Abstract

Protein expression in the nervous system undergoes regulated changes in response to changes in behavioral states, in particular long-term memory formation. Recently, methods have been developed (BONCAT and FUNCAT), which introduce noncanonical amino acids bearing small bio-orthogonal functional groups into proteins using the cells’ own translational machinery. Using the selective “click reaction”, this allows for the identification and visualization of newly synthesized proteins in vitro. Here we demonstrate that noncanonical amino acid labeling can be achieved in vivo in an intact organism capable of simple learning behavior, the larval zebrafish. We show that azidohomoalanine is metabolically incorporated into newly synthesized proteins, in a time- and concentration-dependent manner, but has no apparent toxic effect and does not influence simple behaviors such as spontaneous swimming and escape responses. This enables fluorescent labeling of newly synthesized proteins in whole mount larval zebrafish. Furthermore, stimulation with a GABA antagonist that elicits seizures in the larval zebrafish causes an increase in protein synthesis throughout the proteome, which can also be visualized in intact larvae.
Keywords: Protein synthesis, larval zebrafish, noncanonical amino acid tagging, click chemistry, pentylenetetrazol
Both chemical stimuli and changes in behavioral states alter protein expression in the nervous system. In particular, studies in many different model organisms have shown that protein synthesis, during or shortly after learning, is an essential step in the formation of long-term memory.1 In 1964, Agranoff and co-workers showed that the protein synthesis inhibitor (PSI) puromycin injected intracranially into the goldfish produces impairment of memory for a shock avoidance task and that this impairment is time- and PSI concentration-dependent.2,3 Since then, protein synthesis has been shown to be necessary for long-term memory formation in a variety of learning paradigms, including appetitively and shock-motivated discrimination learning, passive and active avoidance learning, shuttle box learning, and long-term habituation [reviewed in ref (1)].
While it is now clear that long-term memory requires new protein synthesis, the identification of newly synthesized proteins has been sparse and limited to individually identified candidate proteins. Advances in mass spectrometry based approaches now permit the characterization and quantification of proteins, especially when paired with approaches such as stable isotope labeling with amino acids in cell culture (SILAC),4 which allow for comparative quantification between proteomes of differentially stimulated cell populations. However, the proteome of the nervous system is complex and without a chemical handle to enable affinity purification of the newly synthesized proteins specifically, proteins of low abundance will likely be missed.
In addition, the identification of cells or neural circuits that show increased protein synthesis in response to memory formation would allow us to understand the components of memory circuits that undergo long-term modifications after learning. Genetically encoded fluorescent tags, such as GFP, have revolutionized cell biology by permitting visualization of fusion proteins of interest in vivo.5 However, the size of GFP and the requirement for genetic manipulation of the target protein may interfere with its endogenous function, while at the same time only permitting investigation of a small number of candidates at once.
Recently, new techniques for labeling a variety of molecules based on the principle of bio-orthogonal metabolic labeling have been developed.6 Here, small functional groups that are commonly absent in the cellular environment, most prominently ketones and azides or alkynes, are introduced using the cells’ own synthetic machinery. Using this approach, sugars,7 lipids,8 virus particles,9 DNA, and RNA10 have been labeled and subsequently visualized using fluorescent dyes or enriched and identified using affinity reagents. Bertozzi and co-workers, in particular, have demonstrated in vivo labeling of glycans in living organisms ranging from rodents11,12 to larval zebrafish (13−15) and C. elegans.16
Using a similar approach, bio-orthogonal noncanonical amino acid tagging (BONCAT)17,18 and fluorescent noncanonical amino acid tagging (FUNCAT)19 have been used to tag and identify or visualize newly synthesized proteins. BONCAT and FUNCAT utilize noncanonical methionine derivatives, such as the azide-bearing azidohomoalanine (AHA), to bio-orthogonally label newly synthesized proteins. AHA can cross cell membranes and be charged onto methionine tRNAs by the endogenous methionyl-tRNA synthetase (MetRS). During protein synthesis, AHA is introduced in place of methionine, resulting in the introduction of azide groups into the newly synthesized proteins. These azide groups can be used to tag proteins with either an alkyne affinity tag (BONCAT) or an alkyne fluorescent tag (FUNCAT) via selective Cu(I)-catalyzed or strain-promoted [3 + 2] azide–alkyne cycloaddition.20−22 Affinity tagged proteins can be quantified using immunoblot analysis or separated from the preexisting proteome by affinity purification and identified by tandem mass spectrometry. Fluorescent tags can be used to visualize newly synthesized proteins, including those proteins of interest whose identities may not be known. Alternatively, the alkyne moiety may also be introduced into newly synthesized proteins by replacing methionine with the noncanonical amino acid homopropargylglycine (HPG) and subsequently labeled using azide bearing affinity or fluorescent tags. Azides and alkynes are small, so light labeling with AHA or HPG is likely to only cause modest, perhaps even insignificant, perturbations of protein folding, localization,17 and therefore function of the labeled protein in vivo. Furthermore, azides and alkynes are stable under biological conditions and essentially absent from vertebrate cells, which makes the azide–alkyne ligation (“click chemistry”) very selective.
BONCAT and FUNCAT techniques have already successfully been applied to study the proteome of HEK293 cells during a 2 h time window,17 as well as investigate local protein synthesis in dissociated hippocampal cultures.19 Furthermore, metabolic AHA incorporation has been used to identify regions of the drosophila genome that show high levels of histone turnover,23 show that Chlamydia co-opt the functions of the lysosomes of their host cells to acquire essential amino acids,24 as well as demonstrate that treatment of primary sensory neurons with the cytokine interleukin-6 or the neurotrophin nerve growth factor (NGF) increases nascent protein synthesis in axons.25 Recently, these techniques have also been used to indicate that the transmembrane receptor DCC may regulate protein synthesis in a localized manner within the cells as DCC was found to overlap with areas of new protein synthesis at the tips of filopodia in commissural neurons.26 However, these studies have only used the techniques in vitro. Given the role of protein synthesis in learning and memory, developing BONCAT and FUNCAT for use in an intact organism in which simple forms of learning may be investigated is the essential next step.
In this study, we describe the application of these techniques in vivo, in the 7-day-old larval zebrafish. The larval zebrafish is an excellent model organism as it is a genetically tractable, simple vertebrate, which is transparent and therefore ideal for imaging. Furthermore, zebrafish larvae have a well-defined behavioral repertoire,27 and the range of experimental paradigms to test this has recently been expanded to include associative conditioning.28 Larval zebrafish can absorb small chemical compounds directly from their surrounding medium, all of which make them not only amendable to chemical screens and an emerging human disease model but also an excellent system to study the applicability of bio-orthogonal metabolic labeling of newly synthesized proteins in vivo.
Here we show that AHA is metabolically incorporated into newly synthesized proteins, in a time- and concentration-dependent manner, but has no apparent toxic effects and does not influence simple behaviors. This enables fluorescent labeling of newly synthesized proteins in whole mount larval zebrafish. Furthermore, we find that stimulation with the GABA antagonist, pentylenetetrazole (PTZ), causes an increase in protein synthesis throughout the proteome, which can also be visualized in intact larvae.
Results and Discussion
The BONCAT and FUNCAT protocols were adapted to larval zebrafish (Figure 1a). All larvae, unless otherwise noted, were analyzed at 7dpf. We incubated larvae in E3 embryo medium supplemented with the methionine surrogate AHA (Figure 1b) for a period of 0–72 h immediately prior to harvesting, with the aim of incorporating the azide group into newly synthesized proteins throughout the zebrafish proteome. To quantify successful incorporation of AHA into protein, larvae were washed, anesthetized, and homogenized and the resulting lysate was reacted with biotin-alkyne in the presence of CuBr and the triazole ligand (see Methods). This allowed for detection and quantification of newly synthesized biotin-labeled proteins using immunoblot analysis or for affinity purification of the newly synthesized proteins (BONCAT). To visualize newly synthesized proteins following AHA exposure, larvae were washed, anesthetized, fixed, and permeabilized. Whole mounted larval zebrafish were reacted with AlexaFluor-488-alkyne in the presence of CuSO4, the reducing agent tris(2-carboxyethyl)phosphine (TCEP), and the triazole ligand, before imaging using a confocal microscope (FUNCAT). This allowed for visualization of new protein synthesis, in the intact larval zebrafish.
Figure 1.
Labeling of newly synthesized proteins for identification (BONCAT) and visualization (FUNCAT) in larval zebrafish. (a) Scheme depicting metabolic labeling of newly synthesized proteins in 7-day-old larval zebrafish using AHA incorporation and Cu(I)-catalyzed [3 + 2] azide–alkyne cycloaddition. TCEP, tris(2-carboxyethyl)phosphine. (b) Chemical structures of methionine and azidohomoalanine (AHA).
Previously, Dieterich et al. showed that metabolic labeling of mammalian cell culture with AHA does not alter global protein synthesis rates or promote ubiquitin-mediated degradation, indicating that AHA incorporation does not cause severe protein misfolding or degradation.17 To ensure that incubation and incorporation of AHA into newly synthesized proteins is not toxic for the living animal, larvae were exposed to E3 embryo medium supplemented with 0–20 mM AHA, or 10 mM methionine, for 6–72 h. Larvae were scored as healthy, if after incubation they were still responsive to light touch. No significant toxic effects were observed when larvae were incubated with 1–10 mM AHA, even after 72 h incubations (Figure 2a). Only incubations with extremely high (20 mM) concentrations of AHA were toxic beginning around 24 h after onset of incubation. This indicates that incubation with low to moderate concentrations of AHA, even over extended periods of time is not toxic to the living animal. In the remainder of the studies reported here, concentrations of ≤4 mM AHA were used.
Figure 2.
At low concentrations, AHA exposure is not toxic and does not significantly alter simple behaviors. (a) Survival rate of 7-day-old larval zebrafish when incubated with AHA (0–20 mM, 6–72 h) or methionine (10 mM, 6–72 h), n = 20. (b) Quantification of spontaneous swimming behavior of larval zebrafish after AHA incubation (4 mM, 0–48 h). Percentage of larvae that show no spontaneous swimming behavior per 15 min interval. Mean swimming bursts per 15 min interval, n = 10–12. Differences are not statistically significant. (c) Traces depicting the angle of eye rotation during a typical optokinetic response after AHA incubation (4 mM, 0–48 h). (d) Sample startle response upon light flash after AHA incubation (4 mM, 24 h). (e) Mean response percentage to light or dark flash after AHA incubation (4 mM, 0–48 h), n = 5 larvae, flashed three times each. Error bars represent standard deviation of response percentage. Differences are not statistically significant. (f) Mean delay in response to light or dark flash after AHA incubation (4 mM, 0–48 h), n = 5 larvae, flashed three times each. Error bars represent standard deviation of response time. Differences are not statistically significant.
Next, we explored whether incorporation of AHA causes changes in simple behaviors. We conducted a series of behavioral tests after incubation in E3 medium supplemented with 4 mM AHA, for 0–48 h. First we investigated spontaneous swimming behavior. 7-day-old larval zebrafish were incubated in 4 mM AHA for 0–48 h prior to observation, and then placed individually into a 1 cm by 7.5 cm swimming chamber (Supporting Information Figure S1) and their spontaneous swimming bouts were recorded for a period of 15 min. Sample traces of spontaneous swimming behavior are depicted in Supporting Information Figure S1. There was no significant difference in the number of individual spontaneous swimming bouts initiated between 48 h AHA incubated, 24 h AHA incubated and control larvae, although there was a small, not significant decrease in the 48 h and 24 h AHA groups as compared to the control group (Figure 2b). There was also no difference in the number of AHA incubated and control larvae that failed to exhibit spontaneous swimming bouts during the 15-min trial period (Figure 2b).
To study whether AHA incubation causes deficits in visual tracking, 7-day-old larvae were tested for the optokinetic response (29) after incubation in 4 mM AHA for 24–48 h. Larvae were immobilized in 0.4% low-melting point agarose in a circular array of LEDs, which delivered a spot of white light that moved in a horizontal plane around the immobilized larvae. Similar to control larvae, AHA incubated larvae were able to track the light stimulus, producing smooth tracking eye movements and rapid saccades (Figure 2c, Supporting Information video), indicating that neither visual acuity nor neural circuits underlying visual tracking behavior seem to be affected by prolonged incubation with 4 mM AHA. To further test whether AHA incubation altered visual acuity and simple reflexive behaviors, we tested the animal’s startle response to light flash and dark flash. Larvae were placed in a circular array of LEDs, which delivered either a light flash or a dark flash while the response of the larva was monitored. Figure 2d shows a representative startle response to a light flash in an animal following a 24 h incubation with 4 mM AHA. The larva is clearly exhibiting a stereotypical C-bend escape response,30 indicating that AHA has no effect on the motor function associated with escape behavior. Furthermore, incubation with 4 mM AHA for 24–48 h did not alter the percentage of larval zebrafish that responded to either light or dark flash (Figure 2e) nor did it affect the delay in response to either light or dark flash (Figure 2f). Therefore, we conclude that AHA incorporation is not toxic and has no effects on simple behaviors at low concentrations (4 mM), even over prolonged incubation periods, making it suitable for labeling newly synthesized proteins in vivo.
To determine whether AHA is metabolically incorporated into newly synthesized proteins, we tagged lysates prepared from larval zebrafish incubated for 0–72 h with 4 mM AHA with biotin-alkyne in the presence of the Cu(I) catalyst. Subsequent dot blot analysis with a biotin antibody revealed successful incorporation of AHA into proteins in an incubation-time-dependent manner. A sample dot blot is shown in Figure 3a, along with quantification of several experiments. After only a 6 h incubation period with E3 embryo medium supplemented with 4 mM AHA, statistically significant (p < 0.005) AHA incorporation could be detected. After 24, 48, and 72 h incubations, approximately 140 ng (±8 ng), 375 ng (±34 ng), and 699 ng (±72 ng) of biotinylated protein were detected per homogenized larva, respectively. The total soluble protein per larva under the experimental conditions we used was 6.38 μg (±0.53 μg). From this, we can estimate that 24, 48, or 72 h incubation with 4 mM AHA leads to labeling and tagging of approximately 2.2%, 5.9%, and 10.9%, respectively, of the total soluble protein per larval zebrafish. However, as different proteins may show different levels of AHA incorporation, and therefore different biotin signal strength, the analysis given here should be regarded as semiquantitative.
Figure 3.
AHA is metabolically incorporated into larval zebrafish proteins in vivo. Sample immunoblot and quantification of immunoblots of lysates from AHA-treated 7-day-old larval zebrafish reacted with biotin-alkyne (10 μM) for 12 h, probed with antibody against biotin. (a) Larval zebrafish were incubated with 4 mM AHA for 0–72 h, n = 4. (b) Larval zebrafish were incubated with AHA (0 or 4 mM) or 4 mM AHA in the presence of puromycin (2.5–10 μg/mL) for 48 h, n = 3. ***p < 0.001.
To verify the specificity of AHA incorporation into newly synthesized proteins, we incubated larval zebrafish in E3 embryo medium supplemented with AHA along with low concentrations of the protein synthesis inhibitor puromycin. These very low concentrations of PSI did not have a toxic effect on larval zebrafish (data not shown). Although abundant biotin signal was detected in lysates of larval zebrafish incubated with AHA only, no signal was detected when larval zebrafish were incubated without AHA, and a significantly lower signal was detected when larval zebrafish were incubated in AHA in the presence of puromycin (Figure 3b). Furthermore, when the concentration of PSI in the incubation medium was increased from 2.5 to 5 μg/mL, a significant decrease in AHA labeled and biotinylated proteins was observed. However, no further decrease was observed when the PSI concentration was further increased to 10 μg/mL.
The above results confirm that BONCAT labels newly synthesized proteins with high specificity in the larval zebrafish. In addition, we observed that AHA incorporation in larval zebrafish scales nonlinearly with incubation time (Figure 3a) and we assume that an incorporation plateau would be reached after even longer incubation periods. Also, labeling was AHA concentration-dependent (Supporting Information Figure S2). While no signal was detected when 4-day-old larval zebrafish were incubated with 0 mM AHA, increasing the concentration of AHA in the incubation medium from 1 to 4 mM resulted in a detectable signal increase. Furthermore, AHA was incorporated not only into a few select proteins, but into a large variety of newly synthesized proteins throughout the proteome over time, as is shown by the abundance of protein bands on the Western blot of affinity purified biotinylated proteins from whole larval zebrafish lysates reacted with the biotin-alkyne and probed against biotin (Supporting Information Figure S3a). Biotin signal detected in the samples not incubated with AHA are likely a result of endogenous biotinylation.
To examine whether AHA is also incorporated into newly synthesized proteins in deeper structures such as the nervous system, we incubated 4-day-old transgenic HuC::GFP larval zebrafish with 4 mM AHA for 48 h. HuC encodes an RNA-binding protein that serves as an excellent early marker for differentiating neurons and the HuC::GFP line is a stable zebrafish transgenic line in which GFP is expressed specifically in neurons.31 With the exception of a few cells in the olfactory pit and the lateral line, the majority of these neurons are not surface structures. As before, whole zebrafish lysates were labeled with the biotin-alkyne, affinity purified, and then analyzed using Western blot probed against GFP. Only in the sample that was incubated in 4 mM AHA for 48 h were we able to affinity purify AHA-labeled, biotin-tagged GFP, indicating that AHA is not only incorporated into newly synthesized proteins in surface structures of the larval zebrafish, but also in the nervous system, the sole area of GFP expression in the HuC::GFP transgenic line (Supporting Information Figure 3b).
We next optimized the labeling and reaction conditions to maximize specific visualization of newly synthesized proteins (FUNCAT) in the intact larval zebrafish. For this purpose, we used the mutant zebrafish line nacre, which lacks melanophores throughout development32 and thus is relatively transparent and ideal for imaging. Larval zebrafish were, as before, incubated in E3 medium supplemented with 4 mM AHA for 0–72 h. Larvae were anesthetized, fixed, and permeabilized, before whole mount samples were reacted with 5 μM AlexaFluor-488-alkyne, in the presence of CuSO4, TCEP, and the triazole ligand, at room temperature overnight. After several washes in PBDTT buffer, samples were immobilized in 0.4% agarose and imaged using a confocal microscope.
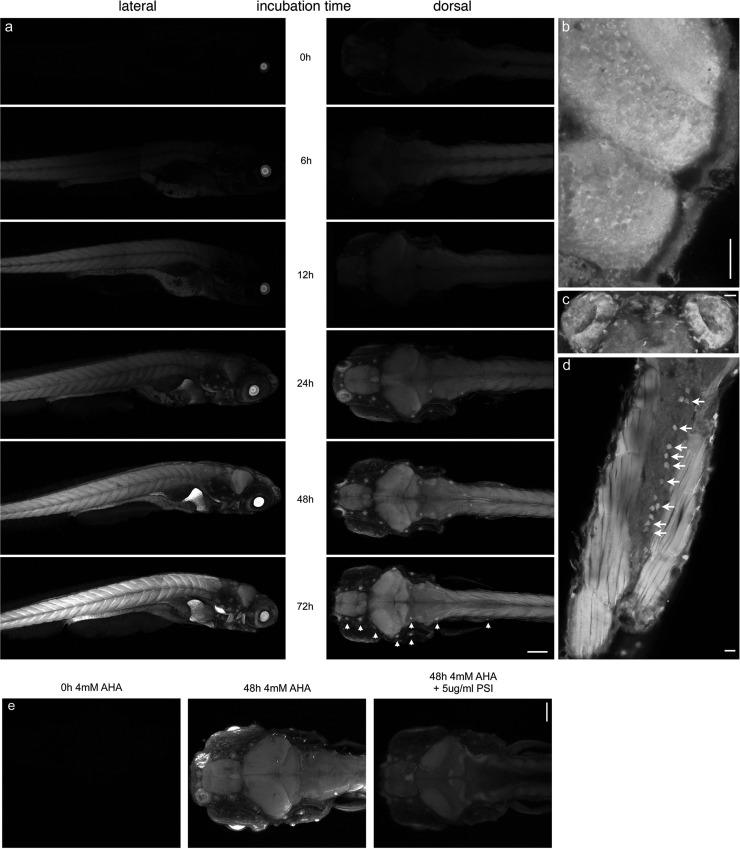
Incubation of larval zebrafish with 4 mM AHA followed by reaction with Alexa-488-alkyne resulted in an incubation-time dependent fluorescent labeling of newly synthesized proteins throughout the larval zebrafish (Figure 4a). Low fluorescent signals, especially in the muscles of the tail, could be detected after as little as 12 h incubation with AHA. Other structures, including the brain, spinal cord, liver, intestines, and heart, could be readily visualized after 24 h incubation with AHA. Specifically, sensory organs such as the neuromasts of the lateral line (indicated by arrow heads in Figure 4a, 72 h incubation dorsal view panel) and the olfactory pit (Figure 4c) seem to be areas of especially high levels of fluorescence. Furthermore, deeper structures such as the optic tectum, cerebellum (Figure 4b), and the spinal cord (Figure 4d) are not only readily labeled and tagged using the AlexaFluor-488 alkyne but show differences in fluorescence intensity on the cellular level. In the case of the spinal cord, we believe this population of brightly labeled cells corresponds to Rohon-Beard neurons33 (Figure 4d, as indicated by arrows). To verify that the fluorescent signal observed in the above experiments represents incorporation of AHA into newly synthesized proteins, larval zebrafish were incubated in E3 medium containing 4 mM AHA in the presence of 5 μg/mL puromycin (Figure 4e). In agreement with previously described results from lysates, abundant fluorescent signal was detected in whole mounts of larval zebrafish incubated with AHA only, while no signal was detected when larval zebrafish were incubated without AHA, and only background signal was detected when larval zebrafish were incubated in AHA in the presence of puromycin. These results suggest that FUNCAT may be used to identify regions of protein synthesis, specific cells, or groups of cells that are metabolically active, during the AHA incubation window in intact larval zebrafish.
Figure 4.
Imaging of newly synthesized proteins after in vivo labeling. (a) 7dpf larval zebrafish were metabolically labeled with 4 mM AHA for 0–72 h prior to fixation and reacted with 5 μM AlexaFluor-488-alkyne tag for 12 h. Left panel, lateral view; right panel, dorsal view. Arrow heads indicate neuromasts of the lateral line. (b, d, e) 7-day-old larval zebrafish labeled with 4 mM AHA for 48 h imaged at higher magnification. Dorsal views of (b) optic tectum and cerebellum, (c) olfactory pits, (d) horizontal cross-section of tail, showing tail muscles and spinal cord. Arrows indicate potential Rohon-Beard neurons. Scale bar in (a), 150 μm; in (b, d, e), 20 μm. (e) Larval zebrafish were metabolically labeled with 4 mM AHA for 0, 48, or 48 h in the presence of 5 μg/mL puromycin. Dorsal view; scale bar is 100 μm; n = 5.
To further investigate whether BONCAT and FUNCAT can be used to identify changes in protein synthesis in vivo, larval zerbafish were exposed PTZ, a GABAergic receptor antagonist that induces epileptic-like neuronal discharges and seizure-like behaviors in rodents and zebrafish.34−36 It has been shown that exposure to PTZ induces expression of immediate early genes in larval zebrafish34 and leads to changes in postsynaptic GABA receptor expression37 and hilar neurogenesis (38) in rodents.
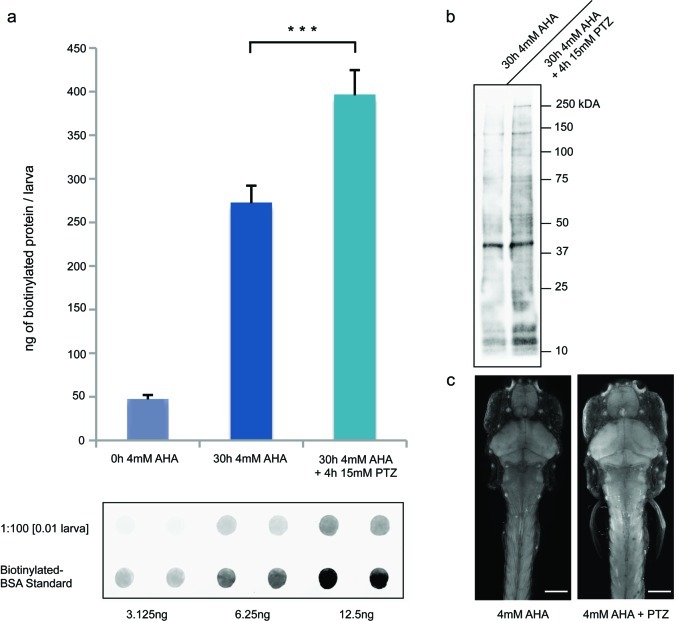
Larval zebrafish were exposed to 15 mM PTZ for two 2 h periods, 24 and 8 h before anesthesia while being incubated in 4 mM AHA for 30 h. The amount of biotinylated protein per larva was detected using dot blot analysis, as previously described. We observed a significant increase in the amount of biotinylated protein in larval zebrafish exposed to PTZ during AHA incubation, as compared to larvae that were not exposed to PTZ (Figure 5a), indicating that PTZ induces an increase in protein synthesis. This increase in biotinylated protein signal is not specific to one or a few protein bands, but it seems to be the result of a general increase of protein synthesis throughout the proteome as detected by Western blot analysis of affinity purified samples (Figure 5b). Furthermore, using the FUNCAT technique, we were able to visualize an increase in fluorescent signal in the brain and tail muscles in larval zebrafish that had been incubated in 4 mM AHA for 48 h and exposed to 15 mM PTZ for two 2 h periods (Figure 5c). These results indicate that chemical stimulation with the GABAergic receptor antagonist PTZ induces an increase in protein synthesis, which can be quantified and localized using the BONCAT and FUNCAT techniques in larval zebrafish.
Figure 5.
GABA antagonist PTZ induces increased protein synthesis in larval zebrafish. (a) Sample immunoblot and quantification of immunoblots of lysates from 7-day-old larval zebrafish reacted with biotin-alkyne tag (10 μM) for 12 h, probed with antibody against biotin. Zebrafish were incubated with 4 mM AHA (0 or 30 h) or with 4 mM AHA for 30 h as well as 15 mM PTZ for two periods of 2 h, 20 and 6 h before harvesting, n = 3. ***p < 0.001. (b) Western blot of biotin affinity-purified lysates of zebrafish incubated with 4 mM AHA for 30 h with or without 4 h 15 mM PTZ exposure. (c) Imaging of 7-day-old larval zebrafish after 48 h 4 mM AHA incubation with or without 4 h 15 mM PTZ exposure, reacted with AlexaFluor-488-alkyne (5 μM, 12 h); dorsal view. Scale bar is 150 μm; n = 6.
In this study we have shown that the BONCAT and FUNCAT techniques, which introduce bio-orthogonal chemical groups into newly synthesized proteins using the endogenous cellular translation machinery, can be applied to live, 7-day-old larval zebrafish. This enables the enrichment and quantification of newly synthesized proteins, when using an affinity tag such as the biotin-alkyne, and the visualization of protein synthesis, when using fluorescent-alkyne tags, such as the AlexaFluor-488-alkyne. Furthermore, we have shown that chemical stimulation with the proconvulsant PTZ increases protein synthesis, which can be detected using the methods developed in this study.
BONCAT and FUNCAT techniques enable labeling of newly synthesized proteins only when methionine is substituted by noncanonical amino acids during translation. However, AHA competes with endogenous methionine for charging onto methionine tRNA by the somewhat promiscuous MetRS. Previous work by the Tirrell group has shown that the charging rate of AHA relative to that of methionine onto methionine tRNA in bacterial cells is 1/390, as indicated by the specificity constant kcat/Km,39 suggesting that not all newly synthesized proteins may incorporate AHA in the presence of endogenous methionine. Furthermore, only proteins that contain at least one methionine residue can be labeled. This, however, is not an important factor in zebrafish, as 97.97% of zebrafish proteins contain at least one nonterminal methionine. Only two of 27 014 currently annotated zebrafish proteins contain no methionine at all (NCBI Danio rerio protein database, 5.17.2011).
Recent work using bacterial cells has opened the door to increasing the specificity of these techniques. A different noncanonical amino acid, azidonorleucine (ANL), can be used for the BONCAT/FUNCAT reaction.40 ANLs’ azide bearing side-chain is too bulky to fit into the binding pocket of wild-type MetRS and can therefore not be charged onto methionine tRNA in wild-type cells. However, introducing specific point mutations into the MetRS sequence enables charging of ANL. This permits genetic restriction of the tagging techniques by selective expression of the mutant MetRS in cell populations of interest. We are currently constructing transgenic fish in which the mutant MetRS is placed under the control of a specific promoter. Subsequent incubation with ANL will enable us to observe labeling and downstream identification of newly synthesized proteins in specific cell populations, as oppose to the whole organism.
Recently, the larval zebrafish has become a model organism for small molecule screens, permitting identification of small neuroactive molecules, which alter motor activity41 or circadian rhythm.42 In the future, the FUNCAT and BONCAT techniques can be paired with different chemical stimuli that cause behavioral changes to investigate underlying adjustments of the proteome in distinct regions of the nervous system. Even complex tasks known to be protein synthesis-dependent, such as long-term memory formation, can now be tackled with these techniques to elucidate which neurons and neuronal circuits are affected or involved.
Methods
Reagents
All chemical reagents were of analytical grade, obtained from Sigma unless otherwise noted, and used without further purification. We prepared AHA as described previously.43 The AlexaFluor-488 alkyne was purchased from Invitrogen (catalog number A10267), while the biotin-alkyne tag was purchased from Jena Biosciences (catalog number TA105).
Zebrafish Stocks and Husbandry
Adult fish strains AB, HuC::GFP and nacre were kept at 28 °C on a 14 h light/10 h dark cycle. Embryos were obtained from natural spawnings and were maintained in E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO444).
Toxicity and Behavioral Tests
To test AHA toxicity, larvae were placed five at a time in a 24-well Falcon culture dish well. Each well contained approximately 2 mL of embryo medium. Medium was replaced with embryo medium supplemented with 0–20 mM AHA or 10 mM methionine at the appropriate time point. Larvae were checked for response to light touch at 7 dpf.
For other behavioral tests, larvae were incubated in 10 mL of embryo medium or embryo medium supplemented with 4 mM AHA for 24–48 h in a 6 cm Petri dish. To monitor spontaneous swimming bouts, larvae were placed individually in a 1 cm by 7.5 cm behavioral chamber and spontaneous swimming was recorded using a webcam for 15 min. Subsequently, swimming bouts were scored. The optokinetic response was measured by immobilizing 7dpf larval zebrafish in a drop of 0.4% low melting point agarose (Promega) in embryo medium. Immobilized larvae were placed in a circular array of LEDs, which delivered a spot of white light that moved in a horizontal plane around the immobilized larvae. The optokinetic response was recorded using a high-speed camera (Redlake MotionScope M3), and eye movements were analyzed using Matlab. The startle response was measured by placing larval zebrafish in 5 cm Petri dish in a circular array of LEDs. LEDs delivered 50 ms light or dark flashes, while a high-speed camera mounted above the arena recorded responses. Response onset was scored.
Copper-Catalyzed [3 + 2] Azide–Alkyne Cycloaddition Chemistry and Detection of Tagged Proteins
Zebrafish larvae were incubated in embryo medium supplemented with AHA after which larvae were washed three times in 25 mL of embryo medium. Larvae were moved into a 1.5 mL Eppendorf tube in ∼1 mL of embryo medium and anesthetized on ice for 1 h. Remaining medium was removed, and anesthetized fish were washed once with 1 mL of ice cold PBS + protease inhibitor (PI; Roche, complete ULTRA Tablets, Mini, EDTA-free Protease Inhibitor cocktail tablets). PBS+PI was removed and replaced with 100 μL of fresh PBS+PI. Zebrafish larvae were homogenized using a Kontes pellet pestle motor. Then 1% SDS and 1 μL of benzonase (≥500 U) were added and the lysate vortexed and heated at 95 °C for 10 min. Lysate was allowed to cool to room temperature, before 400 μL of PBS+PI and 0.2% triton X-100 were added. Then, lysates were centrifuged at 15 000g at 4 °C for 10 min. Supernatant was transferred to a new 1.5 mL Eppendorf tube. For BONCAT, samples were reacted with 10 μM biotin-alkyne in the presence of 200 μM triazole ligand (tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, 97%) and 5 mg/mL CuBr suspension and incubated at 4 °C with agitation overnight. Samples were then centrifuged at 4 °C for 5 min at 5000g to pellet CuBr. Supernatant was moved into a new 1.5 mL Eppendorf tube. To remove excess, unligated biotin-alkyne, samples were applied to a PD MiniTrap G-25 size exclusion column (GE Healthcare). Samples were then analyzed using “dot blots” and affinity purified as described in ref (18). For Western blot analysis of affinity purified samples, 25 μL of washed NeutrAvidin beads (Thermo Scientific) previously incubated with sample were heated at 95 °C for 5 min in 50 μL of LDS sample buffer (Invitrogen) containing reducing agent (Invitrogen). Proteins were separated on precast NuPAGE 4–12% Bis-Tris gels (Invitrogen) and transferred to PVDF membranes and blocked in PBST (PBS+0.1% Tween-20) containing 5% milk. For detection, membranes were probed with goat anti-biotin (Biomol) and mouse anti-goat LI-COR-IR 800 secondary antibody and analyzed using the Odyssey Infrared Imaging system (LI-COR).
To image AHA labeled proteins, larval zebrafish were incubated in embryo medium supplemented with AHA, washed, and anesthetized as described above. Remaining embryo medium was removed and replaced with ∼1 mL of fixation solution (4% PFA, 88 mM sucrose in PBS). Larvae were fixed at room temperature for 3 h, dehydrated in 100% methanol, and stored at −20 °C overnight. Larvae were rehydrated through successive 5 min washes with 75% methanol in PBST, 50% methanol in PBST, 25% methanol in PBST, and finally PBST. This was followed by two washes in PBDTT (PBST + 1% DMSO and 0.5% Triton X-100) and 1 h permeabilization in Protease K (10 μg/mL in PBST). After permeabilization, larvae were briefly washed with PBST and then immediately postfixed for 20 min. Larvae were washed twice for 5 min with PBST and three times for 5 min with PBDTT, before blocking (5% BSA, 10% goat serum in PBDTT) for at least 3 h at 4 °C. Larvae were washed three times in PBST (pH 7.8), before being conjugated to the probe by addition of 200 μM triazole ligand, 5 μM AlexaFluor-488-alkyne, 200 μM CuSO4, and 400 μM TCEP at room temperature overnight with gentle agitation. Samples were washed four times for 30 min in PBDTT + 0.5 mM EDTA, and twice for 1 h in PBDTT, before being rinsed in PBST and immobilized on Matek dishes using 0.4% low melting point agarose. Images were obtained using a Zeiss LSM780 laser scanning confocal microscope with 10× or 20× air lens. AlexaFluor-488 was excited with the 488 nm line of an argon ion laser, and the emitted light was detected between 510 and 550 nm. We performed all postacquisition processing and analysis with ImageJ (NIH). Significance was tested for using the two-tailed t test and error bars represent standard deviation.
Acknowledgments
The authors would like to thank Mark Aizenberg for his help with behavioral tests, Georgi Tushev for his help with calculations of zebrafish proteome methionine content, Jennifer Hodas and John Ngo for general discussions and Stefanie Bunse and Susanne tom Dieck for comments on the manuscript.
Supporting Information Available
S1. Tracking spontaneous swimming behavior of 7-day-old larval zebrafish with AHA incubation (4 mM, 0–48 h). 15 min interval; frame captured every 10 s. S2. AHA incorporation is AHA concentration-dependent. Immunoblot of lysates from 4-day-old larval zebrafish reacted with biotin-alkyne (10 μM) for 12 h, probed with an antibody against biotin. Larval zebrafish were incubated with 0–4 mM AHA for 48 h. S3. AHA incorporation occurs throughout the proteome. (a, b) Western blot analysis of biotin affinity purified lysates of zebrafish incubated with 4 mM AHA for 0 to 72 h. (a) Probed with an antibody against biotin. (b) HuC::GFP larval zebrafish lysates probed with an antibody against GFP. Supplementary video. Optokinetic response of 7-day-old larval zebrafish with 48 h 4 mM AHA incubation. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Notes
This work was supported by the Max-Planck Society. D.A.T. acknowledges support from NIH (GM62523). F.I.H. acknowledges support from NIH/NRSA Institutional training grant 5T32 GM07616.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Davis H. P.; Squire L. R. (1984) Protein Synthesis and Memory: A Review. Psychol. Bull. 96, 518–559. [PubMed] [Google Scholar]
- Agranoff B. W.; Klinger P. D. (1964) Puromycin Effect on Memory Fixation in the Goldfish. Science 146, 952–953. [DOI] [PubMed] [Google Scholar]
- Agranoff B. W.; Davis R. E.; Brink J. J. (1966) Chemical Studies on Memory Fixation in Goldfish. Brain Res. 1, 303–309. [DOI] [PubMed] [Google Scholar]
- Ong S. E.; Blagoev B; Kratchmarova I; Kristensen D. B.; Steen H; Pandey A; Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 5, 376–386. [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. (1998) The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544. [DOI] [PubMed] [Google Scholar]
- Best M. D. (2009) Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry 28, 6571–6584. [DOI] [PubMed] [Google Scholar]
- Laughlin S. T.; Bertozzi C. R. (2009) Imaging the glycome. Proc. Natl. Acad. Sci. U.S.A. 1, 12–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho Y; Kim S. C.; Jiang C; Barma D; Kwon S. W.; Cheng J; Jaunbergs J; Weinbaum C; Tamanoi F; Falck J; Zhao Y. (2004) A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. U.S.A. 34, 12479–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckman M. A.; Kaur G.; Lee L. A.; Xie F.; Sepulvecla J.; Breitenkamp R.; Zhang X.; Joralemon M.; Russel T. P.; Emrick T.; Wang Q. (2008) Surface modification of tobacco mosaic virus with “Click” chemistry. ChemBioChem 9, 519–523. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. H.; Marx A. (2008) Novel strategies for the site-specific covalent labeling of nucleic acids. Chem. Commun. 5675–5658. [DOI] [PubMed] [Google Scholar]
- Prescher J. A.; Dube D. H.; Bertozzi C. R. (2004) Chemical remodelling of cell surfaces in living animals. Nature 7002, 873–877. [DOI] [PubMed] [Google Scholar]
- Chang P. V.; Prescher J. A.; Sletten E. M.; Baskin J. M.; Miller I. A.; Agard N. J.; Lo A; Bertozzi C. R. (2010) Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. U.S.A. 5, 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin S. T.; Baskin J. M.; Amacher S. L.; Bertozzi C. R. (2008) In vivo imaging of membrane-associated glycans in developing zebrafish. Science 5876, 664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin J. M.; Dehnert K. W.; Laughlin S. T.; Amacher S. L.; Bertozzi C. R. (2010) Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 23, 10360–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehnert K. W.; Beahm B. J.; Huynh T. T.; Baskin J. M.; Laughlin S. T.; Wang W; Wu P; Amacher S. L.; Bertozzi C. R. (2011) Metabolic labeling of fucosylated glycans in developing zebrafish. ACS Chem Biol. 6, 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin S. T.; Bertozzi C. R. (2009) In vivo imaging of Caenorhabditis elegans glycans. ACS Chem Biol. 12, 1068–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich D. C.; Link A. J.; Graumann J; Tirrell D. A.; Schuman E. M. (2006) Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U.S.A. 103, 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich D. C.; Lee J. J.; Link A. J.; Graumann J; Tirrell D. A.; Schuman E. M. (2007) Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2, 532–540. [DOI] [PubMed] [Google Scholar]
- Dieterich D. C.; Hodas J. J.; Gouzer G; Shadrin I. Y.; Ngo J. T.; Triller A; Tirrell D. A.; Schuman E. M. (2010) In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat. Neurosci. 7, 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. (2002) A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective ″ligation″ of azides and terminal alkynes. Angew. Chem. 14, 2596–2599. [DOI] [PubMed] [Google Scholar]
- Tornøe C. W.; Christensen C; Meldal M. (2002) Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 9, 3057–3064. [DOI] [PubMed] [Google Scholar]
- Agard N. J.; Prescher J. A.; Bertozzi C. R. (2004) A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- Deal R. B.; Henikoff J. G.; Henikoff S. (2010) Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 5982, 1161–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette S. P.; Dorsey F. C.; Moshiach S; Cleveland J. L.; Carabeo R. A. (2011) Chlamydia species-dependent differences in the growth requirement for lysosomes. PLoS One 3, 16783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian O. K.; Asiedu M. N.; Tillu D. V.; Peebles K. A.; Yan J; Ertz N; Dussor G. O.; Price T. J. (2010) IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J. Neurosci. 45, 15113–15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J; Brittis P. A.; Thomas F; Roux P. P.; Flanagan J. G. (2010) Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell 4, 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney K. B. (2011) Behavioural assessments of neurotoxic effects and neurodegeneration in zebrafish. Biochim. Biophys. Acta 3, 381–389. [DOI] [PubMed] [Google Scholar]
- Aizenberg M; Schuman E. M. (2011) Cerebellar-dependent learning in larval zebrafish. J. Neurosci. 24, 8708–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Y.; Neuhauss S. C. (2008) The optokinetic response in zebrafish and its applications. Front Biosci. 13, 1899–1916. [DOI] [PubMed] [Google Scholar]
- Kimmel C. B.; Patterson J; Kimmel R. O. (1974) The development and behavioral characteristics of the startle response in the zebra fish. Dev. Psychobiol. 1, 47–60. [DOI] [PubMed] [Google Scholar]
- Park H. C.; Kim C. H.; Bae Y. K.; Yeo S. Y.; Kim S. H.; Hong S. K.; Shin J; Yoo K. W.; Hibi M; Hirano T; Miki N; Chitnis A. B.; Huh T. L. (2000) Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 2, 279–293. [DOI] [PubMed] [Google Scholar]
- Clarke J. D.; Hayes B. P.; Hunt S. P.; Roberts A. (1984) Sensory physiology, anatomy and immunohistochemistry of Rohon-Beard neurones in embryos of Xenopus laevis. J .Physiol. 348, 511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister J. A.; Robertson C. P.; Lepage T; Johnson S. L.; Raible D. W. (1999) nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 17, 3757–3767. [DOI] [PubMed] [Google Scholar]
- Baraban S. C.; Taylor M. R.; Castro P. A.; Baier H. (2005) Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience 3, 759–768. [DOI] [PubMed] [Google Scholar]
- Baraban S. C.; Dinday M. T.; Castro P. A.; Chege S; Guyenet S; Taylor M. R. (2007) A large-scale mutagenesis screen to identify seizure-resistant zebrafish. Epilepsia 6, 1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann E. A.; Kampff A. R.; Prober D. A.; Schier A. F.; Engert F. (2010) Monitoring neural activity with bioluminescence during natural behavior. Nat. Neurosci. 4, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks-Kayal A. R.; Shumate M. D.; Jin H; Rikhter T. Y.; Coulter D. A. (1998) Selective changes in single cell GABA(A) receptor subunit expression and function in temporal lobe epilepsy. Nat. Med. 10, 1166–1172. [DOI] [PubMed] [Google Scholar]
- Parent J. M.; Yu T. W.; Leibowitz R. T.; Geschwind D. H.; Sloviter R. S.; Lowenstein D. H. (1997) Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus. J. Neurosci. 10, 3727–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiick K. L.; Saxon E; Tirrell D. A.; Bertozzi C. R. (2002) Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. U.S.A. 1, 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo J. T.; Champion J. A.; Mahdavi A; Tanrikulu I. C.; Beatty K. E.; Connor R. E.; Yoo T. H.; Dieterich D. C.; Schuman E. M.; Tirrell D. A. (2009) Cell-selective metabolic labeling of proteins. Nat. Chem. Biol. 10, 715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokel D; Bryan J; Laggner C; White R; Cheung C. Y.; Mateus R; Healey D; Kim S; Werdich A. A.; Haggarty S. J.; Macrae C. A.; Shoichet B; Peterson R. T. (2010) Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat. Chem. Biol. 3, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel J; Prober D. A.; Arvanites A; Lam K; Zimmerman S; Jang S; Haggarty S. J.; Kokel D; Rubin L. L.; Peterson R. T.; Schier A. F. (2010) Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 5963, 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link J. A.; Vink M. K. S.; Tirrell D. A. (2007) Preparation of the functionalizable methionine surrogate azidohomoalanine via copper-catalyzed diazo transfer. Nat. Protoc. 8, 1879–1883. [DOI] [PubMed] [Google Scholar]
- Brand M., Granato M., and Nüsslein-Volhard C. (2002) Keeping and Raising Zebrafish, in Zebrafish. A Practical Approach (Nüsslein-Volhard, C. and Dahm, R., Eds.), Oxford University Press, Oxford. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.