Abstract
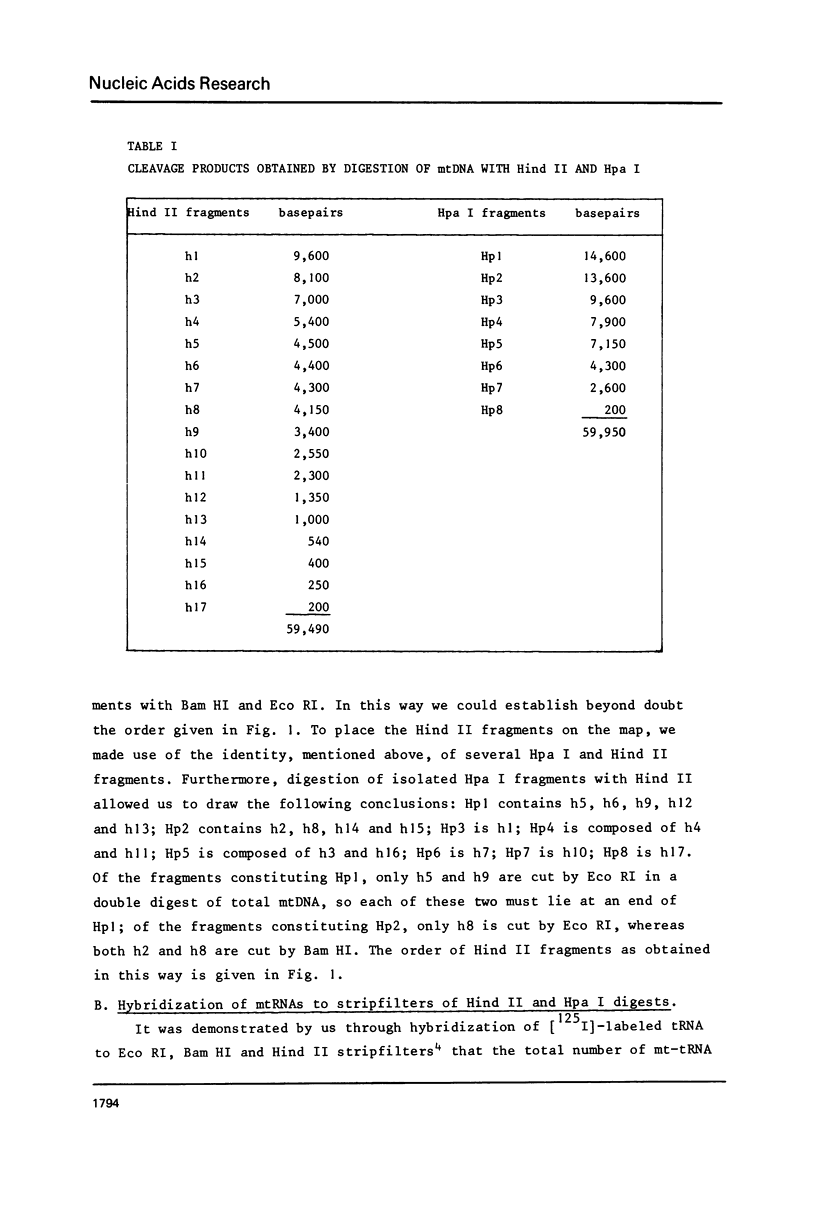
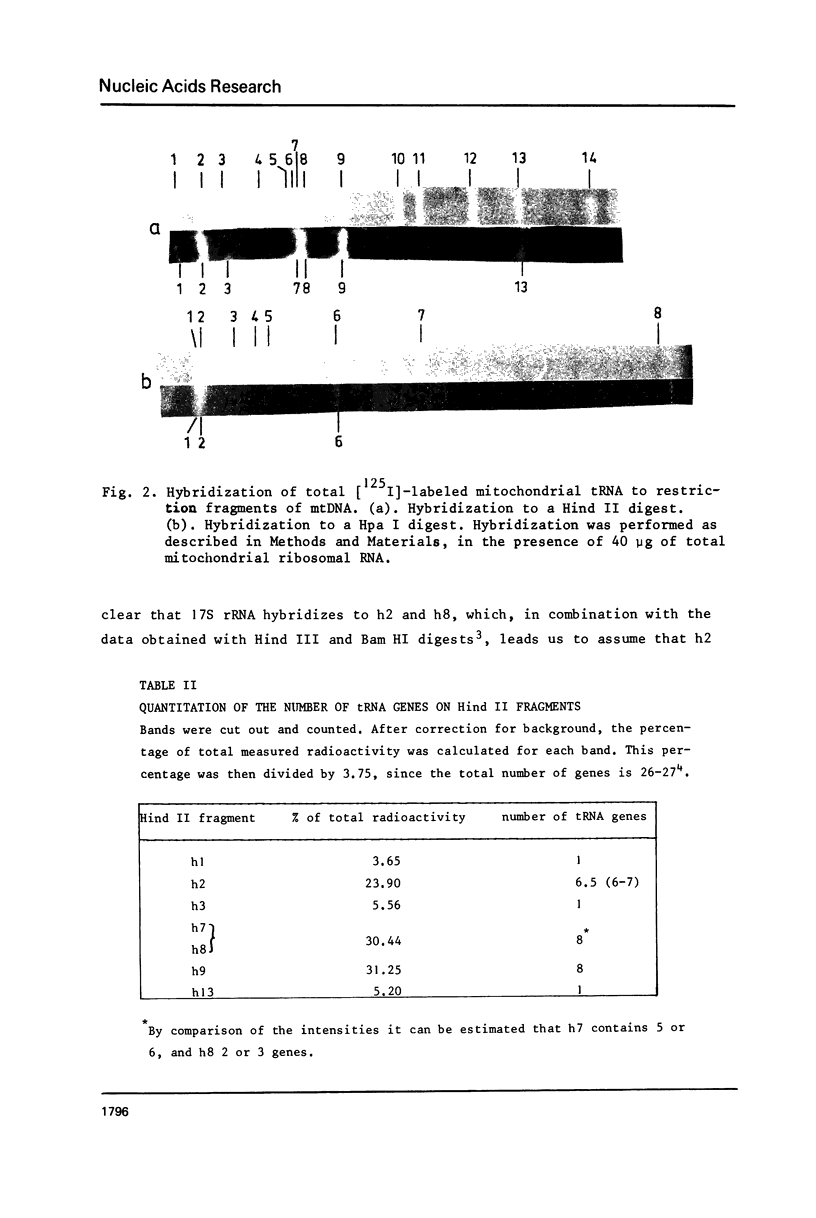
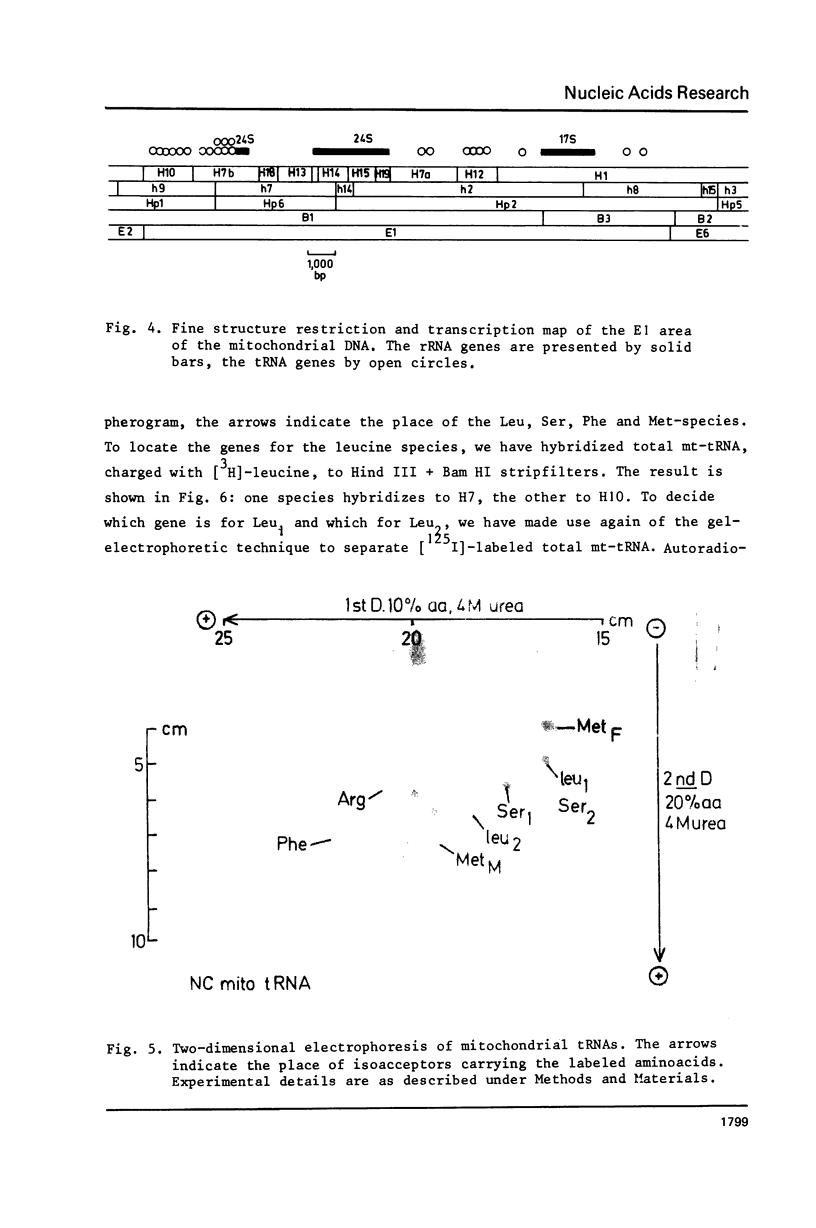
To obtain more information about the arrangement of Hind III restriction fragments in the tRNA-rRNA region of the Neurospora crassa mitochondrial (mt) DNA we have cleaved the mtDNA with Hpa I and Hind II. We could construct additional cleavage maps for these enzymes. Hybridization of rRNAs to Hind II fragments confirmed the existence of an intervening region of about 2,300 basepairs in the 24S rRNA (Hahn et al., Cell, in press). About seven tRNA genes, among which the genes for tRNA1Ser and tRNAMetM, are located in a segment of about 5,000 bp separating the 24S and 17S rRNA genes. Another cluster of 14 tRNA genes is found adjacent to the other end of the 24S gene. The genes for tRNALeu1 and tRNAMetF are located in this cluster.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Jeppesen P. G., Sanger F., Barrell B. G. Nucleotide sequence from the coat protein cistron of R17 bacteriophage RNA. Nature. 1969 Sep 6;223(5210):1009–1014. doi: 10.1038/2231009a0. [DOI] [PubMed] [Google Scholar]
- Angerer L., Davidson N., Murphy W., Lynch D., Attardi G. An electron microscope study of the relative positions of the 4S and ribosomal RNA genes in HeLa cells mitochondrial DNA. Cell. 1976 Sep;9(1):81–90. doi: 10.1016/0092-8674(76)90054-4. [DOI] [PubMed] [Google Scholar]
- Blin N., von Gabain A., Bujard H. Isolation of large molecular weight DNA from agarose gels for further digestion by restriction enzymes. FEBS Lett. 1975 Apr 15;53(1):84–86. doi: 10.1016/0014-5793(75)80688-0. [DOI] [PubMed] [Google Scholar]
- Bos J. L., Heyting C., Borst P., Arnberg A. C., Van Bruggen E. F. An insert in the single gene for the large ribosomal RNA in yeast mitochondrial DNA. Nature. 1978 Sep 28;275(5678):336–338. doi: 10.1038/275336a0. [DOI] [PubMed] [Google Scholar]
- Chiu N., Chiu A., Suyama Y. Native and imported transfer RNA in mitochondria. J Mol Biol. 1975 Nov 25;99(1):37–50. doi: 10.1016/s0022-2836(75)80157-4. [DOI] [PubMed] [Google Scholar]
- De Vries H., De Jonge J. C., Schneller J. M., Martin R. P., Dirheimer G., Stall A. J. Neurospora crassa mitochondrial transfer RNAs. Biochim Biophys Acta. 1978 Sep 27;520(2):419–427. doi: 10.1016/0005-2787(78)90239-3. [DOI] [PubMed] [Google Scholar]
- Faye G., Dennebouy N., Kujawa C., Jacq C. Inserted sequence in the mitochondrial 23S ribosomal RNA gene of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1979 Jan 5;168(1):101–109. doi: 10.1007/BF00267939. [DOI] [PubMed] [Google Scholar]
- Fradin A., Gruhl H., Feldmann H. Mapping of yeast tRNAs by two-dimensional electrophoresis on polyacrylamide gels. FEBS Lett. 1975 Feb 1;50(2):185–189. doi: 10.1016/0014-5793(75)80485-6. [DOI] [PubMed] [Google Scholar]
- Haff L. A., Bogorad L. Hybridization of maize chloroplast DNA with transfer ribonucleic acids. Biochemistry. 1976 Sep 7;15(18):4105–4109. doi: 10.1021/bi00663a029. [DOI] [PubMed] [Google Scholar]
- Kuriyama Y., Luck D. J. Ribosomal RNA synthesis in mitochondria of Neurospora crassa. J Mol Biol. 1973 Feb 5;73(4):425–437. doi: 10.1016/0022-2836(73)90091-0. [DOI] [PubMed] [Google Scholar]
- McCrea J. M., Hershberger C. L. Chloroplast DNA codes for transfer RNA. Nucleic Acids Res. 1976 Aug;3(8):2005–2018. doi: 10.1093/nar/3.8.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R., Merten S., Lewin A., Martin N. C., Rabinowitz M. Physical mapping of genes on yeast mitochondrial DNA: localization of antibiotic resistance loci, and rRNA and tRNA genes. Mol Gen Genet. 1978 Jul 25;163(3):241–255. doi: 10.1007/BF00271954. [DOI] [PubMed] [Google Scholar]
- Rochaix J. D., Malnoe P. Anatomy of the chloroplast ribosomal DNA of Chlamydomonas reinhardii. Cell. 1978 Oct;15(2):661–670. doi: 10.1016/0092-8674(78)90034-x. [DOI] [PubMed] [Google Scholar]
- Schneller J. M., Faye G., Kujawa C., Stahl A. J. Number of genes and base composition of mitochondrial tRNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1975 Jun;2(6):831–838. doi: 10.1093/nar/2.6.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Terpstra P., Holtrop M., Kroon A. M. A complete cleavage map of Neurospora crassa mtDNA obtained with endonucleases Eco RI and Bam HI. Biochim Biophys Acta. 1977 Apr 19;475(4):571–588. doi: 10.1016/0005-2787(77)90318-5. [DOI] [PubMed] [Google Scholar]
- Terpstra P., Holtrop M., Kroon A. M. The ribosomal RNA genes on Neurospora crassa mitochondrial DNA are adjacent. Biochim Biophys Acta. 1977 Sep 20;478(2):146–155. doi: 10.1016/0005-2787(77)90178-2. [DOI] [PubMed] [Google Scholar]
- Van Den Hondel C. A., Schoenmakers J. G. Studies on bacteriophage M13 DNA. 1. A cleavage map of the M13 genome. Eur J Biochem. 1975 May 6;53(2):547–558. doi: 10.1111/j.1432-1033.1975.tb04098.x. [DOI] [PubMed] [Google Scholar]
- Van Ommen G. J., Groot G. S., Borst P. Fine structure physical mapping of 4S RNA genes on mitochondrial DNA of Saccharomyces cerevisiae. Mol Gen Genet. 1977 Sep 9;154(3):255–262. doi: 10.1007/BF00571280. [DOI] [PubMed] [Google Scholar]