Abstract
micoRNAs (miRNAs) are small noncoding RNAs that regulate gene expression by targeting the mRNAs of a large number of human genes. Gliomas are the most common and deadly primary human brain tumors and are thought to originate from transformed stem-like cells (GSCs). microRNAs are frequently deregulated in cancer and gliomas and their deregulation has been associated with various aspects of glioma pathobiology. The present review summarizes the published literature on the role of miRNAs in gliomas with a focus on their role in GSCs.
Keywords: Glioma, Glioblastoma, Stem Cells, microRNA, Review
2. INTRODUCTION
microRNAs (miRNAs) are short non-coding endogenous RNAs that post-transcriptionally regulate the expression of a large number of genes. miRNAs play important roles in a wide variety of physiological and pathological processes including cancer. They are aberrantly expressed in many cancers and can exert tumor suppressive or oncogenic functions by regulating the expressions of mRNAs. miRNAs have also been associated with glioma formation and growth. Gliomas are the most common and deadly malignant primary brain tumors. The origin of gliomas is largely unknown but there is increasing speculation that they might arise from glioma stem cells (GSCs), which might consist of transformed neural stem cells. Similar to other cancers, miRNAs have been shown to regulate various cancer-associated genes and oncogenic functions in gliomas. Some evidence also suggests a role for miRNAs in the regulation of GSC biology. The present review provides a succinct but comprehensive summary of the literature on miRNAs in gliomas with a focus on their role in GSCs.
3. MICRORNAS
miRNAs are 17–25 nucleotide noncoding regulatory RNA molecules, that have a wide impact on the regulation of gene expression (1, 2). More than five hundred human miRNAs have been identified to date and the existence of several hundred more has been predicted (3–5). About two thirds of miRNA encoding genes are located in the intronic regions of protein-coding or -noncoding transcription units (2, 6). One third of miRNAs are organized into clusters, most of which are single transcriptional units (2, 7). miRNAs are first transcribed by RNA polymerase II to yield long transcripts known as pri-miRNAs. In the nucleus, pri-miRNAs are processed to pre-miRNAs by the RNase III enzyme Drosha. The pre-miRNAs are exported to the cytoplasm by exportin-5 and subsequently converted to mature duplex miRNAs by another RNase III enzyme, Dicer (8–10). Mature miRNAs regulate their targets by direct cleavage of the mRNA or by inhibition of protein synthesis, according to the degree of complementarities with their targets’ 3′UTR regions. Perfect or nearly perfect complementarity between miRNAs and their target 3′ UTR lead RNA-induced silencing complex (RISC) to cleave the target mRNA, whereas imperfect base matching induces mainly translational silencing of the target but can also reduce the amount of the mRNA target (11). Computational predictions of miRNA targets suggest that up to 30% of human protein coding genes may be regulated by miRNAs (12).
Many malignant tumors and tumor cell lines have deregulated miRNA expression as compared to normal cells and tissues (13, 14). Various mechanisms of miRNA deregulation in cancer have been identified. These include deregulations at the genetic, epigenetic, transcriptional, and processing levels. Many miRNA genes are located at fragile sites in the genome or regions that are commonly amplified or deleted in human cancer (15). Some miRNAs are repressed by CpG hypermethylation in cancers relative to normal tissue (16). A number of miRNAs are also regulated by transcription factors including p53, c-Myc and E2F (17–19). miRNA levels can also be deregulated via deregulation of Dicer or Drosha as has been observed in many cancers (20). Most miRNAs are downregulated in cancer but a number of miRNAs are also upregulated (2). By targeting the mRNA of oncogenes or tumor suppressors, miRNAs can act as tumor suppressors or oncogenes. Because miRNAs target a large number of mRNAs of genes associated with cancer, they regulate all aspects of cancer biology. They have been shown to regulate the cell cycle and cell proliferation, cell death and apoptosis, cell migration and invasion, metastasis, angiogenesis, tumor microenvironment, tumor immunology as well as many aspects of cancer stem cell biology (2). Clinically, miRNAs could serve as therapeutic targets and as diagnostic, prognostic and therapeutic tools. Global miRNA expression profiles can distinguish between normal and tumor tissue and help classify and grade various cancers (21, 22). Overexpression or inhibition of miRNAs could be used to achieve anti-tumor effects. Synthetic miRNA mimics used to overexpress miRNAs include siRNA-like oligoribonucleotide duplex or chemically modified oligoribonucleotides (23). miRNAs can be inhibited by variously modified antisense oligonucleotides such as 2′-O-methyl antisense oligonucleotide, antagomirs (24). In summary, miRNAs are important and critical regulators of cancer. Their implication in glioma stem cell biology and pathology is therefore predictable and will be discussed below.
4. GLIOMA STEM CELLS
Gliomas are the most common malignant primary brain tumors with an incidence of ~ 5 cases per 100,000 persons (25). Despite the most advanced treatment with combinations of surgery, radiotherapy and chemotherapy, glioblastoma multiforme, the most malignant and most common glioma, is associated with an average life expectancy of only 14 months (25). Accumulating evidence suggests that a subset of cells initiate and maintain the growth of gliomas and are responsible for their resistance to therapy (26). These cells are designated glioma stem cells (GSC) and have been isolated from glioblastoma by several research groups (27–29). Some studies suggest that GSCs form a small fraction of the total cell population within glioblastomas with the bulk of other tumor cells consisting of a mix of partially differentiated cancer progenitor-like cells with limited proliferative capacity and terminally differentiated cancer cells (26). More recent studies have challenged this concept for glioblastoma multiforme. One such study found that the majority of cells within the tumour bulk can be considered as tumour-initiating cells with different degrees of stemness (30). Functionally heterogeneous tumour initiating cell subpopulations therefore probably co-exist within the same tumour mass.
GSCs have similar characteristics to normal neural stem cells, including self-renewal, tumorigenesis and multipotency. They can divide and give rise to daughter stem cells with identical stem cell capabilities of the parent, have the proliferative ability to generate many progeny that can be propagated serially in undifferentiated state, form tumors in animals upon transplantation, and can differentiate into multiple cell types including astrocyte- neuron- and oligodendrocyte-like cells. GSCs are isolated from dissociated tumors, propagated as neurospheres in specific neurobasal medium. A subset of GSCs expresses neural stem cell surface markers such as Nestin and CD133, though the specificity of these as markers of stemness remains controversial (31).
The tumor stem cell concept could have profound implications on the basic understanding of tumor biology as well as on the development of new therapeutic strategies (32). It implies that effective targeting of glioma stem cells is critical for the success of anti-tumor therapies. Glioma stem cells were found to be major contributors to the therapy resistance of gliomas. It was shown that CD133-positive tumor cells, presumably GSCs, represent the cellular population that confers glioma radioresistance and could be the source of tumor recurrence after radiation (31). It was also shown that highly invasive gliomas with a stem-like phenotype are more chemoresistant than angiogenic tumors derived from the same patients (33). Additionally, CD133 positive glioma stem cells were found to be very resistant to chemotherapy. This resistance was probably contributed by the high expression in the stem cells of BCRP1 and MGMT, as well as the anti-apoptosis protein and inhibitors of apoptosis protein families (34).
Several signaling pathways and molecules have been implicated in the regulation of glioma stem cells. The expression of PDGFR on neural stem cells led to the speculation that their dysregulation might link NSCs to GSCs (35). The BMP-BMPR signalling system, which controls the activity of normal brain stem cells, was identified as a key inhibitory regulator of tumour-initiating, stem-like cells from glioblastomas (36). STAT3 was shown to regulate the growth and self-renewal of GSCs (37). OLIG2, a transcription factor that can promote the proliferation of neural progenitors by repressing the p21 tumor suppressor, has similar effects on glioblastoma stem cells (38). Blockage of the Sonic hedgehog (SHH) as well as the NOTCH signaling pathways depletes GSC cell populations in glioblastoma (39, 40). Myc was shown to be an important target for the cooperative actions of p53 and Pten in the regulation of normal and malignant stem/progenitor cell differentiation, self-renewal and tumorigenic potential of experimental glioblastomas (41). Many of the above stem cell regulators have been identified as targets of several miRNAs which therefore could theoretically regulate various aspects of GSC biology. A more detailed discussion of such miRNAs, their targets and functions in GSCs is described below.
5. MICRORNAS IN GLIOMAS
More than one hundred studies on miRNAs in gliomas have been published to date. Most of these studies examined the expressions, targets and functional effects of selected miRNAs in glioma cells and tissues. A few studies also conducted miRNA profiling in gliomas. This section reviews what has been reported to date on the role of miRNAs in gliomas (Table 1). miRNAs that appear in more than one publication are discussed first in separate paragraphs. The rest are discussed afterwards in this section. miRNAs that have been specifically implicated in GSC malignancy are discussed in a subsequent section of the review.
Table 1.
miRNAs that play a role in gliomas
| MicroRfNAs | Expression (Tumor/Cells) | Effects | Targets | Role in stem cells | Reference |
|---|---|---|---|---|---|
| miR-21 | (T/C) (+) | Proliferation (+), cell cycle (+), apoptosis (−), migration (+), invasion (+), VM-26 and TMZ chemoresistance (+) | RECK, TIMP3, TGFBR2/3, HNRPK, TAp63JMY, TOPORS, TP53BP2, DAXX, PDCD4 | 42, 43, 45, 47, 48, 49, 50, 51, 52, 53, 54 | |
| miR-221 | (T/C) (+) | Cell cycle (+), proliferation (+), invasion (+), migration (+), tumor growth (+), angiogenesis (+) | p27 (Kip1), c-Kit | 56, 57, 61, 63 | |
| miR-181 family | (T/C) (−) | Growth (−), apoptosis (+), invasion(−) | Bcl-2 | 57, 64, 65, 59 | |
| miR-26a | Transform cells (+), cell growth (+), proliferatioin (+), apoptosis (−) | PTEN, RB1, MAP3K2/MEKK2 | 66, 67 | ||
| miR-296 | (C) (+) | Angiogenesis (+) | HGS | 71 | |
| miR-15b | (T) (−) | Cell cycle (+) | CyclinE1 | 72 | |
| miR-146b | (T) (−) | Migration (−), invasion (−) | MMP16 | 70 | |
| miR-125b | (C) (−) | Cell cycle (−) | U251 glioma stem cell proliferation (−), cell cycle (−) | 74 | |
| miR-10b | (T) (+), | Invasion (+) | HOXD10 | 75 | |
| miR-153 | (T) (−) | Cell proliferation (−), apoptosis (+) | Bcl-2, Mcl-1 | 76 | |
| miR-17 | (T) (+) | Viability (+), proliferation (+), apoptosis (−), invasion (+) | 46 | ||
| miR-184 | (T) (−) | Viability (−), proliferation (−), apoptosis(+), invasion (−) | Npm1, Akt2 | 46 | |
| miR-196a | Decreased risk of glioma (+) | 77 | |||
| miR-195, miR-455-3p, miR-10a* | (C) (TMZ resistant cells) (+) | 78 | |||
| let-7 | proliferation (−), migration (−) | 79 | |||
| miR-182 | (T/C) (+) | Patient survival (−) | 80 | ||
| miR-7 | (T/C) (−) | Cell cycle(−), cell death(+), invasion(−) | EGFR | Invasion (−), viability (−) | 82 |
| miR-124 | Differentiation (+) | PTBP1, CDK6 | Differentiation (+) | 83, 44 | |
| miR-137 | (T/C) (−) | Cell cycle (−), proliferation (−), differentiation (+) | CDK6 | Differentiation (+) | 44 |
| miR-451 | (C) (+) | Downregulation of miR-451: proliferation (+), cell survival (+), migration (+) | Neurosphere formation (−), SMAD induction | 84, 36, 85 | |
| miR-128 | (T/C) (−) | Proliferation (−), self-renewal (−) | Bmi-1 | Proliferation (−), self-renewal (−) | 86, 87 |
| miR-34a | (T) (−) | Cell proliferation (−), cell cycle (−), cell survival (−), invasion (−) | c-Met, Notch-1, Notch-2 | Glioma stem cell proliferation(−), death (+), differentiation(+) | 89, 90 |
| miR-326 | (C) (neuronal cells) (+),(T) (glioma) (−) | Tumorigenicity (−), apoptosis (+), metabolic activity (−) | Notch, pyruvate kinase M2 | Cytotoxic in established and stem cell-like glioma lines | 95 |
| miR-17-92 cluster | (T) (+) | Astrocytoma progression (+), persistence of tumor-specific T cells (−) | Pold2, CTGF | Differentiation (−), apoptosis (−), cell proliferation (+) | 96 |
T = tumor, C = cells, (+) = increased, (−) = decreased.
5.1. miR-21
miR-21 was the first miRNA to be linked with glioma malignancy and is perhaps the most investigated miRNA in these tumors to date. Most reports describe miR-21 as an oncogenic miRNA. miR-21 levels are elevated in human glioma cells and tissues as compared to normal glial cells and/or brain(42) (43, 44). Also, miR-21 levels in gliomas correlate with tumor grade and low miR-21 levels in human tumors are associated with slightly better survival according to the cancer genome atlas (TCGA) (45, 46). miR-21 regulates several malignancy parameters. Inhibition of miR-21 induces glioma cell apoptosis, inhibits invasion, represses growth, induces chemosensitization and inhibits in vivo xenograft growth (43, 45, 47–53). miR-21 exerts its oncogenic effects by targeting the expression of several genes. miR-21 induces glioma cell migration by inhibiting the matrix metalloproteinase regulators RECK and TIMP3 (45). It affects apoptosis and cell cycle by inhibiting heterogeneous nuclear ribonucleoprotein K (HNPRK), the tumor suppressor homologue of p53 (Tap63), programmed cell death 4 (PDCD4) and possibly also EGFR, cyclin D and Bcl2 (48, 50, 54). Downregulation of miR-21 contributes to the antitumor effects of IFN-beta. miR-21 expression is negatively regulated by STAT3 activation in human glioma cells and xenografts (55). miR-21 is therefore an important overexpressed miRNA in gliomas that exerts potent oncogenic effects by downregulating multiple targets.
5.2. miR-221/222
Several reports have implicated miR-221/222 in glioma malignancy. The genes for miR-221 and miR-222 occupy adjacent sites on the X chromosome. Their expression appears to be coregulated and they largely have the same target specificity (56). A screening study identified miR-221 as one of the most frequently upregulated miRNAs in human glioma tumors and cell lines (57). miR-221 upregulation was confirmed in a subsequent study which also found that miR-221 levels are higher in higher-grade tumors (58). However, one report seemed to contradict previous findings and described a significant downregulation of miR-221/222 in glioblastoma tumors as compared to normal brain (59). The TCGA data show that miR-221/222 downregulation in human tumors is associated with a better patient prognosis. It was subsequently shown that the tumor suppressor and negative regulator of the cell cycle p27 was a direct target of miR-221/222 and that downregulation of p27 mediates the proliferative effects of miR-221/222 in glioma cells (56, 60). Other potential targets of miR-221 are the survivin-1 homolog BIRC1 and the neuronal inhibitor of apoptosis NIAP (61). miR-222 and miR-339 were also found to promote resistance of glioma cells to cytotoxic T-lymphocytes by down-regulation of the cell adhesion molecule ICAM-1 (62). Knock-down of miR-221/222 was found to indirectly lead to STAT1/2 upregulation (63).
5.3. miR-181
miR-181a, miR-181b and miR-181c were originally identified as downregulated miRNAs in glioblastoma cells and tumors by miRNA microarrays (57). miR-181a and to a greater extent miR-181b were subsequently described as tumor suppressors that inhibit growth and induce apoptosis of glioma cells (64). miR-181a overexpression sensitizes glioma cells to radiation treatment concurrent with the down-regulation of Bcl-2 (65). Also, miR-181b and miR-181c were significantly down-regulated in patients who responded to radiation therapy and temozolomide in comparison to patients with progressive disease. It was therefore proposed that expression levels of miR-181b and miR-181c could serve as a predictive marker of response to radiation therapy and temozolomide in glioblastoma patients (59).
5.4. miR-26a
Two high-profile publications identified miR-26a as a regulator of PTEN in gliomas (66, 67). PTEN is a major tumor suppressor that is frequently mutated and deleted in human glioblastoma (68, 69). In the first publication, the authors showed that miR-26a is frequently amplified at the DNA level in human gliomas and that this is associated with monoallelic PTEN loss. They demonstrated that miR-26a-mediated PTEN repression in a mouse glioma model enhances de novo tumor formation and precludes loss of heterozygosity at the PTEN locus. These data therefore described a new epigenetic mechanism for PTEN regulation in glioma via amplification of the miR-26a gene (66). In the second publication, the authors used a multidimensional genomic data set of glioblastoma from TCGA to identify miR-26a as a cooperating component of a frequently occurring amplicon that also contains CDK4 and CENTG1, two oncogenes that regulate the RB1 and PI3K/AKT pathways, respectively. By integrating DNA copy number, mRNA, miRNA, and DNA methylation data, they identified several functionally relevant targets of miR-26a in glioblastoma, including PTEN, RB1, and MAP3K2/MEKK2. They demonstrated that miR-26a alone can transform cells and promote glioblastoma cell growth in vitro and in the mouse brain by decreasing PTEN, RB1, and MAP3K2/MEKK2 protein expression, thereby increasing AKT activation, promoting proliferation, and decreasing c-JUN N-terminal kinase-dependent apoptosis. Overexpression of miR-26a in PTEN-competent and PTEN-deficient glioblastoma cells promoted tumor growth in vivo and increased growth in cells overexpressing CDK4 or CENTG1. Additionally, glioblastoma patients harboring this amplification displayed markedly decreased survival. Therefore, this publication identified miR-26a, CDK4, and CENTG1 as a functionally integrated oncomir/oncogene DNA cluster that promotes aggressiveness in human cancers by cooperatively targeting the RB1, PI3K/AKT, and JNK pathways (67) (70).
5.5. Other miRNAs
Several other miRNAs have been implicated in glioma malignancy.
5.5.1. miR-296
A role for miR-296 in glioma angiogenesis was uncovered (71). miR-296 was induced in endothelial cells isolated from human brain tumors by growth factors secreted by glioma cells. Growth factor-induced miR-296 significantly contributes to angiogenesis by directly targeting the hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) mRNA, leading to decreased levels of HGS and thereby reducing HGS-mediated degradation of the growth factor receptors VEGFR2 and PDGFR-beta. Furthermore, inhibition of miR-296 with antagomirs reduced angiogenesis in tumor xenografts in vivo (71).
5.5.2. miR-15b
miR-15b was implicated in the regulation of cell cycle progression in glioma cells (72). Over-expression of miR-15b resulted in cell cycle arrest at G0/G1, while suppression of miR-15b expression resulted in a decrease of the cell fraction in G0/G1 and a corresponding increase of the cell fraction in S phase. CCNE1, the gene encoding cyclin E1, was found to be one of the downstream targets of miR-15b. However, a direct link between CCNE1 downregulation by miR-15b and cell cycle regulation was not demonstrated (72).
5.5.3. miR-146b
miR-146b was shown to inhibit glioma cell migration and invasion (70). Using microarrays, miR-146b was identified as one miRNAs that is significantly deregulated in human glioblastoma tissue. miR-146b overexpression or knock-down did not affect the growth of human glioblastoma cells but miR-146b significantly reduced the migration and invasion of one glioblastoma cell line while miR-146b inhibition had the opposite effect. The matrix metalloproteinase gene, MMP16, was identified as one of the downstream targets of miR-146b. The authors therefore concluded that MMP16 downregulation mediates the effects of miR-146b on glioma invasion but this was not experimentally proven (70).
5.5.4. miR-125b
miR-125b was shown to induce glioma cell proliferation and inhibit all-trans-retinoic acid-induced apoptosis. Bcl-2 modifying factor (BMF) was identified as direct inhibition target for miR-15b and suggested as a mediator of the effects of miR-125b on apoptosis (73). In apparent contradiction with the above mentioned study, another study found that miR-125b induces cell cycle arrest and inhibit CDK6 and CDC25A expressions in one glioma cell line (U251) (74).
5.5.5. miR-10b
Several lines of evidence suggest that miR-10b plays a role in glioma invasion (75). miR-10b expression was found upregulated in glioma samples as compared to non-neoplastic brain tissues and expression levels of miR-10b were associated with higher grade glioma. mRNA expressions of RhoC and urokinase-type plasminogen activator receptor (uPAR), which were thought to be regulated by miR-10b via HOXD10, statistically significantly correlated with the expression of miR-10b. Also, protein expression levels of RhoC and uPAR were associated with expression levels of miR-10b. Multifocal lesions on enhanced MRI of 7 malignant gliomas were associated with higher expression levels of miR-10b.
5.5.6. miR-153
miR-153 decreased cell proliferation and increased apoptosis in one glioma cell line (DBTRG-05MG). It also directly inhibited Bcl-2 and Mcl-1 (myeloid cell leukemia sequence 1) expressions by directly targeting the 3′UTR regions of their respective mRNAs. It was therefore speculated that miR-153 exerted its pro-apoptotic effects in gliomas by targeting Bcl-2 and Mcl-1 (76).
5.5.7. miR-17 and miR-184
miR-17 and miR-184 were identified as two miRNAs with reduced expression in grade IV secondary glioblastoma tumors as compared to corresponding grade II astrocytomas from which they had progressed (46). Both miRNAs inhibited cell viability, proliferation and invasion and decreases the mRNA expression of several genes including AKT2 (46).
5.5.8. miR-196a
miR-196a was found upregulated in glioblastoma as compared with grade III gliomas and its expression levels correlated with poor prognosis. Multivariate analysis showed that miR-196 expression levels were an independent predictor of overall survival in glioblastoma patients (77).
5.5.9. miR-195, miR-455-3p and miR-10a
miR-195, miR-455-3p and miR-10a* were implicated in temozolomide resistance as they were upregulated in a temozolomide resistant variant of the U251 glioblastoma cell line (78).
5.5.10. Let-7
Although not differentially expressed in gliomas, Let-7 overexpression and its effects were investigated in glioma cells and xenografts (79). Transfection of let-7 reduced expression of pan-RAS, N-RAS, and K-RAS in glioblastoma cells. Let-7 also reduced in-vitro proliferation and migration of the cells, and reduced the sizes of tumors generated by transplantation into nude mice. However, let-7 miRNA exerted no effect on the proliferation of normal human astrocytes. The authors therefore suggested the possible use of let-7 for the therapy of glioblastoma (79).
5.5.11. miR-182
A recent study identified miR-182 as a prognostic marker for glioma progression and patient survival (80). The authors of this study found that miR-182 was up-regulated in glioma cell lines and primary glioma specimens as compared to normal brain. miR-182 expression levels in the tumors statistically significantly correlated with tumor grade and clinical features. The 5-year survival rates of patients with low miR-182 levels were significantly better than the survival rates of patients with high miR-182 levels. The study therefore suggests that miR-182 could serve as a marker of glioma progression and predictor of patient survival (80).
5.6. miRNA profiling
A recent study used genome-wide miRNA profiling to compare miRNA expressions in human glioblastoma (grade IV) with anaplastic astrocytoma (grade III) and normal brain (81). The study found several differentially expressed miRNAs that can differentiate between glioblastoma, astrocytoma and normal brain. Using Prediction Analysis of Microarrays (PAM) software, the authors identified a 23-miRNA expression signature that could distinguish glioblastoma from anaplastic astrocytoma with 95% accuracy. The signature contained miRNAs that had been previously known to be deregulated in glioblastoma cells and stem cells including miR-21, miR-34a, miR-128 and miR-451 as well as miRNAs not previously associated with gliomas such as miR-886-3p, miR-886-5p, miR-16, miR-24 and other. The study also showed that inhibition of two glioblastoma-upregulated miRNAs (miR-21 and miR-23a) and exogenous overexpression of two glioblastoma-downregulated miRNAs (miR-218 and miR-219-5p) resulted in inhibition of soft agar colony formation but did not affect cell proliferation and chemosensitivity (81).
6. MICRORNAS IN GLIOMA STEM CELLS
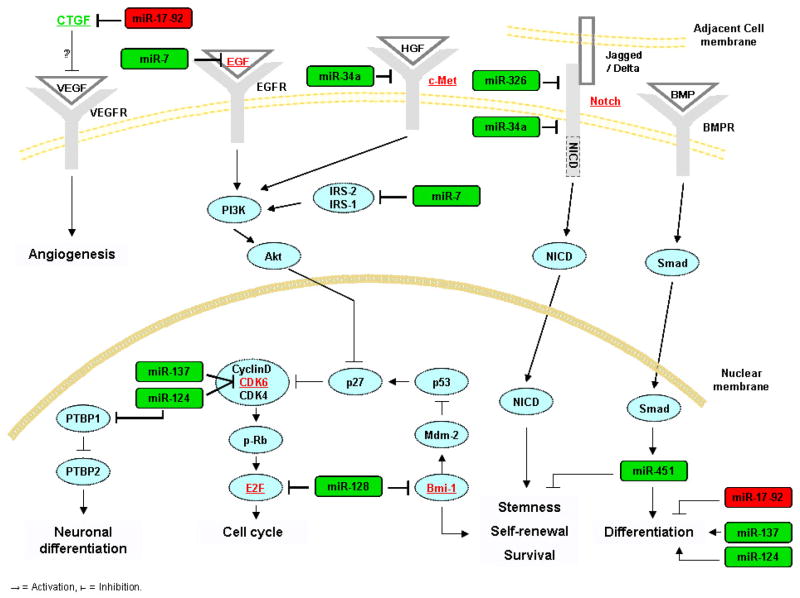
Of more than one hundred published articles on miRNAs in gliomas, only few addressed their roles in glioma stem cells. The findings from these publications are summarized in this section (Figure 1).
Figure 1.
Schematic of miRNAs that have been shown to regulate stem cell targets and functions.
6.1. miR-7
A report implicated miR-7 in the regulation of gene expression and function of glioblastoma stem cells (82). While this report did not specifically investigate the effects of miR-7 on characteristics of stemness, it used a well characterized glioblastoma stem cell line (0308) to examine the targets, functional effects and regulation of miR-7. miR-7 inhibited the expression of EGFR and the activation of Akt in the stem cell line. miR-7 also inhibited glioblastoma stem cell proliferation and invasion. miR-7 was downregulated in human glioblastoma tissue. This report therefore implicated, for the first time, a miRNA in the regulation of critical genes and malignant parameters in glioblastoma stem cells.
6.2. miR-124 and miR-137
miR-124 was originally found to promote neuronal differentiation by targeting PTBP1, which encodes a global repressor of alternative pre-mRNA splicing, and triggering brain-specific alternative pre-mRNA splicing (83). A subsequent study assessed the effects of miR-124 and miR-137 on the differentiation of mouse neural stem cells, mouse oligodendroglioma-derived stem cells and human glioblastoma stem cells, defined based on their CD133 positivity (44). Transfection of miR-124 or miR-137 induced morphological changes and marker expressions consistent with neuronal differentiation in mouse neural stem cells, mouse oligodendroglioma-derived stem cells derived from S100β-v-erbB tumors and cluster of differentiation CD133+ human glioblastoma multiforme-derived stem cells. This study therefore implicated miR-124 and miR-137 in the differentiation of neural and glioma stem cells.
6.3. miR-451 and miR-425
A subsequent report examined the miRNA profiles of glioblastoma stem cell (defined as CD133+ cells) and non-stem cell (defined as CD133− cells) populations and found that several miRNAs including miR-451, miR-486, and miR-425 were upregulated in the CD133− cells (84). Transfection of these miRNAs inhibited glioblastoma neurosphere formation and growth. miR-451 transfection also induced dispersal of glioblastoma neurospheres. Interestingly, the study also found that SMADs, which have been previously associated with glioma stem cell regulation, could upregulate miR-451 by binding to its promoter region (36). The report therefore established a link between miRNAs and well known stem cell regulating proteins. Another recent interesting study found that miR-451 promotes glioma cell growth and that its levels are repressed by low glucose and induced by high glucose concentrations. The effects of miR-451 were mediated by LKB1, which it represses through targeting its binding partner, CAB39 (MO25α). Overexpression of miR-451 sensitized cells to glucose deprivation, suggesting that its downregulation is necessary for robust activation of LKB1 in response to metabolic stress. Thus, miR-451 was proposed as a regulator of the LKB1/AMPK pathway, and as a component of a fundamental mechanism that contributes to cellular adaptation in response to altered energy availability (85).
6.4. miR-128
One study described a link between a stem cell self-renewal factor (Bmi-1) and miR-128 (86). The study first identified miR-128 as downregulated in glioblastoma tissue as compared to normal brain. It found that the polycomb transcriptional repressor Bmi-1 is directly inhibited by miR-128 via binding to its 3′UTR. Bmi-1 had been previously shown to promote normal and cancer stem cell self- renewal and was later also implicated in glioma stem cell regulation (87). Consistent with Bmi-1 downregulation, miR-128 inhibited glioma stem cells self-renewal. This study therefore described the regulation by a miRNA of an important stem cell renewal factor. miR-128 was also found by another study to inhibit the proliferation of established glioma cells by targeting the transcription factor E2F3a (88).
6.5. miR-34a
Two recent studies uncovered critical roles for miRNA-34a in glioblastoma stem cells (89, 90). It was first shown that mir-34a is downregulated in human gliomas and directly inhibits the expressions of c-Met, Notch-1 and Notch-2 by binding to their mRNA 3′-UTR regions in glioma cells and stem cells. Notch is a critical regulator of cell-fate during development and of normal and cancer stem cell maintenance (40, 91, 92). Notch pathway activation enhances the stemness, proliferation and radioresistance of glioma stem cells (40, 92–94). Functionally, miR-34a exerted potent tumor suppressive effects in glioma cells and stem cells. Its expression led to the inhibition of cell proliferation, survival and migration. More importantly, miR-34a induced glioblastoma stem cell differentiation as evidenced by the decrease in stem cell markers and the increase of neuronal, astrocytic and oligodendrocytic markers after miR-34a expression (90). Notch-1 and Notch-2 inhibitions by miR-34a partially mediated the functional effects of this miRNA on glioblastoma stem cells (89). These studies therefore implicated miR-34a in the regulation of glioblastoma stem cells partly via regulation of Notch expression.
6.6. miR-326
miR-326 was identified as a direct regulator of Notch expression in glioblastoma stem cells (95). Interestingly, miR-326 suppressed Notch and was suppressed by Notch. miR-326 was downregulated in gliomas via decreased expression of its host gene. Transfection of miRNA-326 into stem cell-like glioma lines was cytotoxic, and was rescued by Notch restoration. Furthermore, miR-326 transfection reduced glioma cell tumorigenicity in vivo. miR-326 also partially mediated the toxic effects of Notch knockdown. miR-326 was therefore identified as another miRNA that regulates glioblastoma stem cells via regulation of Notch.
6.7. miR-17-92 cluster
The miR-17-92 cluster was also implicated in the regulation of glioblastoma neurosphere (presumably stem cells) differentiation, apoptosis and proliferation (96). It was first shown that expression of several members of miR-17-92 was significantly higher in primary astrocytic tumors than in normal brain and significantly increased with tumor grade. A high-level amplification of the miR-17-92 locus was also detected in one glioblastoma specimen. Inhibition of miR-17-92 induced apoptosis and decreased cell proliferation in glioblastoma neurospheres. miR-17-92 inhibition was also associated with increased mRNA and/or protein expression of CDKN1A, E2F1, PTEN and CTGF. The CTGF gene was shown to be a direct target of miR-17-92 in glioblastoma neurospheres by luciferase reporter assays. The study therefore proposed that miR-17-92 and its target CTGF mediate the effects of differentiation-promoting treatment on glioblastoma cells.
6.8. Glioma vs. neural precursor cell miRNA expression profiles
Glioma vs. neural precursor cell miRNA expression profiles. It has been hypothesized that glioma stem cells originate from transformed neural stem cells. This hypothesis was recently supported by an interesting study that found that gliomas display a miRNA expression profile reminiscent of neural precursor cells (97). The authors compared the miRNA expression profiles of glial tumors, embryonic stem cells (ESCs), neuronal precursor cells (NPCs), and normal adult brains from both human and mouse tissues. They found that human and mouse gliomas shared a miRNA expression profile that is reminiscent of NPCs. About half of the miRNAs expressed in the shared profile clustered in seven genomic regions susceptible to genetic/epigenetic alterations in various cancers. These clusters comprised the miR-17 family, miR-183-182, and the stem cell-specific clusters miR-367-302 and miR-371-373, which are upregulated in gliomas, ESCs, and NPCs. The bipartite cluster of 7 + 46 miRNAs on chromosome 14q32.31, which might represent the largest tumor suppressor miRNA cluster, was downregulated in the shared expression profile. This study therefore provided the first evidence for an association between these clusters and gliomas. Despite the broad similarity in the miRNA expression profiles, 15 miRNAs showed disparate expression between stem cells and gliomas. Ten miRNAs belonged to the two stem cell-specific clusters and the remaining (miR-135b, miR-141, miR-205, miR-200c, and miR-301a) have been previously shown to associate with malignancy. These findings therefore showed that all gliomas displayed NPC-like miRNA signatures, suggesting that transformed NPCs might be the cells of origin of gliomas.
7. CONCLUSIONS
miRNAs are thought to regulate the expression of one third of the human genome. It is therefore not unexpected that they are implicated in many (perhaps all) aspects of glioma malignancy and glioma stem cell biology. The expression of several miRNAs is deregulated in human gliomas, glioma cells and glioma stem cells. Deregulation can be an early event and a result of miRNA gene deregulation or secondary to deregulation of transcription factors that regulate miRNA expression. Deregulation of some miRNAs correlates with patient prognosis. miRNAs positively or negatively regulate tumor cell proliferation, death, migration, invasion, angiogenesis and other by affecting the expressions of numerous target mRNAs. miRNAs have also been implicated in the regulation of glioma stem cell malignancy, differentiation and fate determination, but information on this subject remains incomplete and more research is needed for a better understanding of the role of miRNAs in glioma stem cells and glioma initiation. Nonetheless, the broad and profound involvement of miRNAs in the regulation of gene expression presents an opportunity for a better understanding of the mechanisms of glioma initiation and progression and perhaps also for the use of miRNAs as future agents or targets for glioma therapy.
Acknowledgments
Supported by NIH RO1 NS045209 (R. Abounader) and NIH RO1 CA134843 (R. Abounader).
Abbreviations
- miRNA
microRNA
- GSC
glioma stem cell
References
- 1.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–58. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 2.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, Zhang JW. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349(1):59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 8.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 10.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 12.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–68. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, Spiteri I, Das PP, Caldas C, Miska E, Esteller M. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67(4):1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 17.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282(4):2135–43. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 20.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20(16):2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 22.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101(32):11755–60. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 24.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13(6):496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 25.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 26.Hadjipanayis CG, Van Meir EG. Brain cancer propagating cells: biology, genetics and targeted therapies. Trends Mol Med. 2009;15(11):519–30. doi: 10.1016/j.molmed.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 28.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 30.Mazzoleni S, Politi LS, Pala M, Cominelli M, Franzin A, Sergi Sergi L, Falini A, De Palma M, Bulfone A, Poliani PL, Galli R. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70(19):7500–13. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 31.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 32.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 33.Johannessen TC, Wang J, Skaftnesmo KO, Sakariassen PO, Enger PO, Petersen K, Oyan AM, Kalland KH, Bjerkvig R, Tysnes BB. Highly infiltrative brain tumours show reduced chemosensitivity associated with a stem cell-like phenotype. Neuropathol Appl Neurobiol. 2009;35(4):380–93. doi: 10.1111/j.1365-2990.2008.01008.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kesari S, Stiles CD. The bad seed: PDGF receptors link adult neural progenitors to glioma stem cells. Neuron. 2006;51(2):151–3. doi: 10.1016/j.neuron.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–5. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 37.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27(10):2383–92. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8(2):119–30. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W, Piccirillo S, Vescovi AL, DiMeco F, Olivi A, Eberhart CG. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25(10):2524–33. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, Nikkhah G, Dimeco F, Piccirillo S, Vescovi AL, Eberhart CG. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28(1):5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–33. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conti A, Aguennouz M, La Torre D, Tomasello C, Cardali S, Angileri FF, Maio F, Cama A, Germano A, Vita G, Tomasello F. miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J Neurooncol. 2009;93(3):325–32. doi: 10.1007/s11060-009-9797-4. [DOI] [PubMed] [Google Scholar]
- 43.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 44.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stuhler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20(3):539–50. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67(19):8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 48.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68(19):8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–8. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 50.Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, Zhang W, Kang C. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90(2):144–55. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 51.Ren Y, Zhou X, Mei M, Yuan XB, Han L, Wang GX, Jia ZF, Xu P, Pu PY, Kang CS. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer. 2010;10:27. doi: 10.1186/1471-2407-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren Y, Kang CS, Yuan XB, Zhou X, Xu P, Han L, Wang GX, Jia Z, Zhong Y, Yu S, Sheng J, Pu PY. Co-delivery of as-miR-21 and 5-FU by poly(amidoamine) dendrimer attenuates human glioma cell growth in vitro. J Biomater Sci Polym Ed. 2010;21(3):303–14. doi: 10.1163/156856209X415828. [DOI] [PubMed] [Google Scholar]
- 53.Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010;1352C:255–264. doi: 10.1016/j.brainres.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Liu W, Chao T, Zhang Y, Yan X, Gong Y, Qiang B, Yuan J, Sun M, Peng X. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272(2):197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 55.Ohno M, Natsume A, Kondo Y, Iwamizu H, Motomura K, Toda H, Ito M, Kato T, Wakabayashi T. The modulation of microRNAs by type I IFN through the activation of signal transducers and activators of transcription 3 in human glioma. Mol Cancer Res. 2009;7(12):2022–30. doi: 10.1158/1541-7786.MCR-09-0319. [DOI] [PubMed] [Google Scholar]
- 56.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6(16):2005–9. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 57.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 58.Conrad ME, Barton JC. Factors affecting the absorption and excretion of lead in the rat. Gastroenterology. 1978;74(4):731–40. [PubMed] [Google Scholar]
- 59.Slaby O, Lakomy R, Fadrus P, Hrstka R, Kren L, Lzicarova E, Smrcka M, Svoboda M, Dolezalova H, Novakova J, Valik D, Vyzula R, Michalek J. MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma. 2010;57(3):264–9. doi: 10.4149/neo_2010_03_264. [DOI] [PubMed] [Google Scholar]
- 60.Zhang C, Kang C, You Y, Pu P, Yang W, Zhao P, Wang G, Zhang A, Jia Z, Han L, Jiang H. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol. 2009;34(6):1653–60. doi: 10.3892/ijo_00000296. [DOI] [PubMed] [Google Scholar]
- 61.Lukiw WJ, Cui JG, Li YY, Culicchia F. Up-regulation of micro-RNA-221 (miRNA-221; chr Xp11.3) and caspase-3 accompanies down-regulation of the survivin-1 homolog BIRC1 (NAIP) in glioblastoma multiforme (GBM) J Neurooncol. 2009;91(1):27–32. doi: 10.1007/s11060-008-9688-0. [DOI] [PubMed] [Google Scholar]
- 62.Ueda R, Kohanbash G, Sasaki K, Fujita M, Zhu X, Kastenhuber ER, McDonald HA, Potter DM, Hamilton RL, Lotze MT, Khan SA, Sobol RW, Okada H. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci U S A. 2009;106(26):10746–51. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang C, Han L, Zhang A, Yang W, Zhou X, Pu P, Du Y, Zeng H, Kang C. Global changes of mRNA expression reveals an increased activity of the interferon-induced signal transducer and activator of transcription (STAT) pathway by repression of miR-221/222 in glioblastoma U251 cells. Int J Oncol. 2010;36(6):1503–12. doi: 10.3892/ijo_00000637. [DOI] [PubMed] [Google Scholar]
- 64.Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, You Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–93. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 65.Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23(4):997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- 66.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23(11):1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim H, Huang W, Jiang X, Pennicooke B, Park PJ, Johnson MD. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci U S A. 2010;107(5):2183–8. doi: 10.1073/pnas.0909896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abounader R. Interactions between PTEN and receptor tyrosine kinase pathways and their implications for glioma therapy. Expert Rev Anticancer Ther. 2009;9(2):235–45. doi: 10.1586/14737140.9.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Guessous F, DiPierro C, Zhang Y, Mudrick T, Fuller L, Johnson E, Marcinkiewicz L, Engelhardt M, Kefas B, Schiff D, Kim J, Abounader R. Interactions between PTEN and the c-Met pathway in glioblastoma and implications for therapy. Mol Cancer Ther. 2009;8(2):376–85. doi: 10.1158/1535-7163.MCT-08-0627. [DOI] [PubMed] [Google Scholar]
- 70.Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S, Ge R, Jiang S, Li G, Chen Y, He ML, Kung HF, Lai L, Lin MC. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–65. doi: 10.1016/j.brainres.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 71.Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14(5):382–93. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang M, Li D, Zhao Y, Ge R, Li G, Chen Y, He ML, Kung HF, Lai L, Lin MC. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun. 2009;380(2):205–10. doi: 10.1016/j.bbrc.2008.12.169. [DOI] [PubMed] [Google Scholar]
- 73.Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin XH, Ma X. MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem. 2009;23(4–6):347–58. doi: 10.1159/000218181. [DOI] [PubMed] [Google Scholar]
- 74.Shi L, Zhang J, Pan T, Zhou J, Gong W, Liu N, Fu Z, You Y. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res. 2010;1312:120–6. doi: 10.1016/j.brainres.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 75.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125(6):1407–13. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 76.Xu J, Liao X, Wong C. Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2010;126(4):1029–35. doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- 77.Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, Ma X, Hayashi K, Sasaki T. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16(16):4289–97. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 78.Ujifuku K, Mitsutake N, Takakura S, Matsuse M, Saenko V, Suzuki K, Hayashi K, Matsuo T, Kamada K, Nagata I, Yamashita S. miR-455-3p and miR-10a(*) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells miR-195. Cancer Lett. 2010;296(2):241–8. doi: 10.1016/j.canlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 79.Lee ST, Chu K, Oh HJ, Im WS, Lim JY, Kim SK, Park CK, Jung KH, Lee SK, Kim M, Roh JK. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J Neurooncol. 2010 doi: 10.1007/s11060-010-0286-6. [DOI] [PubMed] [Google Scholar]
- 80.Jiang L, Mao P, Song L, Wu J, Huang J, Lin C, Yuan J, Qu L, Cheng SY, Li J. miR-182 as a prognostic marker for glioma progression and patient survival. Am J Pathol. 2010;177(1):29–38. doi: 10.2353/ajpath.2010.090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rao SA, Santosh V, Somasundaram K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol. 2010;23(10):1404–17. doi: 10.1038/modpathol.2010.135. [DOI] [PubMed] [Google Scholar]
- 82.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68(10):3566–72. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 83.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27(3):435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gal H, Pandi G, Kanner AA, Ram Z, Lithwick-Yanai G, Amariglio N, Rechavi G, Givol D. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem Biophys Res Commun. 2008;376(1):86–90. doi: 10.1016/j.bbrc.2008.08.107. [DOI] [PubMed] [Google Scholar]
- 85.Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, Van Brocklyn J, Ostrowski MC, Chiocca EA, Lawler SE. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37(5):620–32. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 87.Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1(1):87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, Gong Y, Yin B, Liu W, Qiang B, Zhao J, Yuan J, Peng X. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med. 2009;87(1):43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 89.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69(19):7569–76. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, Purow B, Abounader R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9(6) doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, Eberhart CG. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66(15):7445–52. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 92.Shih AH, Holland EC. Notch signaling enhances nestin expression in gliomas. Neoplasia. 2006;8(12):1072–82. doi: 10.1593/neo.06526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, Rich JN, Sullenger BA. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28(1):17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang XP, Zheng G, Zou L, Liu HL, Hou LH, Zhou P, Yin DD, Zheng QJ, Liang L, Zhang SZ, Feng L, Yao LB, Yang AG, Han H, Chen JY. Notch activation promotes cell proliferation and the formation of neural stem cell-like colonies in human glioma cells. Mol Cell Biochem. 2008;307(1–2):101–8. doi: 10.1007/s11010-007-9589-0. [DOI] [PubMed] [Google Scholar]
- 95.Kefas B, Comeau L, Floyd DH, Seleverstov O, Godlewski J, Schmittgen T, Jiang J, diPierro CG, Li Y, Chiocca EA, Lee J, Fine H, Abounader R, Lawler S, Purow B. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29(48):15161–8. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, Reifenberger G, Herold-Mende C, Lichter P, Radlwimmer B. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene. 2010;29(23):3411–22. doi: 10.1038/onc.2010.83. [DOI] [PubMed] [Google Scholar]
- 97.Lavon I, Zrihan D, Granit A, Einstein O, Fainstein N, Cohen MA, Cohen MA, Zelikovitch B, Shoshan Y, Spektor S, Reubinoff BE, Felig Y, Gerlitz O, Ben-Hur T, Smith Y, Siegal T. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol. 2010;12(5):422–33. doi: 10.1093/neuonc/nop061. [DOI] [PMC free article] [PubMed] [Google Scholar]