Abstract
Formaldehyde is an essential metabolic intermediate in human cells and can also enter into the body through environmental exposures. It is classified as a human and animal carcinogen according to the International Agency for Research on Cancer (IARC). Previous research has demonstrated that formaldehyde is genotoxic, causing mutations in multiple genes. However, no exogenous formaldehyde-induced DNA adducts have been detected in animals after inhalation exposure, although formaldehyde can result in N6-deoxyadenosine, N2-deoxyguanosine and N4-deoxycytidine adducts in vitro. This can be partially attributed to the rapid metabolism of formaldehyde by glutathione (GSH)-dependent enzyme systems. Among the intermediates in the pathway of formaldehyde detoxication, S-hydroxymethylglutathione is a reactive species and has the potential to further conjugate with DNA bases. Here, we have demonstrated the formation of S-[1-(N2-deoxyguanosinyl)methyl]glutathione between glutathione and DNA in the presence of formaldehyde. This adduct is unique because of the involvement of S-hydroxymethylglutathione which is a key player during the detoxication of formaldehyde.
Formaldehyde, an essential metabolic intermediate generated endogenously from serine, glycine, methionine and choline and also produced from some metabolites and proteins by demethylation,1 is present in human blood at about 0.1 mM.2 Formaldehyde can also enter the body through environmental exposures. Formaldehyde forms DNA and protein adducts and DNA-protein crosslinks, and its toxicity has been the object of intensive investigation.3-9 Previous studies have shown that formaldehyde is genotoxic.1 Although N6-dA, N2-dG and N4-dC adducts of formaldehyde are found in vitro,10-13 no exogenous formaldehyde-induced DNA adducts have ever been detected in animals exposed by inhalation. The inability to detect DNA adducts may result from rapid binding of formaldehyde by the tripeptide glutathione (GSH), which significantly decreases the chance that exogenous formaldehyde will attack DNA directly.
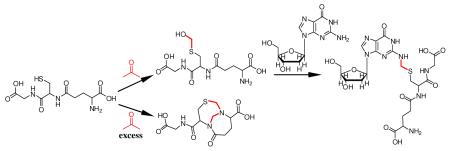
GSH is a major reducing thiol present in all human cells at a concentration around 5 mM. Formaldehyde (1; Scheme 1) reacts spontaneously with GSH (2) to form S-hydroxymethylglutathione (3).14 Formaldehyde dehydrogenase (ADH3) oxidizes 3 to S-formylglutathione, which is then hydrolyzed to formate by S-formylglutathione hydrolase, regenerating free glutathione.1 The S-hydroxymethyl group of 3 is a reactive target for nucleophilic substitution. Recent work in our laboratory on formaldehyde-induced DNA-protein crosslinks shows that the thiol groups of cysteine residues can readily crosslink with DNA bases in the presence of formaldehyde, raising the possibility that 3 can conjugate with DNA as shown in Scheme 1.
Scheme 1.
Formation of S-[1-(N2-deoxyguanosinyl)-methyl]glutathione induced by formaldehyde.
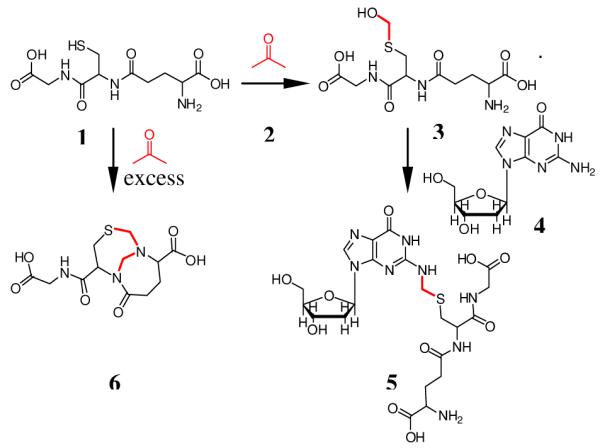
To characterize potential cross-linked products, excess GSH (5 mM) was allowed to react with formaldehyde in 10 mM potassium phosphate buffer (pH=7.2) for 4 h at 37°C, followed by incubation with deoxyguanosine (dG) (4) for another 8 h. A single coupling product eluted at 11.9 min on a C18 reverse phase column, giving a UV spectrum similar to that of dG, with an absorbance maximum at 260 nm (Figure S1). The exact mass of the protonated molecule was 587.1896 Da, consistent with elemental composition C21H30N8O10S expected for S-[1-(N2-deoxyguanosinyl)methyl]glutathione (5).
The ESI-MS/MS of the protonated molecule (Figure 1) shows major product ions corresponding to loss of deoxyribosyl and deoxynucleoside fragments, in accord with structural assignment 5. Definitive structural characterization was provided by NMR analysis of product isolated from a larger-scale reaction, which also served as standard for subsequent quantitation. The formaldehyde-derived methylene linkage between the β methylene carbon of the Cys residue and the exocyclic N2 of dG is established by C-H connectivities in the HMBC spectrum (Figure 2), which shows the expected cross peaks between the diastereotopic Cys β-methylene protons and the formaldehyde-derived carbon of the methylene linker and between the protons of the methylene linker and C2 of dG. Adduct 5 was stable in aqueous solution at room temperature over 16 h at pH 4, approximately 40% loss was observed at pH 7.2 (see Figure S6), supporting the hypothesis that if formed, this adduct would be detectable in DNA.
Figure 1.
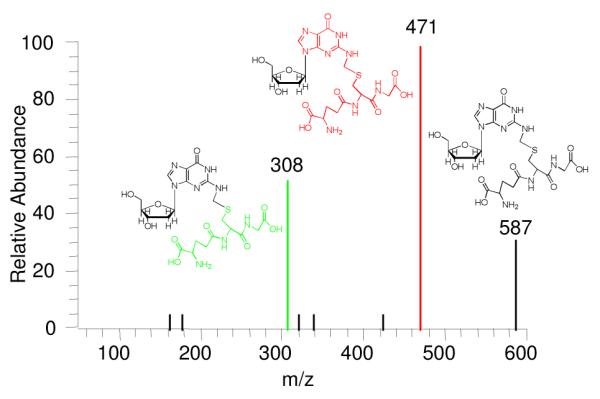
ESI-MS/MS of the protonated molecular ion of 5.
Figure 2.
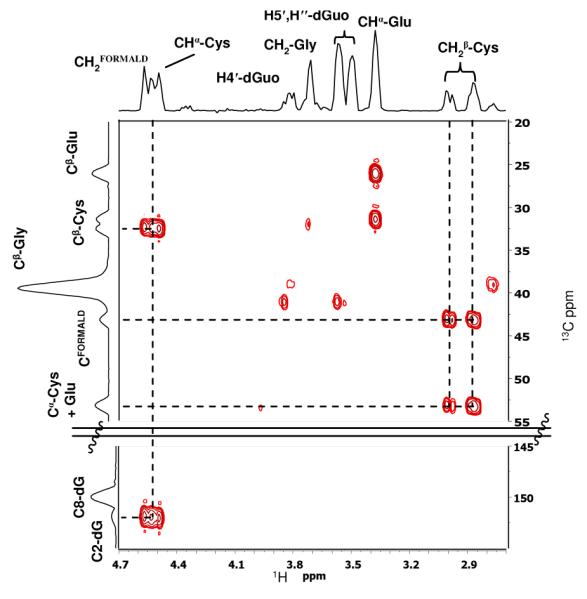
Expansion of the HMBC spectrum of 5 to show the Cys-β-methylene-formaldehyde linker-N2-dG connectivity.
In order to test this hypothesis, DNA was incubated with GSH in the presence of formaldehyde. GSH (5 mM) in 10 mM potassium phosphate buffer (pH=7.2) was treated with different concentrations of formaldehyde (0.1, 0.5, 1, 5, 50 mM) for 4 h at 37°C, followed by incubation with 100 μg of calf thymus DNA for 12 h. After extensive washing and digestion, the resultant adduct 5 was collected by HPLC and quantified by triple quadrupole mass spectrometer using selected reaction monitoring (SRM) mode (m/z 587→ m/z 308) and the previously-generated 5 from the larger-scale reaction as a standard. Figure 3 shows that production of 5 rises with increasing formaldehyde concentration from 0.1 mM to 5 mM, then declines when the formaldehyde concentration is further increased to 50 mM. Mass spectrometry analysis of the 50 mM reaction mixture showed the presence of a species consistent with the bicyclo[4.4.1]undecane structure (6), previously reported to be formed from reaction of GSH with two formaldehyde molecules when formaldehyde is present in excess.15,16
Figure 3.
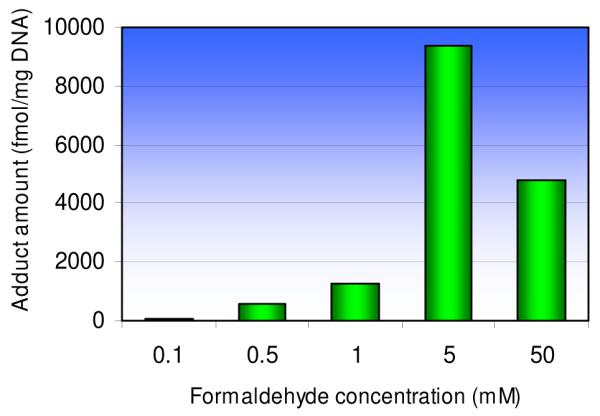
The influence of formaldehyde concentration on the formation of 5 in DNA.
This study establishes that 5 is formed as a consequence of formaldehyde scavenging the cellular antioxidant GSH in vitro. Both formaldehyde and GSH are ubiquitous cellular components, thus, this DNA adduct is expected to form endogenously in cells. The involvement of the cysteine residue of GSH in coupling suggests that other thiols may participate in the formation of this type of DNA damage from formaldehyde.
The formation of 5 from exogenous formaldehyde may serve as a biomarker to evaluate formaldehyde exposure. Concentrations of formaldehyde in the blood of humans and of rats after formaldehyde exposure (1.9 ppm for 40 min and 14.4 ppm for 2 hours) were not different from the pre-exposure concentration,17 consistent with extensive formation of 3. It has also been shown that formaldehyde exposure depletes GSH levels in cells and tissues,1 suggesting that GSH was not completely regenerated. Therefore, the scavenging of formaldehyde-induced 3 by ADH3 may be limited, which would allow opportunity for reaction between 3 and DNA to form 5. Also, adduct 5 is of potential importance for investigating effects of formaldehyde at distat sites. This issue remains one of the biggest challenges for understanding formaldehyde toxicity, carcinogenicity and epidemiology, which have been controversial for many years.1,18,19 Formaldehyde exposure by inhalation results in decreases in cellular GSH concentration in the liver,20 a remote site that inhaled formaldehyde is unlikely to reach by simple diffusion. Detection of 5 in distal tissues will shed light on the intriguing question of whether formaldehyde exhibits systemic toxicity.
In summary, we have demonstrated that formaldehyde can cross-link GSH with DNA by forming S-[1-(N2-deoxyguanosinyl)methyl]glutathione. This adduct may form endogenously since formaldehyde and GSH are ubiquitous in human cells. This adduct is unique because of the involvement of the reactive S-hydroxymethylglutathione intermediate that normally serves for formaldehyde detoxication. Since S-hydroxymethylglutathione is expected to be relatively abundant and highly reactive, and the adduct S-[1-(N2-deoxyguanosinyl)-methyl]glutathione is reasonably stable, the adduct may serve as a biomarker to understand formaldehyde toxicity and to evaluate formaldehyde exposure if coupled with the application of isotope-labeled formaldehyde to differentiate between endogenous and exogenous formaldehyde-derived adducts.
Supplementary Material
Acknowledgement
This work was supported in part by NIH grants P30-ES10126, P42-ES05948 and a grant from the Formaldehyde Council, Inc.
Footnotes
Supporting Information Available: Experimental procedures, Figures S1-S6: HPLC analysis, exact mass, 1H NMR and 2D NMR spectra of 5, calibration curve and evaluation of the stability of 5.
Reference
- (1).International Agency for Research on Cancer. 2008;88:46–331. [Google Scholar]
- (2).Heck HD, White EL, Casanova M. Biomed. Mass Spectrom. 1982;9:347–53. doi: 10.1002/bms.1200090808. [DOI] [PubMed] [Google Scholar]
- (3).Casanova M, Starr TB, Heck H. Toxicol. Appl. Pharmacol. 1987;89:122–134. doi: 10.1016/0041-008x(87)90182-7. [DOI] [PubMed] [Google Scholar]
- (4).Speit G, Schutz P, Merk O. Mutagenesis. 2000;15:85–90. doi: 10.1093/mutage/15.1.85. [DOI] [PubMed] [Google Scholar]
- (5).Quievryn G, Zhitkovich A. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- (6).Kerns WD, Pavkov KL, Donofrio DJ, Gralla EJ, Swenberg JA. Cancer Res. 1983;43:4382–4392. [PubMed] [Google Scholar]
- (7).Nelson N, Levine RJ, Albert RE, Blair AE, Griesemer RA, Landrigan PJ, Stayner LT, Swenberg JA. Environ. Health Perspect. 1986;70:23–35. doi: 10.1289/ehp.867023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lu K, Boysen G, Gao L, Collins LB, Swenberg JA. Chem. Res. Toxicol. 2008;21:1586–1593. doi: 10.1021/tx8000576. [DOI] [PubMed] [Google Scholar]
- (9).Recio L, Sisk S, Pluta L, Bermudez E, Gross EA, Chen Z, Morgan K, Walker C. Cancer Res. 1992;52:6113–6136. [PubMed] [Google Scholar]
- (10).McGhee JD, Hippel PH. Biochemistry. 1975;14:1281–1296. doi: 10.1021/bi00677a029. [DOI] [PubMed] [Google Scholar]
- (11).Huang HF, Solomon MS, Hopkins PB. J. Am. Chem. Soc. 1992;114:9240–9241. [Google Scholar]
- (12).Huang HF, Hopkins PB. J. Am. Chem. Soc. 1993;115:9402–9408. [Google Scholar]
- (13).Zhong W, Hee SQ. J. Anal. Toxicol. 2005;29:182–187. doi: 10.1093/jat/29.3.182. [DOI] [PubMed] [Google Scholar]
- (14).Casanova M, Heck H. Toxicol. Appl. Pharmacol. 1987;89:105–121. doi: 10.1016/0041-008x(87)90181-5. [DOI] [PubMed] [Google Scholar]
- (15).Naylor S, Mason PR, Sanders KMJ, Williams HD. Biochem. J. 1988;249:573–579. doi: 10.1042/bj2490573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bateman R, Rauh D, Shokat KM. Org. Biomol. Chem. 2007;5:3363–3367. doi: 10.1039/b707602a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Heck HD, Casanova M, Dodd PB, Schachter EN, Witek TJ, Tosun T. Am. Ind. Hyg. Assoc. J. 1985;46:1–3. doi: 10.1080/15298668591394275. [DOI] [PubMed] [Google Scholar]
- (18).Zhang L, Steinmaus C, Eastmond DA, Xin XK, Smith MT. Mutat. Res. 2008 doi: 10.1016/j.mrrev.2008.07.002. in press. [DOI] [PubMed] [Google Scholar]
- (19).Heck H, Casanova M. Regul. Toxicol. Pharmacol. 2004;40:92–106. doi: 10.1016/j.yrtph.2004.05.001. [DOI] [PubMed] [Google Scholar]
- (20).Söğüt S, Songur A, Özen OA, Özyurt H, Sarsılmaz M. Eur. J. Gen. Med. 2004;1:26–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.