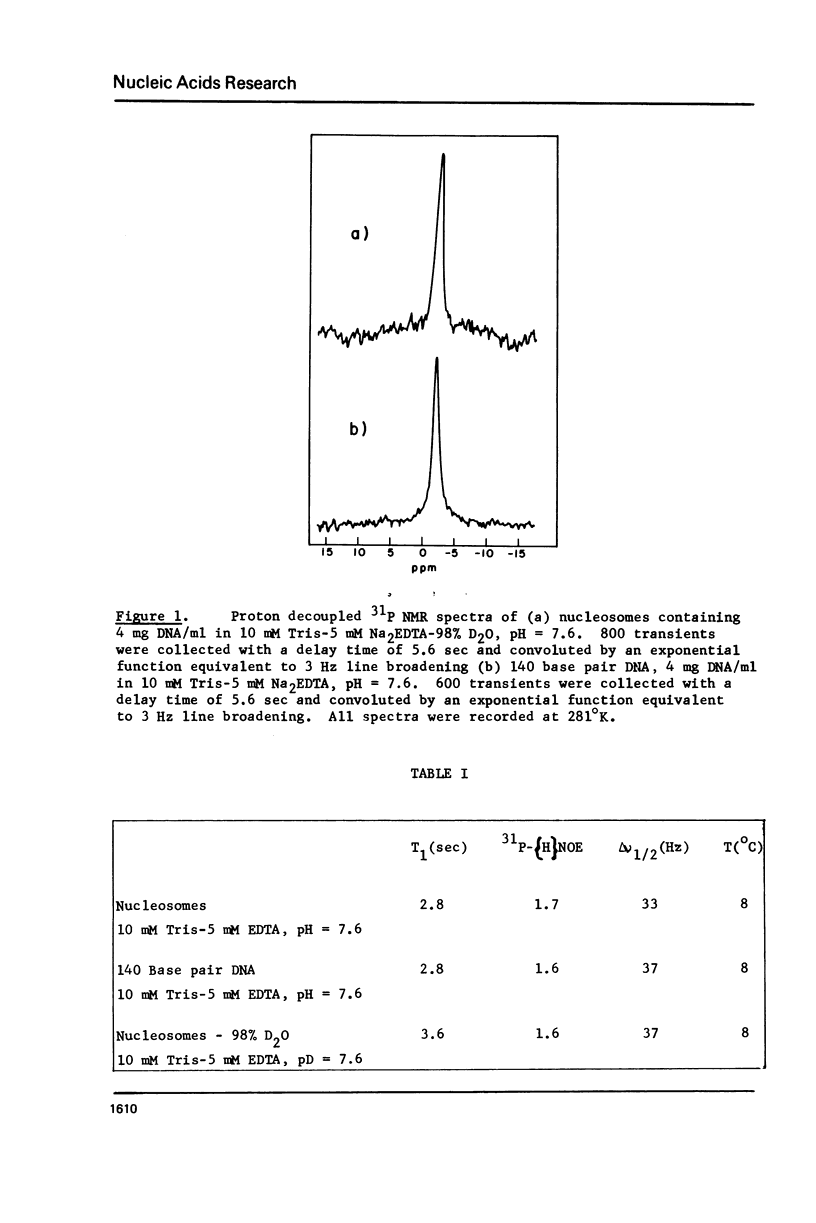
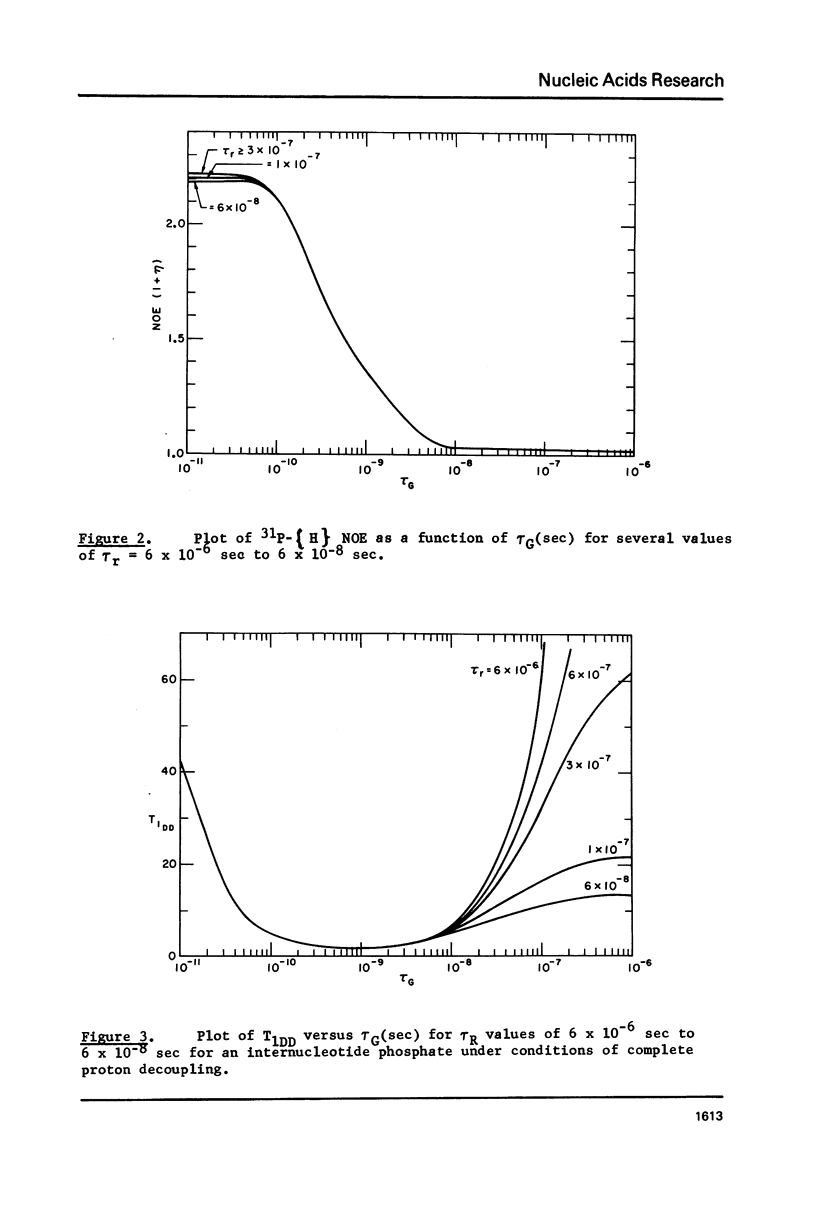
31P NMR studies of 140 base pair DNA fragments in nucleosomes and free in solution show no detectable change in the internucleotide 31P chemical shift or linewidth when DNA is packaged into nucleosomes. Measurements of 31P spin-lattice relaxation times T1 and 31P-[H] nuclear Overhauser enhancements revealed internal motion with a correlation time of about 4 x 10(-10) sec in double helical DNA, both free in solution and bound to nucleosomal core proteins. This result implies greater dynamic mobility in double helical DNA than has previously been supposed.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akasaka K. Proton and phosphorus magnetic relaxation studies on the dynamic structure of single-stranded polyriboadenylic acid. Biopolymers. 1974 Nov;13(11):2273–2280. doi: 10.1002/bip.1974.360131109. [DOI] [PubMed] [Google Scholar]
- Barry C. D., Glasel J. A., Williams R. J., Xavier A. V. Quantitative determination of conformations of flexible molecules in solution using lanthanide ions as nuclear magnetic resonance probes: application to adenosine-5'-monophosphate. J Mol Biol. 1974 Apr 25;84(4):471–409. doi: 10.1016/0022-2836(74)90110-7. [DOI] [PubMed] [Google Scholar]
- Barry C. D., North A. C., Glasel J. A., Williams R. J., Xavier A. V. Quantitative determination of mononucleotide conformations in solution using lanthanide ion shift and broadenine NMR probes. Nature. 1971 Jul 23;232(5308):236–245. doi: 10.1038/232236a0. [DOI] [PubMed] [Google Scholar]
- Cotter R. I., Lilley D. M. The conformation of DNA and protein within chromatin subunits. FEBS Lett. 1977 Oct 1;82(1):63–68. doi: 10.1016/0014-5793(77)80886-7. [DOI] [PubMed] [Google Scholar]
- Cozzone P. J., Jardetzky O. Phosphorus-31 Fourier transform nuclear magnetic resonance study of mononucleotides and dinucleotides. 1. Chemical shifts. Biochemistry. 1976 Nov 2;15(22):4853–4859. doi: 10.1021/bi00667a016. [DOI] [PubMed] [Google Scholar]
- Cozzone P. J., Jardetzky O. Phosphorus-31 Fourier transform nuclear magnetic resonance study of mononucleotides and dinucleotides. 2. Coupling constants. Biochemistry. 1976 Nov 2;15(22):4860–4865. doi: 10.1021/bi00667a017. [DOI] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G., Findlay J. B., Momii R. K., Luxon B. A., Kar D. Temperature dependence of the 31P chemical shifts of nucleic acids. A prode of phosphate ester torsional conformations. Biochemistry. 1976 Aug 24;15(17):3796–3803. doi: 10.1021/bi00662a023. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G., Kar D. 31-P chemical shifts in phosphate diester monoanions. Bond angle and torsional angle effects. Biochem Biophys Res Commun. 1975 Aug 4;65(3):1073–1080. doi: 10.1016/s0006-291x(75)80495-5. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G., Kar D., Luxon B. A., Momii R. K. Conformational study of cyclic and acyclic phosphate esters. CNDO/2 calculations of angle strain and torsional strain. J Am Chem Soc. 1976 Mar 31;98(7):1668–1673. doi: 10.1021/ja00423a005. [DOI] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guéron M., Shulman R. G. 31P magnetic resonance of tRNA. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3482–3485. doi: 10.1073/pnas.72.9.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M., Dattagupta N., Crothers D. M. Transient electric dichroism of rod-like DNA molecules. Proc Natl Acad Sci U S A. 1978 Jan;75(1):195–199. doi: 10.1073/pnas.75.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach N. R., Appleby D. W., Bradley C. H. 31P magnetic resonance of DNA in nucleosome core particles of chromatin. Nature. 1978 Mar 9;272(5649):134–138. doi: 10.1038/272134a0. [DOI] [PubMed] [Google Scholar]
- Klevan L., Crothers D. M. Isolation and characterization of a spacerless dinucleosome from H1-deleted chromatin. Nucleic Acids Res. 1977 Dec;4(12):4077–4089. doi: 10.1093/nar/4.12.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevan L., Hogan M., Dattagupta N., Crothers D. M. Electric dichroism studies of the size and shape of nucleosomal particles. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):207–214. doi: 10.1101/sqb.1978.042.01.023. [DOI] [PubMed] [Google Scholar]
- Kondo N. S., Danyluk S. S. Conformational properties of adenylyl-3' leads to 5'-adenosine in aqueous solution. Biochemistry. 1976 Feb 24;15(4):756–768. doi: 10.1021/bi00649a006. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. Nuclear magnetic resonance studies of the helix-coil transition of poly (dA-dT) in aqueous solution. Proc Natl Acad Sci U S A. 1976 Mar;73(3):674–678. doi: 10.1073/pnas.73.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J. Proton and phosphorus NMR studies of d-CpG(pCpG)n duplexes in solution. Helix-coil transition and complex formation with actinomycin-D. Biopolymers. 1976 Mar;15(3):533–558. doi: 10.1002/bip.1976.360150310. [DOI] [PubMed] [Google Scholar]
- RICH A., DAVIES D. R., CRICK F. H., WATSON J. D. The molecular structure of polyadenylic acid. J Mol Biol. 1961 Feb;3:71–86. doi: 10.1016/s0022-2836(61)80009-0. [DOI] [PubMed] [Google Scholar]



