Abstract
Metabolomics aims at detection and quantitation of all metabolites in biological samples. The presence of metabolites with a wide variety of physicochemical properties and different levels of abundance challenges existing analytical platforms used for identification and quantitation of metabolites. Significant efforts have been made to improve analytical and computational methods for metabolomics studies.
This review focuses on the use of liquid chromatography with tandem mass spectrometry (LC-MS/MS) for quantitative and qualitative metabolomics studies. It illustrates recent developments in computational methods for metabolite identification, including ion annotation, spectral interpretation and spectral matching. We also review selected reaction monitoring and high-resolution MS for metabolite quantitation. We discuss current challenges in metabolite identification and quantitation as well as potential solutions.
Keywords: High-resolution mass spectrometry, Identification, Ion annotation, Liquid chromatography, Metabolite, Metabolomics, Selected reaction monitoring, Spectral interpretation, Spectral matching, Tandem mass spectrometry
1. Introduction
Metabolomics is primarily concerned with identification and quantitation of small-molecule metabolites (<1500 Da) in the metabolome [1]. It facilitates understanding of the mechanisms of biological and biochemical processes in complex systems. It also provides a diagnostic aid to diseases.
Current metabolomics investigations can be categorized as two complementary approaches: targeted and untargeted. The targeted approach focuses on the analysis of specific group of metabolites related to certain metabolic pathway or a class of compounds. The untargeted approach is a global analysis of metabolic changes in response to disease, environmental or genetic perturbations. Untargeted approach is typically carried out for hypothesis generation, followed by targeted profiling for more confident quantitation of relevant metabolites.
Metabolomics has been applied in various research areas including environmental and biological-stress studies [2], biomarker discovery [3], functional genomics [4] and integrative systems biology [5]. Due to the complexity and dynamic nature of the metabolome, multiple analytical platforms are needed to cover the full spectrum of metabolites. Among them, mass spectrometry (MS), nuclear magnetic resonance (NMR) spectroscopy, and Fourier transform infrared (FT-IR) spectroscopy are the most commonly employed.
MS offers quantitative analysis of metabolites with high sensitivity and selectivity and potential to identify metabolites. For example, the availability of various atmospheric pressure ionization (API) methods in both positive and negative modes [e.g., electrospray ionization (ESI), atmospheric pressure chemical ionization (APCI), and atmospheric pressure photoionization (APPI)] enables ionization of diverse classes of metabolites. Among them, ESI is often preferred for profiling “unknown” metabolites, since this “soft” ionization approach forms intact molecular ions and aids initial identification. Similar to ESI, APCI and APPI typically induce little or no fragmentation and are considered robust and relatively tolerant to high buffer concentrations. These ionization approaches can be complementary to ESI for the analysis of non-polar and thermally-stable compounds (e.g., lipids). A single ionization source containing combinations of ESI and APCI or ESI and APPI currently has become the trend in source configuration.
Meanwhile, versatile mass analyzers working in tandem or hybrid configuration can further aid metabolite identification by acquiring highly resolved and accurate MS/MS spectra. This is achieved through ion fragmentation by collision-induced dissociation (CID) in either quadruple-based tandem in-space instruments [e.g., triple quadrupole (QqQ) or quadrupole time-of-flight (QTOF)], or ion-trap-based tandem in-time instruments [e.g., quadrupole-ion trap (QIT), linear trap quadrupole (LTQ)-Orbitrap, or linear trap quadrupole Fourier-transform ion cyclotron resonance (LTQ-FT-ICR)]. Among them, QqQ has been considered as a reference tool for absolute quantitation of small molecules due to its sensitivity and specificity using selected reaction monitoring (SRM), which has been applied for quantitation of trace-level metabolites with detection limit of ng/mL in sample matrices (e.g., plasma, serum, or cellular media) [6–8].
The coupling of liquid chromatography (LC) to MS (LC-MS) facilitates metabolite identification and quantitation by reducing sample complexity and allowing metabolite separation prior to detection. High-performance LC (HPLC), as the most versatile separation method, allows separation of compounds of a wide range of polarity. LC coupled to electrospray-ionization MS (LC-ESI-MS) is becoming a method of choice for detecting metabolites in complex biological samples [1]. In LC-ESI-MS, reversed-phase LC (RPLC), normally using C18 columns, can separate semi-polar compounds (phenolic acids, flavonoids, glycosylated steroids, alkaloids and other glycosylated species). However, hydrophilic interaction LC (HILIC) can use polar columns (e.g., aminopropyl) to separate polar compounds (e.g., sugars, amino sugars, amino acids, vitamins, carboxylic acids and nucleotides). Although normal-phase LC (NPLC) can also separate polar compounds, the use of non-polar organic mobile phases makes it more compatible with APCI-MS instead of ESI-MS. NPLC-APCI-MS has been applied for the analysis of non-polar lipids (e.g., triacylglycerols, sterols and fatty-acid esters) and other lipidomics studies [9].
The advantages of coupling HPLC separation with MS detection include improved MS sensitivity and signal reproducibility by reducing sample complexity, thereby alleviating matrix interferences in the ionization process. In addition, good chromatographic separation will result in better quality MS data due to reduced background noise. Recent developments in LC [e.g., capillary monolithic chromatography and ultra-performance LC (UPLC)] have achieved significant progress to improve peak resolution and expedite analysis [10].
In this review, we not only focus on aspects relevant to identification and quantification of metabolites in terms of developing trends in analytical and computational methods, but also provide our personal views on recent issues of high-throughput quantitative MS and the latest developments in qualitative and quantitative metabolomics. Section 2 presents a computational framework for metabolite identification by LC-MS/MS, and reviews a variety of computational tools that can reduce the number of putative identifications and prioritize them for further verification. Although comparing MS/MS spectrum of individual metabolites with authentic compounds is still considered as the “gold standard” for metabolite identification, the use of computational and informatics tools can accelerate the identification process and reduce the cost. Section 3 focuses on metabolite quantitation by LC-SRM-MS/MS and LC-high-resolution MS (LC-HRMS). We discuss their limitations and advantages as quantitative approaches to metabolomics in detail. Finally, Section 4 discusses current challenges in metabolite identification and quantitation as well as future ways to address these challenges.
2. LC-MS/MS-based metabolite identification
One of the key challenges in metabolomics studies is identification of metabolites. Compared to peptides, which comprise 20 amino acids repeatedly arranged in linear orders, metabolites are random combinations of elements (e.g., C, H, O, S, N, and P). The chemical and physical diversities of metabolites make them difficult to identify based on MS data.
At present, metabolite identification in untargeted metabolomics analysis is mainly achieved through mass-based search followed by manual verification. First, the m/z value of a molecular ion of interest is searched against database(s) [11–13]. The metabolites having molecular weights within a specified tolerance range to the query m/z value are retrieved from the databases as putative identifications. Then, authentic compounds of these putative identifications are subjected to a tandem MS (MS/MS) experiment side-by-side with the sample. By comparing the MS/MS spectra and retention times of the authentic compounds with the molecules of interest in the sample, the identities of the molecules can be confirmed. However, putative identifications from mass-based search are rarely unique, due to the existence of isomers and the limited accuracy of mass spectrometers [14]. In some cases, one molecule ion can have more than 100 putative identifications, which makes the manual verification costly and laborious. As a result, this approach is practically applicable only to a limited number of molecules.
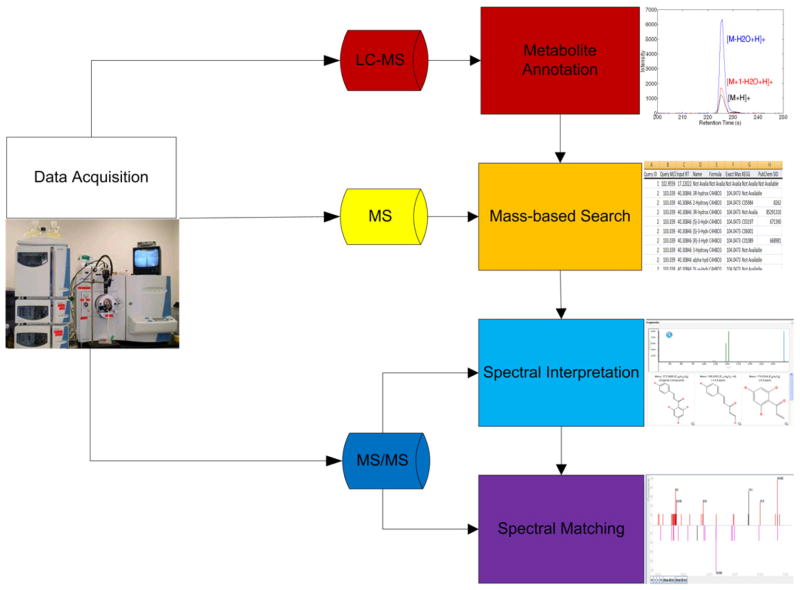
To improve the efficiency of metabolite identification for a large number of metabolites, we suggest a computational framework that can reduce the number of putative identifications and prioritize them (Fig. 1). The framework primarily focuses on untargeted endogenous metabolomics studies. Here, we do not consider metabolite-identification tasks in pharmacokinetic/pharmacodynamic (PK/PD) analysis, although some techniques that we discuss below will also benefit drug-metabolite identification. In the following, we discuss the components illustrated in Fig. 1.
Figure 1.
The metabolite identification framework, which uses EICs, m/z values, and MS/MS spectra. EICs are used for ion annotation. The m/z values are used in mass-based search to obtain a list of putative identifications. MS/MS spectra are used for spectral interpretation, spectral matching, and metabolite verification.
2.1. Data acquisition
Both LC-MS and MS/MS data provide useful information for identification purpose. A typical LC-MS-based metabolomics study often starts with an untargeted LC-MS experiment, where full-scan LC-MS data are acquired. Statistical methods are then used to select a list of molecular ions whose levels are significantly altered between case and control samples. These molecules are then subjected to precursor-ion (PI) scans to acquire MS/MS data by manually setting the m/z values of the PIs. The MS/MS data, in combination of precursor m/z and retention time, are used to derive the structural information about these molecules.
The MS/MS information can also be acquired “on the fly” by either data-dependent acquisition (DDA) or data-independent acquisition (DIA).
DDA has been used in both proteomics and metabolomics [15]. It includes a survey scan followed by MS/MS acquisition. During the survey scan, MS automatically selects PIs above a pre-set abundance threshold and triggers the instrument to perform fragmentation on those PIs, followed by MS/MS full scan on the product ions.
DIA subjects all the ions within an m/z window to fragmentation instead of selecting a PI. One example of DIA is the MSE mode in the Waters QTOF instrument, where the mass spectrometer alternates between modes – low collision energy and high collision energy. In DIA, all compounds that elute at the same retention time go through fragmentation without selection. MSE was used [16] to study the endogenous metabolites in rat urine and showed spectra comparable to those obtained using conventional PI-scan mode. A lipidomics study performed using a Thermo Orbitrap instrument was reported [17], where qualitative and semi-quantative information about mitochondrial lipid cardiolipins were acquired by alternating MS full scans with high-energy collisional dissociation (HCD) scans. In mitochondrial samples harvested from rats, 28 unique cardiolipins species were identified. DIA covers a broader intensity range of analytes than DDA. However, the acquired fragmentation spectra are relatively difficult to analyze due to the lack of PI selection. As a result, the acquired fragmentation spectra need to be decomposed into individual MS/MS spectrum corresponding to PIs. In combination with UPLC, this decomposition is usually accomplished by grouping together the product ions (from the high-collision-energy scan) and PIs (from the low-collision-energy scan) on the basis of retention time [18].
In the Waters SYNAPT G2 HDMS mass spectrometer, in addition to UPLC, ion-mobility separation (IMS) is used between the quadrupole and the time-of-flight (TOF) analyzer to provide another dimension of separation for MSE experiments, thereby simplifying the acquired fragmentation spectra and making them easier to decompose.
Although DDA and DIA provide the capability to acquire MS/MS spectra, it should be noted that the successful identification of metabolites requires high-quality MS/MS spectra, which often involves specific experimental consideration (e.g., selection of collision energy when using CID, and the use of multiple collision energies). MS/MS spectra acquired with DDA or DIA should be treated with necessary caution.
2.2. Ion annotation
Ion annotation is a procedure to recognize a group of ions likely to originate from the same compound. In LC-MS-based metabolomics, one metabolite is often represented by multiple peaks in LC-MS data with distinct m/z values but, at similar retention times, due to the presence of isotopes, adducts and neutral loss fragments. As long as the scan rate of mass spectrometer is properly adjusted and enough scanning points are acquired to define the chromatographic peaks, the ions from the same compound share similarly shaped elution profiles, which can be represented by their extracted ion chromatograms (EICs). Thus, ion annotation can be achieved by clustering similar elution profiles together, thereby facilitating metabolite identification.
In one approach to ion annotation [19], the ions were grouped based on the Pearson correlation of their EICs. If the correlation between two ions is above a pre-defined threshold, and the m/z difference between the two ions can be explained as adducts/isotopes/neutral-loss products, the two ions are considered to originate from the same compound. In this method, the choice of the Pearson-correlation threshold is largely empirical without statistical interpretation. Also, when the elution profiles of two ions have a large overlap, Pearson correlation is generally high and not sensitive enough to capture the subtle difference in EICs.
A statistically rigorous approach was proposed [20] to test if two ions measured by TOF-MS are derived from the same compound. In this approach, the TOF-MS signal observed is modeled as a Poisson process. If two ions are derived from the same compound, the conditional distribution of observed intensity, given the summed intensity, should have a binomial distribution with a constant success rate across the retention time.
The Pearson chi-square test was used to evaluate the goodness-of-fit of the observation to binomial distribution, from which an associated p-value was derived. It was shown that this approach can reduce the false-positive rate of ion annotation to 6% compared to the false-positive rate of the Pearson-correlation approach while maintaining the same sensitivity level [20]. However, the approach is limited to data acquired using time-to-digital converter (TDC) detectors. Also, when the ion intensity is high, the acquired signal will deviate from the Poisson process due to detector saturation, so an inflated p-value is obtained.
2.3. Mass-based identification
After grouping peaks together by ion annotation, the monoisotopic exact masses of these compounds can be calculated based on the mass differences of adducts/isotopes from their monoisotopic neutral forms. The calculated masses can be used to search against metabolite databases [e.g., HMDB (Human Metabolome Database), Metlin and MMCD (Madison Metabolomics Consortium Database)] or more general chemical databases (e.g., PubChem or ChemSpider). Metabolites having molecular masses within the pre-specified tolerance of the query masses are retrieved from these databases. However, mass-based identification seldom results in unique identification of these ions. By acquiring MS/MS spectra of these ions, the results from mass-based identification can be further refined through following steps.
2.4. Spectral interpretation
Spectral interpretation deduces the possible structure or sub-structure of an unknown molecular ion by comparing its MS/MS spectrum with hypothetical spectra predicted through in-silico fragmentation approaches.
There are two major ways to predict the fragment ions of a given molecule. One way is to use a rule-based predictor, which resorts to fragmentation pathways collected from the literature. Such predictors include ACD Fragmenter (ACD/Labs) and Mass Frontier (HighChem, Ltd.), both of which are commercially available. Currently, there are about 5000 manually-collected fragmentation rules involving around 19,000 reactions in the Mass Frontier Fragmentation Library. These fragmentation rules are used to predict possible product ions in MS/MS experiments. The advantage of a rule-based approach is its potentially high specificity. Not only are the general fragmentation rules used in in-silico fragmentation, but also those specific for a particular class of compounds. However, if a fragmentation rule is missed from the knowledge base, the corresponding product ion can never be predicted.
Other in-silico fragmentation tools {e.g., Fragment Identificator (FiD) [21] or MetFrag [22]} are developed under a different rationale, which is that they generate a list of possible fragments through combinatorial disconnection of chemical bonds in the compound. The internal energy of each cleaved bond is computed. Generally, low-energy cleavages are preferred over high-energy cleavages. The combinatorial approach does not depend on any knowledge base, so it eliminates the labor-intensive rule collection step and avoids the potential bias derived from a limited set of fragmentation rules. However, the combinatorial calculation itself is time consuming when the possible structure has a large number of chemical bonds. Also, it lacks the specificity of the rule-based prediction.
After a hypothetical MS/MS spectrum is generated by in-silico fragmentation, it is compared against the experimental spectrum to calculate a similarity score. The candidate structures are ranked according to similarity scores. However, as suggested [23], identification based on in-silico fragmentation needs to be checked with caution. The prediction of low-resolution electron-ionization (EI) spectra is found to exhibit bias towards certain types of structure, depending on the program setting. Although we would expect results to improve with high-resolution mass spectrum or with combinatorial approaches, it remains to be proved in future studies.
2.5. Spectral matching
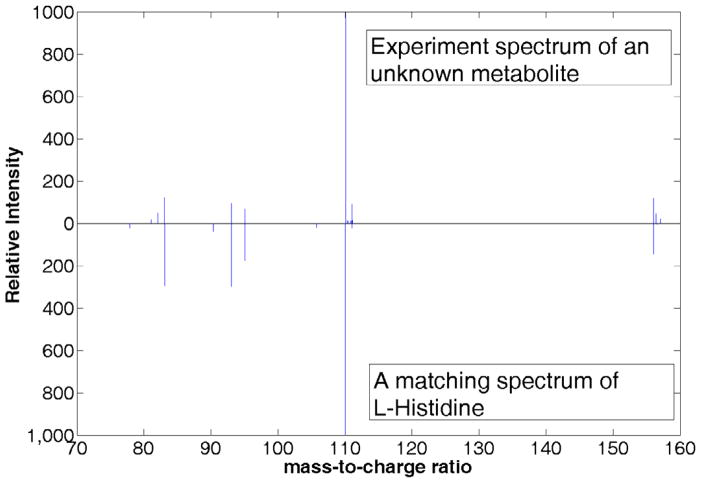
Spectral matching mimics the manual verification of metabolite identity using the MS/MS spectrum. Instead of acquiring the MS/MS spectrum of the authentic compound each time, previously acquired MS/MS spectra of authentic compounds are assembled in a spectral library and used to compare with the spectra acquired from biological samples. Several spectral libraries have already been constructed and open to public [12,13,24]. Fig. 2 shows an example of spectral matching using the MassBank database. An appropriate scoring function that can measure the similarity between two MS/MS spectra is the key for any mass spectral matching algorithm. Spectra with a high similarity are considered to represent the same metabolites. Previously, several spectral matching approaches have been developed for gas chromatography (GC) with MS (GC-MS) spectra and MS/MS spectra acquired in proteomics studies. Most of these algorithms calculate the similarity between the query spectrum and a library spectrum by treating the two spectra as vectors and calculating their modified dot-product of [24,25], e.g.:
where WL and WQ are scaled and mass-weighted intensities of the library spectrum and the query spectrum, respectively.
Figure 2.
An example of spectra matching using MassBank database. A spectrum of unknown metabolite is searched against the database and the library spectrum of L-histidine gives the best matching score.
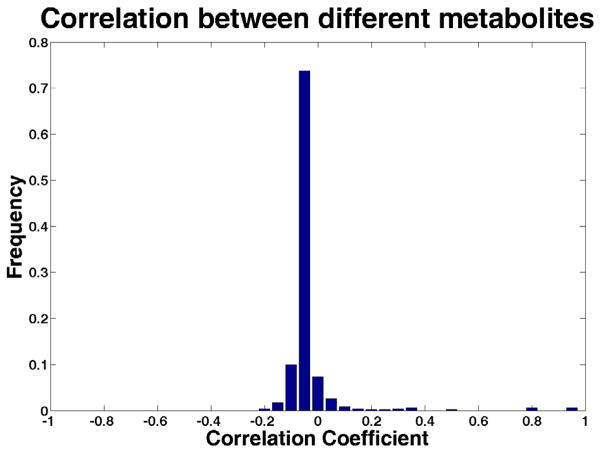
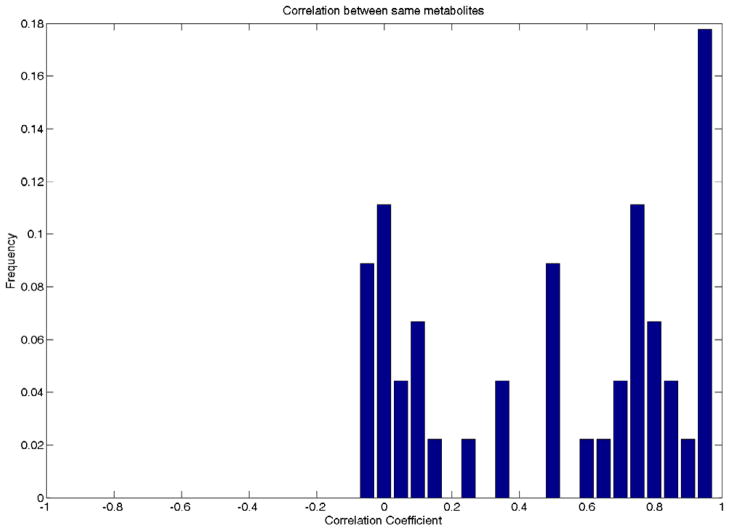
One of the major difficulties for MS/MS spectral matching is that the acquired spectra greatly depend on the machines and the acquisition settings, as API technique is more often used in LC-MS/MS experiment. Fig. 3 demonstrates that, although the spectra of different metabolites are generally uncorrelated, the correlation coefficients of the spectra derived from the same metabolite can span a large range in the presence of data heterogeneity. Using the dot-product-based approach, MassBank reported the maximum F-measure less than 0.4 when the query data and the spectral library data were from different types of machine [24].
Figure 3.
The Pearson correlation coefficients between a set of MS/MS spectra from the same metabolites (A) and various spectra from different metabolites (B). The data in this analysis include in-house datasets generated by UPLC-QTOF (Waters Premier) and LC-QqQ (Waters Micromass Quattro) datasets from the HMDB database.
One solution is to expand the spectral libraries with spectra acquired under different experimental settings (e.g. different collision energies or instrument types) as in MassBank. The other possible solution is to design better scoring functions by considering the different aspects of “spectral similarity”. Previously, in a proteomics study, linear discriminant analysis was applied to gauge the relative importance of multiple SEQUEST matching parameters [26]. A similar method was also used in HMDB database for spectral matching of metabolites. We proposed a spectral matching approach that uses support vector machines to combine profile similarity and peak similarity of two spectra [27]. The approach led to a 7–10% improvement in identification performance over dot-product spectral matching approaches.
The other limitation of spectral matching is the limited coverage of spectral libraries. One effort is to retrieve those metabolites that are not exactly identical to the compounds of the query spectra but share similar (sub-)structures. It is known that metabolites with similar structure or sub-structure may share similar spectral characteristics (e.g., common product ions or neutral losses). Thus, by allowing metabolites with similar (sub-)structures to be matched, this approach can help to address the limited library coverage problem. MassLib (MSP Kofel), a spectral matching software tool, has utilized this concept to retrieve compounds of similar structures.
2.6. Metabolite verification
Metabolite identification, especially structure elucidation, must be performed cautiously. The computational approaches discussed above are to assist identification of metabolites. However, they cannot replace strict experimental verification of the identities of metabolites.
The main purpose of computational approaches is to reduce the search space and prioritize putative identifications. Some ions may have up to hundreds of putative identifications obtained from a mass-based search. Since the availability of authentic compounds is limited, it will be preferable to prioritize putative identifications, so that more effort can be concentrated on the few most likely candidates. However, to verify the identity of an unknown metabolite confidently, the authentic standard still needs to be obtained and injected into the same instrument with the biological sample to compare their MS/MS spectra and retention times.
For some metabolites, MS/MS spectra may not be adequate to verify them uniquely. In this case, MSn (e.g., MS3 or MS4) is often used to acquire further fragmentation information from the desired fragment ions. MSn can only be achieved with ion-trap based mass spectrometers which provide fragmentation mass spectra of both PI and some of fragment ions. MSn helps to discriminate between very, and helps to discriminate between very similar metabolites and provides more confidence for metabolite verification.
3. LC-MS/MS-based metabolite quantitation
One of the aims in metabolomics is quantitation of metabolites in order to evaluate changes in response to disease, treatment, environmental and genetic perturbations. In the following, we present metabolite -quantitation methods using QqQ-based SRM and ion trap, HRMS-based full-scan MS analysis.
3.1 Metabolite quantitation by LC-SRM-MS/MS
In QqQ instrument, the PI selected in the first quadruple (MS1) is dissociated to fragment ions in the collision cell and only a specific fragment ion (daughter ion) is selected in the second quadruple (MS2). This two-stage ion-selection method called selected reaction monitoring (SRM) is sensitive to molecular weight and specific to structure. SRM can measure the real concentration of metabolites through absolute quantitation by correlating signal intensities of the analytes with a calibration curve set up using spiked stable isotope-labeled analogues. This approach has been used for quantitation of small molecules for more than three decades [28].
SRM also has been applied to screen for a large number of metabolites that display certain common structural motifs and might represent a selective metabolomics profile (e.g., glucuronidation, sulfation and glutathione-conjugate formation in endogenous and exogenous molecules during biotransformation). For example, SRM has been used to screen glucuronide metabolites as potential surrogate biomarkers in human urine [29]. Such methodology has been also applied in screening study for glucuronosyl conjugates and glutathione conjugates in biological fluids [30]. SRM-based approach has been used for screening newborn disorders in amino acid, fatty acid, and carbohydrate metabolism [31].
After combination with effective sample preparation and chromatographic separation, modern QqQ mass spectrometers with faster spectral scan rate (e.g., SRM dwell time of 2 ms in Thermo Quantum TSQ) and higher ionization efficiency (e.g., heated ESI) can achieve simultaneous quantitation of tens to hundreds of metabolites. Because of its high sensitivity, high specificity, and excellent quantitation capability, QqQ-based LC-SRM-MS/MS has become ideal for targeted metabolomics studies. The greatest challenge in such studies comes from the diversity of the chemical properties of metabolites and 7–9 orders of magnitude differences in their concentrations [32], so it is currently impossible to quantitate all metabolites simultaneously with any platform.
Modern QqQ instruments (e.g. API 5000, TSQ Vantage) with fast SRM dwell time allow large numbers of SRMs in a duty cycle and numerous data points across a chromatographic peak for accurate, precise quantitation of up to hundreds of metabolites without compromising sensitivity. Meanwhile, acquisition of standards from the Human Metabolome Database for confirmation of targeted metabolite identity has helped select SRM transitions for metabolites and develop SRM-based targeted metabolomics.
Although coupling LC to MS facilitates metabolite quantitation, no single LC method is ideal for separating all classes of metabolites [33]. Many efforts have been made to improve LC-separation capacity in order to detect a broader range of metabolites from various biological samples. For example, Bajad et al. compared the performance of nine approaches to chromatography using seven different column chemistries in recent study [34]. They concluded that HILIC using an aminopropyl column is an effective method for separating a wide range of 141 cellular metabolites, including amino acids, nucleosides, nucleotides, coenzyme A and derivatives, carboxylic acids, and sugar phosphates.
Luo et al. demonstrated that using the tributylamine as ion-pairing agent in reversed-phase chromatography is a useful method for separating negatively-charged metabolites, including nucleotides, sugar phosphates, and carboxylic acids [35].
Based on the above LC-method developments, Rabinowitz et al. adopted an approach by running two separate LC-MS systems: one for positive-ionization mode using HILIC chromatography and one for negative-ionization mode using reversed-phase ion-pairing chromatography. This dual-chromatography methodology enables quantitation of approximately 250 water-soluble metabolites of verified identities, including amino acids and derivatives, sugar phosphates, nucleotides, coenzyme A and derivatives, and carboxylic acids [33,36].
Such a dual-chromatography methodology was further examined by Büscher et al. in a cross-platform comparison study, where they found that LC is a better quantitative approach than GC and CE, due to its robustness and better coverage of more metabolites [37].
More recently, Wei et al. reported usage of three HPLC columns in reversed-phase mode under different separation conditions to separate 205 endogenous metabolites of amino acids, sugar and nucleic acids, and organic acids in a short run (10 min) [38]. Low-picogram sensitivity and 3–4 orders of linearity have been achieved for more than half the metabolites. In Table 1, we summarize these and other LC developments for targeted metabolomics quantitation.
Table 1.
Application of LC-SRM-MS/MS and LC-HRMS in quantitative metabolomics
| MS instrument | LC separation mode | Biological medium | Ref. |
|---|---|---|---|
| AB Sciex 4000 Qtrap: ESI (−) SRM | RP: Luna C18(2) (150 mm × 2 mm, 3 μm) | Rat blood and urine | [30] |
| Agilent G3250AA LC/MSD TOF: ESI(−); AB Sciex 4000 QTRAP: ESI (−) SRM | Ion-pairing RP: end-capped C18 column Synergi Hydro RP (2.1 mm × 150 mm, 4 μm); HILIC : aminopropyl column Luna NH2 (2 mm × 250 mm, 5 μm). | Standard mixture | [37] |
| AB Sciex 3200 QTrap: ESI (+)/(−) SRM | RP: Hypersil Gold (100 mm × 2.1 mm, 1.9 μm) | Human plasma | [41] |
| AB Sciex 3200 QTrap: ESI (+) SRM | RP: pentafluorophenyl Restek Allure PFP Propyl (50 mm × 2.1 mm, 5 μm) | Human urine | [42] |
| Sciex API 3000 QqQ: APCI (−) SRM | RP : Zorbax XDB C18 (30 × 4.6 mm i.d., 3.5 μm) | Human plasma | [69] |
| API 4000 QqQ: ESI (+) SRM | RP: ACQUITY BEH phenyl column (2.1 × 100 mm, 1.7 μm ) | Rat prefrontal cortex microdialysate | [56] |
| Agilent SL ion trap: ESI (+) full scan | RP: Atlantis C18 (1.0 mm × 150 mm, 3 μm) | Human cell line | [52] |
| Thermo LTQ-Orbitrap: ESI (+) full scan | RP: Max RP (2 mm × 5 cm, 5 μm) | Rat plasma | [43] |
| Thermo LTQ-FT: ESI (−) full scan | RP: C18 column Waters XBridge (2.1 mm × 50 mm, 2.5 μm) | Mice liver tissue and serum | [70] |
| Thermo LTQ-Orbitrap: ESI (+)/(−) full scan | ZIC-HILIC column (150 × 4.6 mm, 5 μm) | Fetal calf serum | [48] |
| Thermo LTQ-Orbitrap: ESI (+)/(−) full scan | HILIC: ZIC-HILIC column (150 × 4.6 mm, 5 μm) and ACE silica gel column (150 × 3 mm, 3 μm) | Mouse liver tissue | [51] |
| Thermo LTQ-Orbitrap: ESI (+) full scan | HILIC: ZIC-HILIC column (150 × 4.6 mm, 5 μm) | Drosophila melanogaster | [49] |
| Thermo QqQ Quantum Ultra: ESI (+) SRM and Thermo LTQ-Orbitrap: ESI (+) full scan | RP: BioSuite C18 (2.1 × 150 mm, 3 μm) and Discovery HS-F5 (2.1 × 250 mm, 5 μm) | Human cerebrospinal fluid | [59] |
| Thermo LTQ FT-ICR Ultra: ESI (+) full scan | RP : UPLC HSS T3 C18 (100 × 2.1 mm i.d. 1.8 μm) | Plant extracts | [71] |
| Bruker 12 T apex-Qe Qq- FTICR: ESI (+) full Scan | Aqueous Phase: Alltech Solvent Miser Silica (2.1 × 150 mm, 5 μm) | Mouse serum | [72] |
| Bruker 9.4 T Apex-Qe FT- ICR: ESI (+) full scan | RP: ACQUITY BEH C18 column (2.1 mm × 50 mm, 1.7 μm) and RP :Agilent Zorbax XDB C18 column (1.0 mm × 150 mm, 3.5 μm) | Human urine | [58] |
| Bruker 9.4 T Apex-Qe FT- ICR: ESI (+) full scan | Ion pairing RP: Agilent Zorbax Rx-C18 (9.4 mm × 250 mm, 5 μm); RP: Eclipse plus C18 (2.1 mm × 100 mm, 1.8 μm) | Human urine | [55] |
| Bruker 9.4T Apex-Qe FT- ICR :ESI (+)/(−) full scan | RP: ACQUITY BEH C18 (1.0 mm × 150 mm, 1.8 μm) and HILIC : ACQUITY BEH C18 (1.0 mm × 150 mm, 1.8 μm) | Yeast cell | [54] |
| Thermo Exactive Orbitrap: ESI (+) full scan | RP: Thermo Hypersil Gold C18 (2.1 × 100 mm, 1.9 μm) | Rat liver microsome and plasma | [60] |
| Thermo Exactive Orbitrap: ESI (+) | RP: UPLC Hypersil Gold C18 column (5.0 × 2.1 mm, 1.9 μm) | Human plasma | [50] |
| Thermo Exactive Orbitrap: ESI (+) full scan | Ion-pairing RP: synergy Hydro-RP (100 mm × 2 mm, 2.5 μm) | Escherichia coli | [61] |
| AB QSTAR Pulsar: ESI (+) full scan | HILIC: hydrophilic coated TC-WAX capillary (150 μm i.d. Primesep A 5 μm,) | Cerebrospinal fluid | [73] |
| Agilent 6220 TOF/QTOF: ESI/APCI (−) full scan and MS/MS | RP : Waters XBridge C18 (2.1 mm × 150 mm, 5 μm) | Mycobacterium cell | [74] |
| Micromass QTOF Premier: ESI (−) | RP: ACQUITY BEH C8 (2.1 mm × 100 mm I.D., 1.7 μm) | Plant | [75] |
| Micromass QTOF2:ESI (+) full scan | RP: C18 (100 × 2.1 mm, 3 μm) | Human urine | [53] |
| Bruker Micro-TOF: ESI (+) full scan | HILIC: poly(hydroxyethyl) aspartamide capillary column (150 × 0.320 mm, 5 μm) | Arabidopsis seed tissue | [57] |
Despite the development of LC-SRM-MS/MS in targeted metabolomics, several drawbacks limit its application for accurate metabolite quantitation and identification. For example, pre-defined SRM transitions lack the flexibility of using a different product ion for quantitation, which could be affected by cross-talking among analytes with similar structures and masses (same RT and same fragment ions) or subject to interference by endogenous isobaric interference from matrix. To address this drawback, other dominant product ions, which are unique to the specific analyte, have to be chosen in the SRM. Current software tools for MS data acquisition have the function of “scrambling” the transitions with identical product ions to avoid monitoring these transitions one after another. The tools can also increase inter-channel delay between each SRM transition to allow the collision cell to be emptied prior to loading ions for the next SRM transition. Another solution is to perform adequate chromatographic separation between analytes. Meanwhile, in SRM acquisition, most ions are filtered out along with the loss of qualitative information that is needed for recognition and structural elucidation of metabolites. This limits the application of QqQ for target identification based on full-scan MS/MS spectrum. The triple quadruple-LIT hybrid instrument (QLIT also called ABI QTRAP) addresses this issue by using the Q3 analyzer as both quadruple and LIT. The latter has a fast duty cycle as LIT, allowing the full scan on product ions. The QTRAP mass spectrometer provides the same capabilities of neutral loss (NL) scanning, PI scanning, and SRM acquisition that QqQ has for unknown metabolite screening and known metabolite quantitation. QTRAP can also provide a survey scan to trigger information-dependent acquisition (IDA) of enhanced product ion (EPI) spectra. Among them, MRM-EPI [here multiple-reaction monitoring (MRM) is used by Applied Biosystems as the synonym of SRM) provides better selectivity and more sensitivity than NL-EPI and PI-EPI [39, 40]. MRM-EPI can be set to follow up to 100 SRM transitions and also conserve the quantitative performance of SRM methods without significant loss of sensitivity. MS/MS spectra generated from MRM-EPI are useful for recognizing false-positive peaks displayed in the SRM ion chromatograms. QTRAP can therefore work as a good alternative to QqQ-based SRM methodology that allows simultaneous quantitation of expected metabolites and verification of their identities by MS/MS. Such a technique has been used for multi-target screening analysis of hundreds of metabolites in various biological samples [29,30,41,42]. Some of these applications are presented in Table 1.
3.2. Metabolite quantitation by LC-IT-MS and LC-HRMS
The QqQ-based SRM quantitative method is limited to targeted metabolites and often neglects the information of other metabolites that are invisible due to the specificity of the analysis. Some analytes have non-specific transitions that are common for matrix interferences (e.g., the neutral loss of H2O or CO2). This compromises the specificity of SRM acquisition and causes inaccurate quantitation.
Recent developments in LC provide well-resolved peaks with narrow peak width. These developments also challenge the compatibility of the acquisition rate of mass spectrometers (dwell time for SRM transitions) with chromatographic elution of such short duration, since accurate quantitation by LC-MS requires enough data points (>20) across the peak. Insufficient data points will result in poor temporal peak resolution and compromise the sensitivity of SRM.
Mass spectrometers with higher acquisition speed (e.g., LIT and 3D ion trap) and high mass resolution can help as alternative tools for simultaneous quantitation of target analytes and identification of off-target analytes. 3D-ion-trap MS with unit-mass resolution can provide reasonable quantitative results by extracting selected ions from full-scan data. However, the sensitivity of such a quantitative approach is not comparable with true SRM and selectivity is also limited [43].
Modern LIT-MS (e.g., LTQ) shows improved sensitivity and was recently reported for screening more than 320 different pesticides and metabolites in blood [44].
A recent comparison study between LC-LIT-MS and LC-QqQ-MS showed that QqQ-based LC-SRM-MS/MS is still a better choice for small-molecule analysis with respect to limit of detection, lower limit of quantitation and precision [45].
HRMS [e.g., FT-MS (e.g., FT-ICR and Orbitrap) and TOF or QTOF can provide global MS detection and offer better solutions to the limitations in SRM analysis. HRMS, especially FT-MS in full-scan mode, can determine virtually all compounds present in a sample because of its high resolving power, high mass accuracy and broad dynamic range.
Modern HRMS with its fast scan rate allows acquisition of enough data points across a chromatographic peak and uses EICs for accurate quantitation by centering a narrow mass window (e.g. ±5–10 mmu) on the theoretical m/z value of the analyte. This approach to quantitation avoids pre-selection of SRM transitions for target compounds and offers identification of off-target compounds at the same time.
The hybrid configuration in HRMS (e.g., LIT-FT-ICR, LTQ-Orbitrap or QTOF) provides information-dependent MS/MS acquisition on full-scan product-ion spectra to help in the verification of compound identity.
FT-MS (e.g., FT-ICR and Orbitrap-based mass spectrometers) offers high mass resolution and mass accuracy (e.g., above 1,000,000 FWHM at m/z 400 and sub-ppm for FT-ICR, 100,000 FWHM at m/z 400 and 1–2 ppm for Orbitrap) [46]. High resolving power and high mass accuracy of FT-MS facilitate metabolite identification, as accurate mass measurement can help determine the elemental formula and high mass resolution can generate precise isotopic pattern. These merits of FT-MS can be useful to eliminate some putative identification with similar mono-isotopic mass but different isotopic distributions. They also allow the quantitation o0f metabolites using EICs by centering the narrow mass window on the theoretical m/z value of target analyte and excluding overlapping isobaric signals while the mass accuracy is maintained throughout the acquisition [47–51].
In HRMS-based untargeted metabolomics quantitative studies, efforts have been made in sample preparation and LC separation in order to increase detection sensitivity, coverage of metabolites and quantitation accuracy. For example, the differential isotope-labeling technique has been used to derivatize large numbers of unknown metabolites in complex samples followed by absolute or relative quantitation using LC-FT-MS [52–57]. For example, Guo et al. recently reported several studies of using isotope labeling to derivatize several hundreds of metabolites in human biological fluids containing amines and phenols and quantitating them simultaneously by UPLC-FT-ICR [55,58].
LC developments, similar to those mentioned previously for SRM-based studies, have been made in LC-FT-MS-based quantitative metabolomics (e.g., utilizing multiple column chemistries in several LC platforms to achieve broad range of metabolites detection), and details are in Table 1.
FT-ICR presents unsurpassedly high mass accuracy and high resolving power. However, it is not widely used for metabolite identification and quantitation because its cost is high, it is hard to maintain and it is difficult to couple with LC, compared with Orbitrap and TOF mass spectrometers (e.g., 15,000 FWHM at m/z 400 and 5–10 ppm for TOF) [47].
Most recently, Orbitrap-based HRMS has become the platform of choice to perform integrated qualitative and quantitative analysis in the full-scan mode [48,49,51,59]. Originally, Orbitrap suffered from slow acquisition speed in MS/MS scan, limited sensitivity and dynamic range. Modern Orbitraps, especially the new bench-top Exactive Orbitrap, have shown more competitive advantages in term of cost, sensitivity, mass accuracy and linear dynamic range [50,60,61].
Although there are only a few papers reporting TOF/QTOF-based HRMS for quantitative analysis, modern TOF technology could offer a new alternative. The limited dynamic range in typical TOF instruments relying on TDC detectors has been improved by analog-to-digital converter-detector technology. Significant improvements have been made in mass resolution and mass accuracy of TOF. Currently, 40,000 in resolution (FWHM, m/z 922) and accuracy of <1 ppm in TOF are possible [62]. Meanwhile, modern state-of-the-art QTOF instruments (e.g., Agilent 6540 Ultra High Definition Accurate-Mass QTOF) not only allow accurate-mass measurements for compound confirmation and molecular formula generation, but also provide accurate isotope ratios. Such capabilities help users narrow down the list of plausible molecular formulas and increase confidence in the result, rendering these instruments increasingly competitive with Orbitrap on this dimension.
For each type of major mass analyzers, its overall performance in quantitative analysis of small molecules was recently reviewed by Krauss et al. [63], to which readers are encouraged to refer for detailed evaluations on dynamic range, sensitivity, resolving power and mass accuracy. Although the specifications given in that paper generally apply to most instruments, some new mass spectrometers may achieve better performance in dynamic range and sensitivity. For example, it is reported that AB Sciex TripleTOF 5600 possesses equivalent dynamic range and limit of quantitation to high-performance QqQ instruments. The sensitivity of each mass analyzer also strongly depends on the ionization efficiency of the compounds in the ion source. Furthermore, mass spectrometers may provide higher resolution and mass accuracy depending on the m/z range and scan speed of the specific experiment. Based on evaluations {[63] and other comparison studies [33,43,64]}, it appears that QqQ-based SRM gives the best performance in dynamic range and sensitivity for targeted quantitative analysis. However, some HRMS instruments with high mass accuracy offer comparable quantitative performance. Also, HRMS provides promising performance in untargeted quantitative studies.
4. Challenges and future perspectives
There are certain issues that need to be addressed to realize fully the potential of LC-MS in metabolomics studies, as follows.
Identification of metabolites is the current bottle-neck in metabolomics studies. The most reliable way to identify a metabolite unambiguously and confidently is to compare its mass, retention time and fragmentation spectrum with those of authentic standards. As outlined in [65], at least two independent, orthogonal measurements analyzed under identical experiment conditions are needed to confirm the identity of a metabolite. However, various computational tools can be utilized to reduce the search space and prioritize the putative identifications, thereby improving efficiency and reducing cost.
Although novel metabolites continue to be discovered, many metabolites that we face in practice have already been found and identified in other studies. Collection and utilization of information on these “known unknowns” pose major challenges for computational and informatics tools. Partly, this is because the information is often scattered in different sources where spectra were acquired under different conditions.
Mass information alone can give us limited knowledge about elemental composition and possible structure of the metabolite, which seldom results in unambiguous identification of the metabolite. Adducts, isotopes and fragments of the same metabolite can be preliminarily identified on the basis of retention times and correlation of their EICs. With the help of isotopic pattern of the MS spectrum, we can make more confident deduction about its elemental composition, but still with limited knowledge about its structures, particularly when it has isomers.
By acquiring MS/MS spectra, the structures of the metabolites of interest can be further confirmed, as in-silico fragmentation helps deduction of the possible structures of metabolites during spectra interpretation. If MS/MS spectra of different candidates (putative identifications) are present in databases or spectral libraries, MS/MS spectra acquired from experimental samples are compared with library spectra to confirm the identities of the metabolites. Although this approach is not as rigorous as that achieved with authentic compounds, it provides important guidance for metabolite verification by reducing the number of putative identities or prioritizing them for subsequent verification.
Challenges exist in almost all steps of metabolite identification. The quality of acquired MS/MS spectra is of great importance for successful identification. However, it is often affected by experimental factors (e.g., instrument type and collision energy). A good-quality MS/MS spectrum is often acquired through iterative adjustment of experimental parameters.
Automated acquisition of high-quality MS/MS spectra is needed to increase the throughput of metabolite identification. Spectral libraries for GC-MS demonstrate great success in small-molecule identification. LC-MS/MS spectral libraries for metabolomics studies (e.g., HMDB, Metlin and MassBank) continue to evolve and to expand to increase metabolome coverage.
The heterogeneity of spectral data poses a major challenge to effective usage of spectral libraries. In recent studies, promising results showed a certain degree of reproducibility of MS/MS spectra using different instruments from different laboratories [66]. Through carefully designed experiments and appropriate spectral matching algorithms, performance in compound identification can be greatly improved.
Better in-silico fragmentation models are needed to recognize complex ion-molecular interactions encountered in metabolites fragmentation. With improved specificity, such models will assist identification of unknown metabolites with no spectral library coverage. Also, because metabolites do not exist alone but within certain biological context (e.g., metabolic networks and pathways), integration of contextual information into identification can potentially reduce the ambiguity in metabolite identification [67].
As to the experimental considerations, several issues, which need to be addressed in sample preparation, chromatographic separation, and MS-data acquisition, directly affect the final outcome of LC-MS-based metabolite quantitation. First, matrix components interfere with the ionization of analytes by co-eluting in ESI and APCI, resulting in ion suppression or ion enhancement. These compromise accuracy and precision in metabolite quantitation. Solutions to alleviate those effects include performing adequate sample preparation prior to LC separation. For example, adopting a more selective solid-phase extraction clean-up step, changing the ionization mode (e.g., switching from ESI to APCI, APPI, reducing LC flow rate in conventional ESI or using nano-ESI). The most effective way to circumvent matrix effects is to use stable isotope-labeled analogues as internal standards to establish a more reliable calibration curve, in which both analyte and its stable isotope-labeled analogue show the same retention behavior and go through the same matrix effects during ionization, so the relative ratio of their MS signals within the dynamic range also remains the same. Second, the matrix effect can also cause retention-time shift due to build up of contamination in the column and peak broadening by unresolved interference. As a result, more efficient approaches to separation are needed to reduce sample complexity by separating metabolites from isobaric interferences and to increase peak capacity by separating more metabolites with high peak resolutions.
Usage of multidimensional LC allows the combination of two or more independent separation steps to increase the peak capacity and to improve the separation of metabolites in complex samples. Besides the practice of combining RPLC and HILIC for quantitation of metabolites, as mentioned previously, there are other LC combinations that will contribute to better separation and wider coverage of metabolites (e.g., ion exchange-RPLC, size-exclusion-RPLC and strong cation exchange-HILIC).
Recent developments in monolithic capillary columns, high temperature LC, and UPLC (i.e. pressure >400 bar in columns packed with <2-μm-diameter particles) provide at least comparable quantitative precision and accuracy to conventional LC. Among them, UPLC showed the best gain by offering better chromatographic resolution (e.g., peak width of 1–3 s) and shorter analysis time (run <10 min) [10]. Despite the challenges that the narrow peak width of UPLC imposes on the acquisition rate of the mass spectrometer, as the development of modern mass spectrometers proceeds, combining UPLC with MS can be advantageous for better assignment of metabolites through improved peak resolution and detecting more metabolites by increasing peak capacity.
Meanwhile, sensitivity becomes an issue when metabolites of interest are at very low abundance or have poor ionization. Heated drying gas in the ESI source has become common in modern mass spectrometers (e.g., AB Sciex and Waters instruments). A recent study reported that the heated ESI source enhanced sensitivity compared with the unheated ESI source on the TSQ Quantum Ultra [33].
Application of nanoLC/nano-ESI-MS in metabolomics could also enhance sensitivity and dynamic range. Recently, chemical derivatization on metabolites was reported to help LC separation of metabolites, increase their MS signals and improve quantitation accuracy in untargeted metabolomics studies{e.g., using S-methyl methanethiosulfonate to derivatize thiol compounds [33] and heavy and light isotopic forms of cholamine to label carboxylic acid-containing metabolites [68]}.
Other isotope-labeling techniques have also been developed to enhance the effectiveness of and confidence in metabolite identification and accuracy in quantitation [57]. As new mass analyzers evolve with faster acquisition rates, higher resolving powers, better mass accuracy and broader dynamic range, we believe accurate simultaneous quantitation of a large number of metabolites can be achieved by coupling the mass analyzers with advanced LC separation methods.
At present, QqQ-based SRM is the leading choice for targeted metabolite quantitation. For less targeted analyses involving larger numbers of analytes, use of high mass-resolution and high mass-accuracy full-scan HRMS hold promise for future quantitative metabolomics studies.
Highlights.
A review of LC-MS/MS based quantitative and qualitative metabolomics studies.
A framework summarizing computational methods for improved metabolite identification.
LC-SRM-MS/MS and LC-HRMS-based metabolite quantitation.
Challenges and future perspectives in metabolite identification and quantitation.
Acknowledgments
This work was supported by NCI Grant R21 CA153176. We would like to thank the anonymous reviewers for their constructive comments that improved the manuscript considerably.
Abbreviations
- API
Atmospheric pressure ionization
- APCI
Atmospheric pressure chemical ionization
- APPI
Atmospheric pressure photoionization
- CID
Collision-induced dissociation
- DDA
Data-dependent acquisition
- EI
Electron ionization
- EIC
Extracted ion chromatogram
- EPI
Enhanced product ion
- ESI
Electrospray ionization
- FiD
Fragment Identificator
- FT-ICR
Fourier-transform ion cyclotron resonance
- FT-IR
Fourier-transform infrared
- FT-MS
Fourier-transform mass spectrometry
- FWHM
Full width at half maximum
- GC-MS
Gas chromatography–mass spectrometry
- HCD
High-energy collisional dissociation
- HILIC
Hydrophilic interaction liquid chromatography
- HMDB
Human Metabolome Database
- HPLC
High-performance liquid chromatography
- HRMS
High-resolution mass spectrometry
- IMS
Ion-mobility separation
- IDA
Information-dependent acquisition
- IS
Internal standard
- LC
Liquid chromatography
- LIT
Linear ion trap
- LTQ
Linear trap quadrupole
- MMCD
Madison Metabolomics Consortium Database
- MS
Mass spectrometry
- MS/MS
Tandem mass spectrometry
- NL
Neutral loss
- NPLC
Normal-phase liquid chromatography
- NMR
Nuclear resonance spectroscopy
- PI
Precursor ion
- PK/PD
Pharmacokinetic/Pharmacodynamic
- QLIT
Quadrupole linear ion trap
- QqQ
Triple quadrupole
- QTOF
Quadrupole time-of-flight
- RPLC
Reversed-phase liquid chromatography
- S/N
Signal-to-noise ratio
- SRM
Selected reaction monitoring
- TDC
Time-to-digital converter
- TOF
Time of flight
- UPLC
Ultra performance liquid chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bowen BP, Northen TR. J Am Soc Mass Spectrom. 2010;21:1471. doi: 10.1016/j.jasms.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Rosenblum ES, Viant MR, Braid BM, Moore JD, Friedman CS, Tjeerdema RS. Metabolomics. 2005;1:199. [Google Scholar]
- 3.Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner HP. Angew Chem, Int Ed Engl. 2010;49:5426. doi: 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- 4.Saito K, Matsuda F. Annu Rev Plant Biol. 2010;61:463. doi: 10.1146/annurev.arplant.043008.092035. [DOI] [PubMed] [Google Scholar]
- 5.Oliver SG, Winson MK, Kell DB, Baganz F. Trends Biotechnol. 1998;16:373. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 6.Ding S, Schoenmakers I, Jones K, Koulman A, Prentice A, Volmer D. Anal Bioanal Chem. 2010;398:779. doi: 10.1007/s00216-010-3993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djukovic D, Baniasadi HR, Kc R, Hammoud Z, Raftery D. Rapid Commun Mass Spectrom. 2010;24:3057. doi: 10.1002/rcm.4739. [DOI] [PubMed] [Google Scholar]
- 8.Wei C, Zhu P, Shah SJ, Blair IA. Mol Pharmacol. 2009;76:516. doi: 10.1124/mol.109.057489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Jiang X. Eur J Lipid Sci Technol. 2009;111:39. doi: 10.1002/ejlt.200800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillarme D, Nguyen DTT, Rudaz S, Veuthey JL, Chromatogr J. A. 2007;1149:20. doi: 10.1016/j.chroma.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Nat Biotechnol. 2008;26:162. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 12.Smith CA, Maille GO, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. Ther Drug Monit. 2005;27:747. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 13.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Nucleic Acids Res. 2009;37:D603. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kind T, Fiehn O. BMC Bioinform. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K. Plant J. 2009;57:555. doi: 10.1111/j.1365-313X.2008.03705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. Rapid Commun Mass Spectrom. 2006;20:1989. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- 17.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Anal Chem. 2011;83:940. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geromanos SJ, Vissers JPC, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, Langridge JI. Proteomics. 2009;9:1683. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 19.Tautenhahn R, Böttcher C, Neumann S. In: Bioinformatics Research and Development. Hochreiter S, Wagner R, editors. Springer; Berlin, Heidelberg, Germany: 2007. p. 371. [Google Scholar]
- 20.Ipsen A, Want EJ, Lindon JC, Ebbels TMD. Anal Chem. 2010;82:1766. doi: 10.1021/ac902361f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinonen M, Rantanen A, Mielikäinen T, Kokkonen J, Kiuru J, Ketola RA, Rousu J. Rapid Commun Mass Spectrom. 2008;22:3043. doi: 10.1002/rcm.3701. [DOI] [PubMed] [Google Scholar]
- 22.Wolf S, Schmidt S, Muller-Hannemann M, Neumann S. BMC Bioinform. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schymanski EL, Meringer M, Brack W. Anal Chem. 2009;81:3608. doi: 10.1021/ac802715e. [DOI] [PubMed] [Google Scholar]
- 24.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. J Mass Spectrom. 2010;45:703. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 25.Stein SE. J Am Soc Mass Spectrom. 1994;5:316. doi: 10.1016/1044-0305(94)85022-4. [DOI] [PubMed] [Google Scholar]
- 26.Dworzanski JP, Snyder AP, Chen R, Zhang H, Wishart D, Li L. Anal Chem. 2004;76:2355. doi: 10.1021/ac0349781. [DOI] [PubMed] [Google Scholar]
- 27.Zhou B, Cheema AK, Ressom HW. Engineering in Medicine and Biology Society (EMBC), 2010. Annu Int Conf IEEE. 2010:756. doi: 10.1109/IEMBS.2010.5626337. [DOI] [PubMed] [Google Scholar]
- 28.Baty JD, Robinson PR. Biol Mass Spectrom. 1977;4:36. doi: 10.1002/bms.1200040104. [DOI] [PubMed] [Google Scholar]
- 29.Lutz U, Bittner N, Lutz RW, Lutz WK. J Chromatogr, B. 2008;871:349. doi: 10.1016/j.jchromb.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 30.van de Wetering K, Feddema W, Helms JB, Brouwers JF, Borst P. Gastroenterology. 2009;137:1725. doi: 10.1053/j.gastro.2009.06.052. [DOI] [PubMed] [Google Scholar]
- 31.Wilcken B, Wiley V. Pathology. 2008;40:104. doi: 10.1080/00313020701813743. [DOI] [PubMed] [Google Scholar]
- 32.Dunn WB, Ellis D. Trends Anal Chem. 2005;24:285. [Google Scholar]
- 33.Lu W, Bennett BD, Rabinowitz JD. J Chromatogr, B. 2008;871:236. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. J Chromatogr, A. 2006;1125:76. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 35.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. J Chromatogr, A. 2007;1147:153. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 36.Lu W, Kwon YK, Rabinowitz JD. J Am Soc Mass Spectrom. 2007;18:898. doi: 10.1016/j.jasms.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Büscher JRM, Czernik D, Ewald JC, Sauer U, Zamboni N. Anal Chem. 2009;81:2135. doi: 10.1021/ac8022857. [DOI] [PubMed] [Google Scholar]
- 38.Wei R, Li G, Seymour AB. Anal Chem. 2010;82:5527. doi: 10.1021/ac100331b. [DOI] [PubMed] [Google Scholar]
- 39.Wen B, Ma L, Nelson SD, Zhu M. Anal Chem. 2008;80:1788. doi: 10.1021/ac702232r. [DOI] [PubMed] [Google Scholar]
- 40.Zheng J, Ma L, Xin B, Olah T, Humphreys WG, Zhu M. Chem Res Toxicol. 2007;20:757. doi: 10.1021/tx600277y. [DOI] [PubMed] [Google Scholar]
- 41.Viette V, Guillarme D, Mylonas R, Mauron Y, Fathi M, Rudaz S, Hochstrasser D, Veuthey JL. Clin Biochem. 2011;44:32. doi: 10.1016/j.clinbiochem.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 42.Dresen S, Ferreirós N, Gnann H, Zimmermann R, Weinmann W. Anal Bioanal Chem. 2010;396:2425. doi: 10.1007/s00216-010-3485-2. [DOI] [PubMed] [Google Scholar]
- 43.Zhang NR, Yu S, Tiller P, Yeh S, Mahan E, Emary WB. Rapid Commun Mass Spectrom. 2009;23:1085. doi: 10.1002/rcm.3975. [DOI] [PubMed] [Google Scholar]
- 44.Dulaurent S, Moesch C, Marquet P, Gaulier JM, Lachâtre G. Anal Bioanal Chem. 2010;396:2235. doi: 10.1007/s00216-009-3443-z. [DOI] [PubMed] [Google Scholar]
- 45.Dai SY, Herrman TJ. Rapid Commun Mass Spectrom. 2010;24:1431. doi: 10.1002/rcm.4533. [DOI] [PubMed] [Google Scholar]
- 46.Cortés-Francisco N, Flores C, Moyano E, Caixach J. Anal Bioanal Chem. 2011:1. doi: 10.1007/s00216-011-5046-8. [DOI] [PubMed] [Google Scholar]
- 47.Junot C, Madalinski G, Tabet JC, Ezan E. Analyst (Cambridge, UK) 2010;135:2203. doi: 10.1039/c0an00021c. [DOI] [PubMed] [Google Scholar]
- 48.Kamleh A, Barrett MP, Wildridge D, Burchmore RJS, Scheltema RA, Watson DG. Rapid Commun Mass Spectrom. 2008;22:1912. doi: 10.1002/rcm.3564. [DOI] [PubMed] [Google Scholar]
- 49.Kamleh MA, Hobani Y, Dow JAT, Watson DG. FEBS Lett. 2008;582:2916. doi: 10.1016/j.febslet.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 50.Koulman A, Woffendin G, Narayana VK, Welchman H, Crone C, Volmer DA. Rapid Commun Mass Spectrom. 2009;23:1411. doi: 10.1002/rcm.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacIntyre L, Zheng L, Scullion P, Keating P, Watson DG. Metabolomics. 2011:1. [Google Scholar]
- 52.Abello N, Geurink PP, Toorn M, Oosterhout AJM, Lugtenburg J, Marel GA, Kerstjens HAM, Postma DS, Overkleeft HS, Bischoff R. Anal Chem. 2008;80:9171. doi: 10.1021/ac801215c. [DOI] [PubMed] [Google Scholar]
- 53.Eggink M, Wijtmans M, Ekkebus R, Lingeman H, Esch IJP, Kool J, Niessen WMA, Irth H. Anal Chem. 2008;80:9042. doi: 10.1021/ac801429w. [DOI] [PubMed] [Google Scholar]
- 54.Godat E, Madalinski G, Muller L, Heilier JF, Labarre J, Junot C. Methods Enzymol. 2010;473:41. doi: 10.1016/S0076-6879(10)73002-0. [DOI] [PubMed] [Google Scholar]
- 55.Guo K, Peng J, Zhou R, Li L. J Chromatogr, A. 2011;1218:3689. doi: 10.1016/j.chroma.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 56.Ji C, Li W, Ren X, El-Kattan AF, Kozak R, Fountain S, Lepsy C. Anal Chem. 2008;80:9195. doi: 10.1021/ac801339z. [DOI] [PubMed] [Google Scholar]
- 57.Shortreed MR, Lamos SM, Frey BL, Phillips MF, Patel M, Belshaw PJ, Smith LM. Anal Chem. 2006;78:6398. doi: 10.1021/ac0607008. [DOI] [PubMed] [Google Scholar]
- 58.Guo K, Li L. Anal Chem. 2009;81:3919. doi: 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- 59.Wishart DS, Lewis MJ, Morrissey JA, Flegel MD, Jeroncic K, Xiong Y, Cheng D, Eisner R, Gautam B, Tzur D, Sawhney S, Bamforth F, Greiner R, Li L. J Chromatogr, B. 2008;871:164. doi: 10.1016/j.jchromb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 60.Bateman KP, Kellmann M, Muenster H, Papp R, Taylor L. J Am Soc Mass Spectrom. 2009;20:1441. doi: 10.1016/j.jasms.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 61.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Anal Chem. 2010;82:3212. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stroh JG, Petucci CJ, Brecker SJ, Huang N, Lau JM. J Am Soc Mass Spectrom. 2007;18:1612. doi: 10.1016/j.jasms.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 63.Krauss M, Singer H, Hollender J. Anal Bioanal Chem. 2010;397:943. doi: 10.1007/s00216-010-3608-9. [DOI] [PubMed] [Google Scholar]
- 64.Kaufmann A, Butcher P, Maden K, Walker S, Widmer M. Rapid Commun Mass Spectrom. 2011;25:979. doi: 10.1002/rcm.4952. [DOI] [PubMed] [Google Scholar]
- 65.Sumner L, Amberg A, Barrett D, Beale M, Beger R, Daykin C, Fan T, Fiehn O, Goodacre R, Griffin J, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane A, Lindon J, Marriott P, Nicholls A, Reily M, Thaden J, Viant M. Metabolomics. 2007;3:211. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oberacher H, Pavlic M, Libiseller K, Schubert B, Sulyok M, Schuhmacher R, Csaszar E, Köfeler HC. J Mass Spectrom. 2009;44:485. doi: 10.1002/jms.1545. [DOI] [PubMed] [Google Scholar]
- 67.Rogers S, Scheltema RA, Girolami M, Breitling R. Bioinformatics. 2009;25:512. doi: 10.1093/bioinformatics/btn642. [DOI] [PubMed] [Google Scholar]
- 68.Lamos SM, Shortreed MR, Frey BL, Belshaw PJ, Smith LM. Anal Chem. 2007;79:5143. doi: 10.1021/ac062416m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Majumdar TK, Vedananda S, Tse FLS. Biomed Chromatogr. 2004;18:77. doi: 10.1002/bmc.295. [DOI] [PubMed] [Google Scholar]
- 70.Bobeldijk I, Hekman M, de Vries-van der Weij J, Coulier L, Ramaker R, Kleemann R, Kooistra T, Rubingh C, Freidig A, Verheij E. J Chromatogr, B. 2008;871:306. doi: 10.1016/j.jchromb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Giavalisco P, Köhl K, Hummel J, Seiwert B, Willmitzer L. Anal Chem. 2009;81:6546. doi: 10.1021/ac900979e. [DOI] [PubMed] [Google Scholar]
- 72.Han J, Danell RM, Patel JR, Gumerov DR, Scarlett CO, Speir JP, Parker CE, Rusyn I, Zeisel S, Borchers CH. Metabolomics. 2008;4:128. doi: 10.1007/s11306-008-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Myint KT, Aoshima K, Tanaka S, Nakamura T, Oda Y. Anal Chem. 2009;81:1121. doi: 10.1021/ac802259r. [DOI] [PubMed] [Google Scholar]
- 74.Bhamidi S, Scherman MS, Jones V, Crick DC, Belisle JT, Brennan PJ, McNeil MR. J Biol Chem. 2011 doi: 10.1074/jbc.M110.210534. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou Y, Lee S, Choi FFK, Xu G, Liu X, Song JZ, Li SL, Qiao CF, Xu HX. Anal Chim Acta. 2010;678:96. doi: 10.1016/j.aca.2010.08.010. [DOI] [PubMed] [Google Scholar]