SUMMARY
Ligand binding to proteins is not a static process, but rather involves a number of complex dynamic transitions. A flexible ligand can change conformation upon binding its target. The conformation and dynamics of a protein can change to facilitate ligand binding. The conformation of the ligand, however, is generally presumed to have one primary binding mode, shifting the protein conformational ensemble from one state to another. We report solution NMR studies that reveal peroxisome proliferator-activated receptor γ (PPARγ) modulators can sample multiple binding modes manifesting in multiple receptor conformations in slow conformational exchange. Our NMR, hydrogen/deuterium exchange and docking studies reveal that ligand-induced receptor stabilization and binding mode occupancy correlate with the graded agonist response of the ligand. Our results suggest that ligand and receptor dynamics affect the graded transcriptional output of PPARγ modulators.
INTRODUCTION
Activation of receptors by small molecule ligands is generally described in terms of their ability to agonize (activate) or antagonize (block) receptor function. Nuclear receptors (NRs) are multidomain ligand-dependent transcription factors for which the molecular basis for ligand-mediated receptor activation is often described by the “helix 12” or AF-2 (activation function-2) conformational model (Heldring et al., 2007). This model proposes a switch between an active and inactive conformation of the receptor. Specifically, ligand binding induces stabilization of the conformational mobility of the C-terminal helix, helix 12, of the ligand binding domain (LBD) and helix 12 is referred to as the master regulator of transcriptional activation of NRs. This model has been viewed as a molecular roadmap for NR drug discovery, detailing how to turn the receptor “on” or “off” via structure-function mechanisms (Moore et al., 2006). NR ligands can induce a spectrum or graded response such as full agonism, partial agonism, antagonism, and inverse agonism (Germain et al., 2006). For some NRs, such as the estrogen receptors (ERs), the structural basis of the extremes of these responses, full agonism or antagonism, is well described by the helix 12 conformational model (Heldring et al., 2007). Full agonist ligands position helix 12 in a conformation favoring interaction with coactivator proteins. Antagonist ligands block receptor activation either by promoting corepressor interaction or by positioning helix 12 to preclude coactivator binding. Antagonists have also been shown to destabilize coreceptor (in the case of homodimeric and heterodimeric receptors) and other cofactor binding.
In contrast, the molecular mechanisms providing partial agonism are less well understood. Crystal structures of the apo LBD of the NR peroxisome proliferator-activated receptor γ (PPARγ) as well as co-crystal structures of PPARγ LBD bound to ligands with partial agonist activity all reveal helix 12 in the canonical agonist position (Bruning et al., 2007), providing little insight into the mechanism of action of full vs. partial agonists. In fact, all crystal structures of the PPARγ LBD whether apo or bound to agonist or antagonist, have similar backbone folds with little variation in helix 12 positioning (Bruning et al., 2007; Chandra et al., 2008; Uppenberg et al., 1998). Similar observations were made in the crystal structures of full length PPARγ in complex with its coreceptor retinoid X receptor α (RXRα), DNA, ligands and coactivator NR box peptides (Chandra et al., 2008). Thus, while crystal structures of PPARγ reveal atomic differences in the ligand contacts within the receptor ligand-binding pocket (LBP), they provide no insight into the graded activity of ligands. Based on these observations, we hypothesized that differences in PPARγ ligand binding modes and activity were largely controlled by receptor dynamics (Berger et al., 2003; Bruning et al., 2007; Johnson et al., 2000). Such dynamic effects are not easily resolved within the scope of and time scales sampled by x-ray crystallography (Brunger, 1997). In support of this notion, hydrogen/deuterium exchange (HDX) kinetic profiles implicate ligand subtype-specific differences in receptor conformational dynamics in the mechanism of graded PPARγ agonism. NMR studies also reveal that a selective PPARγ modulator can alter dynamics of the receptor in a manner that leads to selective gene expression profiles (Berger et al., 2003). However, the mechanism is more complex as ligands not only regulate helix 12/AF-2-mediated interactions with coregulators, but also modulate posttranslational modifications and other protein/coregulator binding sites (Choi et al., 2011; Choi et al., 2010). For simplicity, we refer to helix 12-mediated agonism here as “classical agonism”, whereas other modes of modulation of specific NR target genes are helix 12-independent and involve more global alterations in receptor dynamics that alter specific post-translational modifications.
Here we applied solution NMR spectroscopy to further examine the mechanisms contributing to graded PPARγ agonism. NMR is sensitive to structure and dynamics on time scales ranging from ps-s and slower, and in the slower cases NMR is able to observe multiple populations, even those that are lowly populated (Henzler-Wildman and Kern, 2007; Kleckner and Foster, 2011). In contrast, HDX and crystallography report on slower time scales and, in the case of crystallography, the information gleaned typically describes a low energy conformation averaged over all molecules within the crystal lattice (Brunger, 1997). From a functional point of view, fast motions (ps-ns) indeed impact the function of proteins. However, slower motions (µs-s and slower) play an important role in many biological processes, including enzyme catalysis and biomolecular interactions. We show that PPARγ modulators with graded response profiles not only differentially stabilize distinct regions of the receptor surface important for function, but also sample multiple binding modes within the LBP of the receptor in slow conformational exchange. The ability of the ligands to sample multiple binding modes propagates these dynamic effects to the protein surfaces important for its function. Our findings support the view that both ligand and protein dynamics contribute to the mechanism of action of PPARγ modulators.
RESULTS
Graded response of PPARγ modulators is not apparent from PPARγ LBD co-crystal structures
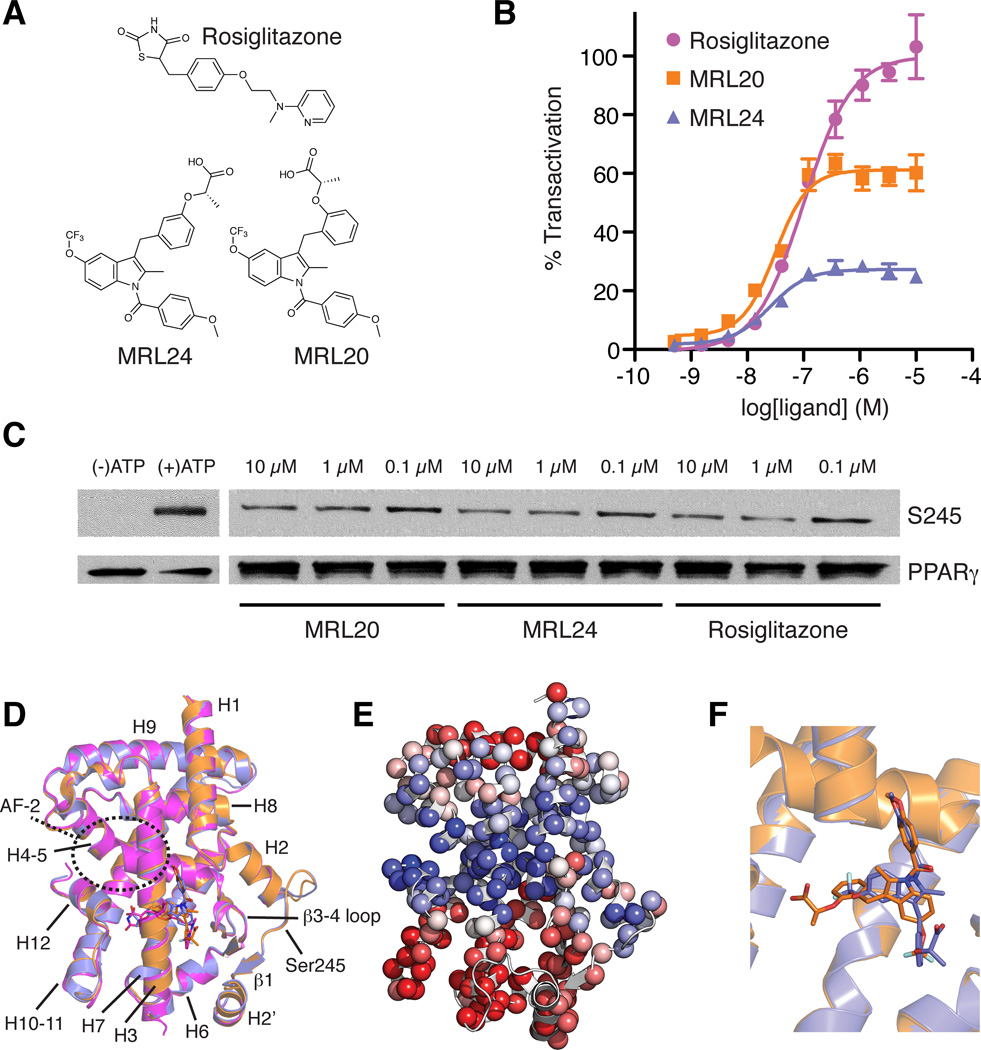
We focused on three potent PPARγ modulators (Acton et al., 2005) with low nM binding affinities that afford graded agonism profiles (Figure 1A): (a) the prototypical full agonist rosiglitazone; (b) MRL20, a full agonist that exhibits ~60% transactivation activity compared to rosiglitazone; and (c) MRL24, a partial agonist that exhibits ~25% activity compared to rosiglitazone. The degree of transactivation in a cell-based receptor:promoter-reporter co-transfection assay (Figure 1B) is concordant with the ability of the ligands to induce interaction of a NR receptor interaction domain (RID) from the steroid receptor coactivator-1 (SRC1) protein with PPARγ LBD as determined in a homogeneous time resolved fluorescence (HTRF) assay (Bruning et al., 2007). More recently, we determined that the anti-diabetic efficacy of PPARγ modulators correlates with ligand binding affinity as well as their ability to block phosphorylation of PPARγ at Ser245, but not with the degree of classical agonism (Choi et al., 2010) (Ser273 in the PPARγ2 sequence; PPARγ1 numbering is used in structural studies and thus used here). The potency of blockage of Ser245 (Ser273:PPARγ2) phosphorylation by ligands appears to be associated with the strength of interaction of the ligand with the backbone amide at Ser342, with the carboxylic acid moiety of MRL24 being in closest proximity to this amide. Consistent with our previous findings for MRL24 and rosiglitazone (Choi et al., 2010), all three modulators blocked Ser245 (Ser273:PPARγ2) phosphorylation (Figure 1C), and densitometry analysis suggest an efficacy rank order of MRL24 > rosiglitazone > MRL20. However, crystal structures of the PPARγ LBD bound to these ligands reveal largely unchanged backbone conformations (Figure 1D) (Bruning et al., 2007), though principal component analysis reveals some propensity for structural differences originating from the LBP (Figure 1E). Although MRL20 and MRL24 are regioisomers, and thus nearly chemically equivalent, they flip binding mode within the LBP of PPARγ (Figure 1F) (Bruning et al., 2007).
Figure 1. Graded response of PPARγ ligands is not explained by ligand-receptor crystal structures.
(A) Chemical structures of rosiglitazone, MRL20 and MRL24. (B) Effect of ligands on PPARγ transactivation in a cotransfection assay in 293T cells using a Gal4-PPARγ expression vector and UAS-luciferase reporter (n=4). (C) In vitro biochemical assay showing the ability of PPARγ ligands studied to block Cdk5-mediated phosphorylation of PPARγ. (D) Cartoon diagram overlay of PPARγ LBD crystal structures bound to rosiglitazone (pink; PDB 2PRG), MRL24 (blue; PDB 2Q5P) and MRL20 (orange; PDB 2Q59). (E) The first principal component from the THESEUS (Theobald and Wuttke, 2006, 2008) correlation matrix for the ML superposition of the structures in (D) mapped onto the PPARγ LBD (PDB 1FM6). Regions colored similarly (red or blue) are self-correlated, whereas regions colored differently (red versus blue) are anti-correlated. (F) PPARγ LBD structures bound to MRL24 (blue; PDB 2Q5P) and MRL20 (orange; PDB 2Q59) illustrate these structurally similar ligands bind in distinct ligand conformations under the crystallization conditions (Bruning et al., 2007).
NMR analysis of the PPARγ LBD
NMR data collected on apo PPARγ LBD reveals approximately half of the expected NMR resonances (Figure 2A). The absence of resonances here is likely due to conformational exchange on an intermediate NMR time scale (kex ≈ |Δv|, or the chemical shift differences between states, which is on the order of 103/sec) (Johnson et al., 2000; Kleckner and Foster, 2011). Backbone chemical shift assignments were obtained for the PPARγ LBD bound to the ligands rosiglitazone, MRL24 and MRL20. Binding of the PPARγ full agonist, rosiglitazone, restores nearly all NMR resonances (Figure 2B), suggesting that it fully stabilizes the receptor conformation on an intermediate time scale and is consistent with published NMR and HDX studies of PPARγ full agonists (Bruning et al., 2007; Hamuro et al., 2006; Johnson et al., 2000; Lu et al., 2008). The majority of residues for which NMR data is missing in the apo form are located within the ligand-binding pocket and AF-2 coregulator-binding surface (Figure 2A), which comprises H3, H4–5 and H12.
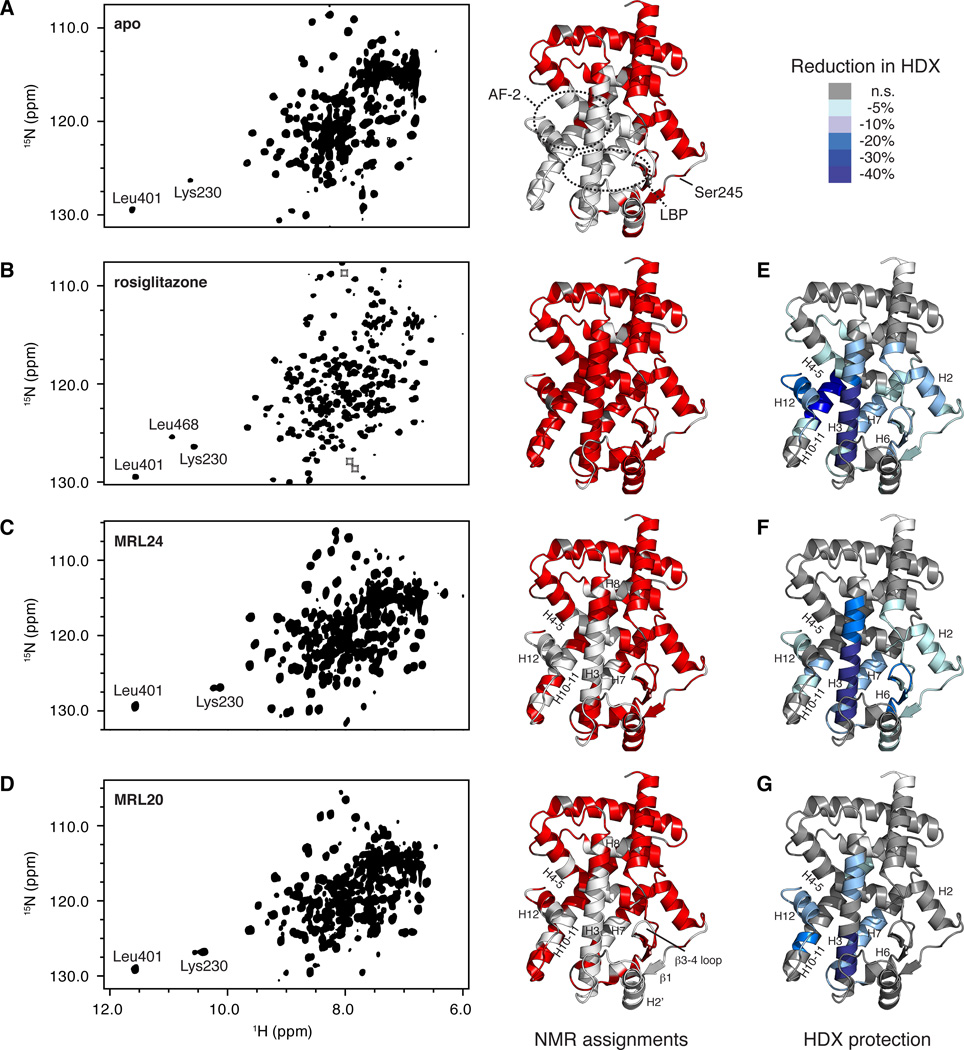
Figure 2. NMR and HDX-MS analysis reveals patterns consistent with graded responses of PPARγ ligands.
(A–D) 2D overlay of 3D TROSY-HNCO spectra and PPARγ LBD structure (PDB 1FM6) colored by the presence (red) or lack of (white) assigned NMR chemical shifts for (A) apo PPARγ LBD, (B) PPARγ LBD bound to rosiglitazone, (C) PPARγ LBD bound to MRL24 and (D) PPARγ LBD bound to MRL20. Proline residues, which are not assigned using traditional NMR methods, are colored grey. Regions boxed indicate locations of resonances that are aliased/folded. The backbone amides of the three most downfield shifted resonances for Leu401, Leu468 and Lys230 make strong hydrogen bonds to other side-chain nuclei in crystal structures and correspond well to ligand activity profiles. Leu468 is part of helix 12, and is only fully stabilized when bound to rosiglitazone; a very weak peak near the noise level is observed for Leu468 in some data in the presence of MRL20 but not for MRL24 (not shown). Lys230 displays two populations in the presence of a graded/partial agonist. (E–G) HDX-MS data for peptides listed in Table S1 in the presence of (E) rosiglitazone, (F) MRL24 and (G) MRL20, mapped onto the PPARγ LBD structure (PDB 1FM6). Color key displays percentage reduction in HDX relative to apo PPARγ LBD.
Compared to apo receptor, the partial agonist MRL24 and near full agonist MRL20 only partially stabilized the receptor conformation on an intermediate time scale (Fig 2c,d, respectively). For MRL24, the missing resonances populate the AF-2 surface, including H3, H4–5, H7, H8, H10–H11, and H12 (Figure 2C), and residues assigned in these regions show very broad resonances, suggestive of conformational exchange on the µs-ms time scale. These observations are in general agreement with those observed for the PPARγ partial agonist nTZDpa (Berger et al., 2003). For MRL20, the regions stabilized differ compared to the partial stabilization afforded by MRL24, including missing resonances in β1, H2’, and H3, and to a lesser degree on H4–5, loop β3–β4, H7, H8, H10–11, and H12 (Figure 2D); resonances in the latter regions also show some degree of line broadening. These observations are consistent with the functional profile of these ligands in that MRL20 better stabilizes the AF-2/helix 12 region through a ligand-receptor hydrogen bond (Bruning et al., 2007) and displays a higher transactivation profile (Figure 1B) and a higher degree of interaction with SRC-1 RID compared to MRL24 (Bruning et al., 2007). In contrast, MRL24 better stabilizes the β-sheet region in the LBP and Ser245 (Ser273:PPARγ2) surface and is more efficacious at blocking Cdk5-mediated phosphorylation of Ser245 (Figure 1C). Furthermore, our NMR data suggests that helix 12 is conformationally dynamic on an intermediate NMR time scale in the presence of MRL24, and to a lesser degree for MRL20, in contrast to the single conformation implied in ligand-receptor co-crystal structures (Figure 1D) (Bruning et al., 2007).
HDX analysis of the PPARγ LBD
For the studies reported here, we obtained HDX-MS data on a high resolution mass spectrometer offering improved sequence coverage compared to our previous analysis (Bruning et al., 2007). Compared to apo receptor, ligand binding induces stabilization of several regions of the PPARγ LBD (Table S1). Rosiglitazone protected hydrogen exchange in H2, H3, H4–5, β2–β4, H6, L6–7, H7, H10–11 and H12 (Figure 2E). The level of protection was robust for H3, H10–11 and H12 and correlates well to residues with missing apo NMR resonances that are observed in the presence of rosiglitazone. The level of protection from hydrogen exchange induced by MRL24 and MRL20 is also consistent with our NMR observations. In particular, MRL24 afforded robust protection of hydrogen exchange for residues in the β-sheet region and H6 (Figure 2F), which are regions where NMR chemical shift assignments were obtained (Figure 2C). Other regions that did not show robust alterations of HDX kinetics in the presence of MRL24 had many missing resonances in the NMR data, including H4–5, H10–11, and H12. MRL20 induces moderate protection from hydrogen exchange for residues within H10–11 and H12 (Figure 2G), more so than MRL24 but less than rosiglitazone. This is consistent with the NMR data, where we observed NMR resonances for some, but not all, residues within H10–11 and H12 in the presence of MRL20 (Figure 2D). Thus, the degree of ligand-induced stabilization observed by NMR via the number of observed resonances within H10–11 and H12, as well as the degree of ligand-induced protection from hydrogen exchange, is correlated with the graded response of PPARγ agonists in a manner that is not apparent from co-crystal structures.
Two receptor populations in slow conformational exchange
A number of residues within the PPARγ LBD were observed with two populations of NMR chemical shifts when bound to MRL24, and to a lesser extent MRL20 (Figure 3A). In contrast, the rosiglitazone-bound receptor primarily afforded only one major population of NMR chemical shifts (Figure S1). For example, Asp380 displays a single peak in the 3D TROSY-HNCO data for all forms studied, apo or bound to ligand (Figure 3A). However, Met348, Lys230, and others display two populations of NMR resonances in the presence of MRL24, suggestive of slow conformational exchange on a time scale of ms or slower. Similarly, when bound to MRL20, several residues also display two populations of NMR resonances, such as Lys230. In addition, the line shapes of other residues display significant line broadening, including Met348 (Figure 3A), which is suggestive of conformational exchange on the µs-ms time scale. Notably, the two populations of NMR resonances are present through all NMR data used to assign backbone chemical shifts (Figure 3B) and when heterodimerized to the RXRα LBD complexed with 9-cis retinoic acid (Figure 3C). We attempted to use ZZ-exchange spectroscopy (EXSY) to quantify the rate of slow exchange, which was limited due to spectral overlap. No exchange peaks were observed for resonances with resolved signals using relaxation delays ranging from 5 to 900 ms. This suggests the rate of slow exchange is likely on the order of seconds, either near the limit (near 0.2/s) or outside of the slow exchange regime detectable by EXSY (kex ≈ 0.2–100/s) (Kleckner and Foster, 2011). In addition, no exchange peaks were observed in 15N-NOESY-TROSY data (τmix = 120 ms). A similar slowly exchanging process, undetectable by EXSY, has been observed for the uncatalyzed cis/trans proline isomerization in CypA (Bosco et al., 2002).
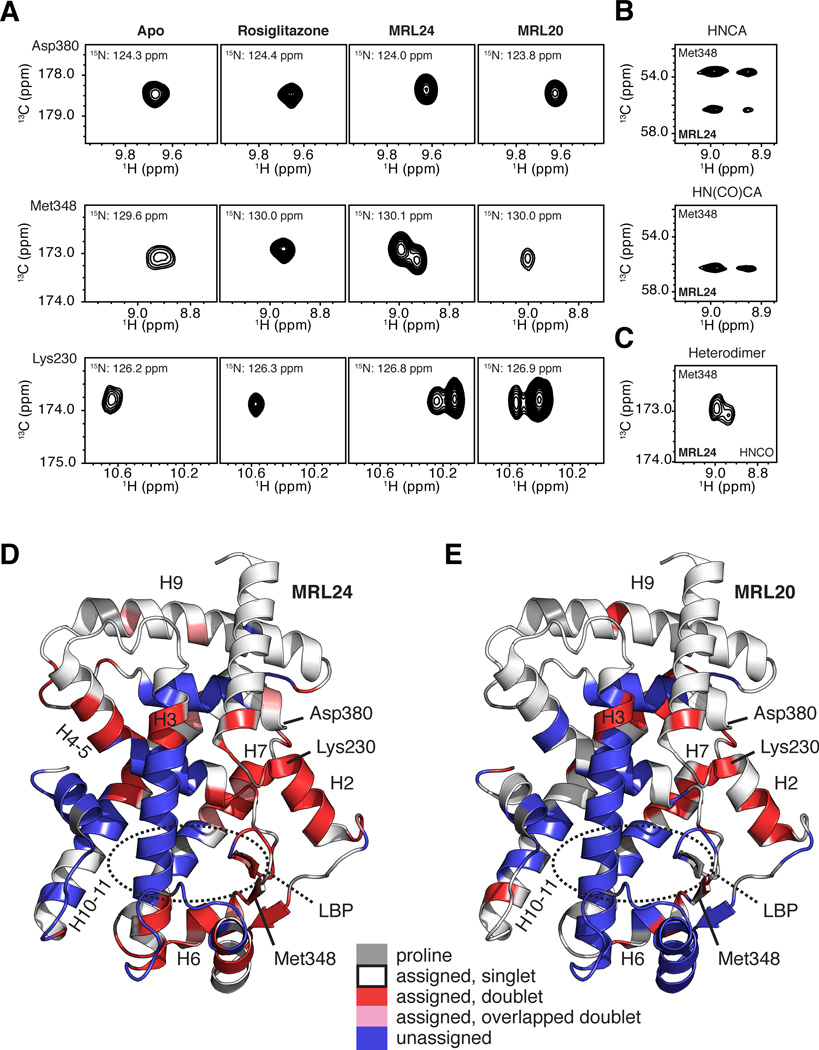
Figure 3. NMR data implicates slow conformational exchange between two receptor conformations in the mechanism of action of graded PPARγ modulators.
(A) Regions of 3D TROSY-HNCO spectra for PPARγ LBD in the apo form or bound to rosiglitazone, MRL24 and MRL20. NMR resonances for Asp380, Met348 and Lys230 are fully stabilized when bound to rosiglitazone, but display varying degrees of two resonance populations when bound to MRL24 and MRL20. (B–C) The two resonance populations were present in (B) data used for backbone assignments, including 3D TROSY-HNCA and TROSY-HN(CO)CA, and (C) in 3D TROSY-HNCO data for [2H,13C,15N]-PPARγ/[unlabeled]-RXRα LBD heterodimer bound to MRL24 and 9-cis-retinoic acid, respectively. (D,E) PPARγ LBD structure (PDB 1FM6) colored by residues with two chemical shift populations (red), line shapes suggesting two populations that have significant overlap (pink), or with missing assignments likely from intermediate conformational exchange (blue) in the presence of MRL24. Proline residues are colored grey. See also Figures S1 and S2.
The average population fractions for a group of well-resolved resonances are Pa≈0.7 and Pb≈0.3 when bound to MRL24. Residues with two populations when bound to MRL24 comprise the LBP and surrounding regions, including the β-sheet surface, H2, H3, H4–5, H6, H7 and H10–11 (Figure 3D). These residues are contiguous with residues missing resonances likely due to intermediate conformation exchange and thus do not have NMR chemical shift assignments. For MRL20, the two populations of resonances are less abundant compared to MRL24, in particular in H2, H4–5, H6 and H10–11, some of which are missing likely due to intermediate exchange, but nonetheless populate similar regions within the ligand-binding pocket (Figure 3E) with average population fractions for a group of well-resolved resonances of Pa≈0.85 and Pb≈0.15. Comparison of NMR data for the PPARγ LBD bound to MRL24 and MRL20 vs. apo receptor (Figure 2A,C,D and 3A) suggests that the two populations of NMR chemical shifts observed in the presence of MRL24 and MRL20 are not a consequence of sampling apo and holo (ligand-bound) receptor states, but rather two distinct ligand-bound conformations. This is particularly notable for Lys230, which displays a different amide proton chemical shift in the apo form compared to the two populations observed for receptor-bound MRL24 and MRL20. These observations are supported by circular dichroism (CD) thermal melt analysis, which reveals our samples are fully saturated under our experimental conditions and confirm a 1:1 ligand-receptor stoichiometry (Figure S2).
Multiple ligand binding modes for MRL20 and MRL24
Residues displaying two populations of NMR chemical shifts when bound to MRL24 or MRL20 map primarily to the LBP of the PPARγ (Figure 3D,E). We posited that slow conformational exchange between different ligand conformations could be the origin of the two receptor populations. To investigate this hypothesis, we used 1H and 19F ligand-observed NMR techniques to directly monitor receptor-bound MRL20 and MRL24. For the 19F NMR studies, there is one CF3 group in the regioisomers MRL20 and MRL24, giving rise to one 19F NMR peak (Figure 4A,F). When bound to the PPARγ LBD, MRL20 (Figure 4B) shows several peaks in the 19F NMR data, some of which are low intensity signals. These data suggest a primary chemical shift environment (a primary conformation) for the CF3 group (δ = −55.1 ppm) and up to four minor chemical shift environments (δ = −56.4, −57.4, −8.5, −59.6 ppm; four minor conformations). In contrast, when bound to the PPARγ LBD, MRL24 (Figure 4G) shows one primary peak in the 19F NMR data, suggesting a single unique chemical environment for the CF3 group in MRL24. These trends generally hold within the context of the PPARγ/RXRα heterodimer (Figure 4C,H), although three of the four minor MRL20 19F chemical shift populations are not observed (Figure 4C), suggesting that heterodimerization with RXRα could influence the conformation of MRL20 bound to PPARγ. This can be rationalized as previous NMR studies have shown that the binding of the RXRα heterodimer partner to PPARγ causes chemical shift perturbations within H12 (Lu et al., 2008). Thus, because the acid group of MRL20 makes a hydrogen bond contact with Y473 in H12, and because the binding of RXRα influences the conformation of H12, heterodimerzation could have an impact on the conformational dynamics of receptor-bound MRL20.
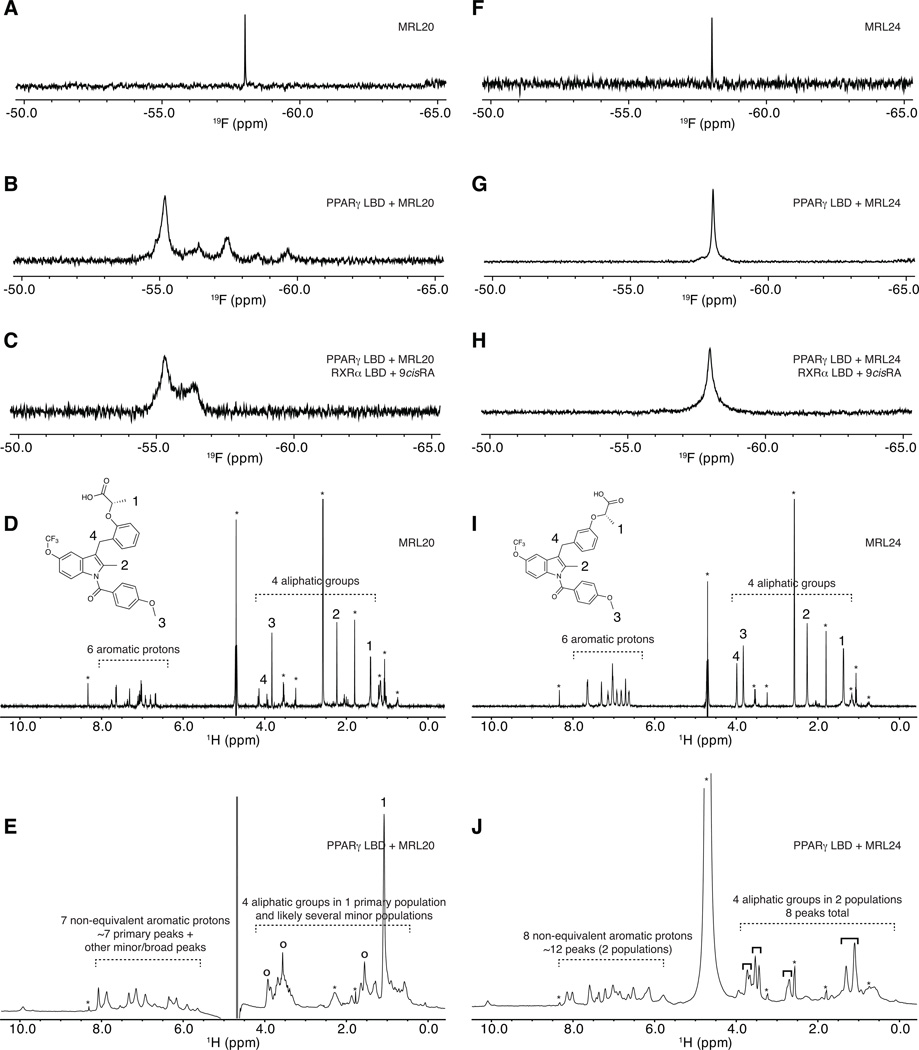
Figure 4. Receptor-bound ligand-observe NMR data reveals multiple binding modes for graded PPARγ modulators.
Ligand-observe 1D NMR data for MRL20 (A–E) and MRL24 (F–J), including 19F (A–C,F–H) and 1H (D,E,I,J) NMR data. 19F NMR data for MRL20 reveals that the single CF3 group (A) displays multiple populations when bound to PPARγ (B) and the PPARγ/RXRα heterodimer (C), including one primary state and one-to-several lowly populated states. 1H NMR data for MRL20 also reveals the aromatic and aliphatic groups (D) display a similar profile when bound to PPARγ (E). 19F NMR data for the single CF3 group of MRL24 (F) reveals a single resonance population when bound to PPARγ (G) or the PPARγ/RXRα heterodimer (H). 1H NMR data for MRL24 reveals that the aromatic and aliphatic NMR resonances (I) are in two populations when bound to PPARγ (J). Buffer signals are marked with an asterisk in (D,E,I,J); primary MRL20 signals are marked with open circles in (E); four groups of two MRL24 populations are marked with solid black brackets in (J). Figure S3 shows a zoomed in view of the aliphatic region for MRL24 in (J).
For the 1H NMR studies, 1D 1H spectra for MRL20 (Figure 4D) and MRL24 (Figure 4i) were used to assign aliphatic proton resonances (~1–4 ppm; inset structures), which are better resolved and fewer in number compared to the aromatic region (~6–8 ppm), after considering peaks arising from the buffer and bulk water (Figure S3). When bound to the PPARγ LBD, MRL20 (Figure 4E) shows a large signal likely corresponding to the methyl near carboxylic acid group (group 1 in the inset). There are three other peaks that likely correspond to the remaining aliphatic signals (groups 2–4, marked with ○), however several minor populations are observed. When considered with the MRL20 19F NMR data (Figure 4A–C), the data suggest a primary conformation of MRL20 when bound to PPARγ, as well as several other chemical states (conformations) that are lowly populated. However, the methyl group near the carboxylic acid (group 1 in the inset), which associates near helix 12 in the PPARγ LBP (ligand-binding pocket) likely exists in one unique chemical environment due to the nature of the dynamics (vide infra); this is further described in Figure S4.
MRL24 (Figure 4I) shows two populations of 1H resonances when bound to the PPARγ LBD (Figure 4J). For the more easily resolved aliphatic groups (Figure S3), the largest chemical shift change is observed for the methyl near carboxylic acid group (group 1 in the inset; Δδ = 0.2 ppm), followed by the methylether group (group 3; Δδ = 0.09 ppm), methylene group (group 4; Δδ = 0.07) and 2-methylindole group (group 2; Δδ = 0.05 ppm; resonances are broader and less intense compared to the others). When considered with the MRL24 19F NMR data (Figure 4F–H), the data suggest that the aliphatic groups of MRL24, which comprise two of the three ligand side chains, sample two conformations. However, the CF3 group of MRL24, which associates near helix 12 in the PPARγ LBP on the third ligand side chain, exists in one unique chemical environment (conformation); this is further described in Figure S4. For each of the four observable MRL24 protons in this aliphatic region (Figure 4I,J; between 1–4 ppm), two peaks are observed suggesting two primary ligand conformations in slow exchange with average population fractions of Pa≈0.6 and Pb≈0.4.
When considered with structures of the PPARγ LBD co-crystalized with these ligands (Bruning et al., 2007), the receptor-bound ligand-detected NMR data reveals mechanistic insight into the binding mode dynamics of the ligand (Figure 5A; Figure S4). NMR resonances originating from the ligand side chain near helix 12 within the LBP gives rise to one NMR chemical shift population (Figure 4; Figure S4). In contrast, NMR resonances for the ligand side chains that associate within the LBP near the β-sheet, which is larger in volume compared to the region near helix 12 (Sheu et al., 2005), display multiple populations (Figure 4; Figure S4). A model describing the dynamics of receptor-bound MRL24 consistent with our NMR observations would suggest that the ligand flips its binding mode around an axis of rotation positioned parallel to the 2-methylindole and CF3 groups. These groups display the smallest chemical shift change among all MRL24 aliphatic groups, whereas the methyl near carboxylic acid group and methylether group at the ends of two ligand side chains display the largest chemical shift changes.
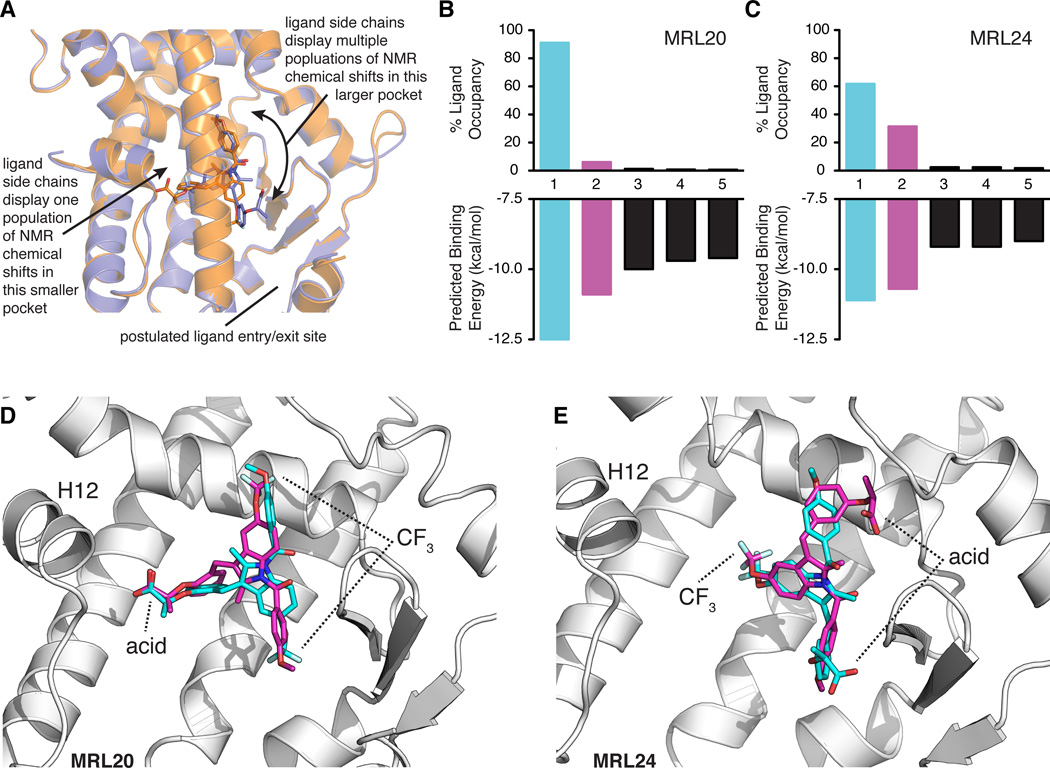
Figure 5. Model of MRL20 and MRL24 ligand dynamics within the ligand-binding pocket and corroboration via ligand molecular docking.
(A) The ligand-observe NMR data suggest that the acid and nearby methyl groups of MRL20 and the CF3 group of MRL24 bind in the same smaller pocket within the PPARγ LBP. In contrast, the region of the LBP where the other ligand side chains associate constitutes the largest cavity within the LBP, and NMR resonances corresponding to the ligand atoms in this region display multiple populations. This suggests the ligands sample multiple conformations/binding modes in this region of the LBP and is supported by protein NMR studies indicating multiple receptor conformations (Figure 3). (B–C) Average predicted binding energies and % ligand occupancy according to the canonical Boltzmann distribution from the molecular docking of MRL20 (B) and MRL24 (C). Data for the primary and secondary modes are colored blue and pink, respectively. (D–E) Primary (blue) and secondary (pink) binding modes obtained in the molecular docking results for MRL20 (D) and MRL24 (E). Primary binding modes (blue) closely match the binding mode captured in PPARγ crystal structures (Bruning et al., 2007), whereas the secondary binding modes (pink) display a ligand orientation flipped ~180° relative to the primary orientation in a manner concordant with our NMR data (A; Figure S4)
Molecular docking reveals the propensity for multiple ligand binding modes
To provide additional insight into the ability of MRL20 and MRL24 to adopt multiple binding modes, we performed molecular docking simulations using the program AutoDock Vina (Trott and Olson, 2010). Analysis of the top five binding models for the docking of MRL20 revealed one significant primary binding mode with ~91% occupancy (Figure 5B), according to the canonical Boltzmann distribution at the temperature used in the NMR experiments. Several other lowly populated binding modes were reported, including a secondary binding mode with ~6% occupancy, and three additional binding modes with occupancy values around or less than 1%. For MRL24, analysis of the top five binding modes (Figure 5C) revealed a primary binding mode with ~62% occupancy, a secondary binding mode with ~31% occupancy, whereas the remaining binding modes had occupancy values of ~2% or less. For both ligands, the primary binding mode (Figure 5D,E; blue) closely resembles the binding mode captured in PPARγ crystal structures (Bruning et al., 2007). The secondary binding mode (Figure 5D,E; pink) displays a ligand orientation flipped ~180° in a manner concordant with our NMR data (Figure S4) around an axis of rotation positioned parallel the ligand side chain associated near H12 within a smaller region of the LBP (Figure 5A). For MRL24, the axis of rotation is approximately parallel to the 2-methylindole and CF3 groups. For both ligands, the remaining three binding modes displayed the ligand associating near the LBP entry/exit site (Figure 5A). None of the docked binding modes predicted for MRL20 correspond to the crystallized MRL24 binding mode and vice versa. Boltzmann distribution analysis of the energy separation between the primary/crystal structure orientation, the secondary (flipped) binding mode and other modes indicates that the flipped mode could be significantly populated for both ligands, with MRL24 occupying the flipped state to a larger degree than MRL20 (Figure 5B,C). As schematically detailed in Figure S4 and discussed above, these docking results agree with our NMR observations and are consistent with the topology of the PPARγ LBP. Namely, the portion of the ligand that flips binding mode orientation in the molecular docking is located within a larger pocket of the LBP, and atoms of the ligand within this region of the pocket display multiple populations in the NMR data.
DISCUSSION
Much of what is known about NR structure-function relationships has been derived from hundreds of x-ray crystallography studies, which have captured static conformational “snapshots” of many ligand-receptor complexes. However, biological complexes are dynamic and undergo constant flexing throughout a hierarchy of motional time scales, which plays an important role in biological function. The concept of structural dynamics in the NR field has been derived primarily from the structural plasticity of H12 in the receptor LBD switching between an “on” (agonist) and “off” (antagonist) conformation in ligand-receptor co-crystal structures. More recently, HDX studies have revealed that the receptor dynamic profiles of some NR-ligand complexes correlate with the pharmacological phenotype of the ligand in a manner that could not be inferred from static crystal structures (Bruning et al., 2007; Dai et al., 2009; Dai et al., 2008; Zhang et al., 2010). However, one key observation lacking from these studies relates to the dynamics of ligand binding, in particular the ability to bind in different binding modes and the effect on receptor conformation. Although crystallography provides high-resolution atomic insight into the molecular details of NR ligand-receptor interactions and HDX is sensitive to detecting ligand-induced changes in receptor dynamics, these techniques are not suitable for observing multiple conformational populations in solution. Here, our solution NMR studies provide the first insight describing a potential role for both ligand and receptor dynamics in the mechanism of action of graded PPARγ modulators.
Our data suggest that the degree of partial agonism exhibited by MRL20 and MRL24 is associated with (1) the presence of two receptor populations in slow conformational exchange, (2) the degree of receptor intermediate conformational exchange within the LBP and AF-2 region, (3) the degree of receptor stabilization observed by NMR and HDX, and (4) the ability of the ligand to sample multiple binding modes and the relative binding mode occupancy. It is well accepted that the ability of a ligand to interact with H12 via a hydrogen bond with Tyr473 plays an important role in the stabilization of receptor conformation and transcriptional agonism. Our studies implicate a role for intermediate time scale dynamics in the H12/AF-2 surface as a likely origin for reduced transactivation response. This type of general phenomenon, where the dynamics of a protein negatively affects its function, has been previously described as “dynamic dysfunction” (Mauldin et al., 2009; Peng, 2009). In the case of MRL20 and MRL24, which are less efficacious agonists compared to rosiglitazone, sampling more than one ligand binding mode allosterically propagates a conformational disorder to surfaces of the receptor that are important for function, contributing to their reduced degree of agonism while independently contributing to their ability to stabilize the Ser245 (Ser273:PPARγ2) phosphorylation surface.
Our ligand-observe NMR data suggest that MRL20 prefers one conformation and samples several other lowly populated conformations. On the other hand, MRL24 samples two primary conformations with relative populations of ~60% and ~40%. This can be structurally rationalized by the observation that the MRL24 acid group forms a hydrogen bond with Ser342 on β3 within the large cavity of the PPARγ LBP. The formation of this hydrogen bond may lock the ligand into one of these two preferred conformations and slow the conformational exchange of MRL24 to the point where two significant populations are observed. In contrast, although MRL20 may prefer one binding mode, it samples several other binding modes at lower occupancy, perhaps as it does not form a hydrogen bond with Ser342. These observations and relative populations are corroborated by molecular docking studies on MRL20 and MRL24 that reveal these ligands can flip binding mode orientation in the large pocket within the LBP that displays ligand-specific differences in receptor intermediate and slow time scale dynamics. Two different interpretations of our ligand-observe NMR data would suggest that either the ligand rotates while bound in the pocket, or that it binds in two different orientations as a result of ligand on-off exchange. In terms of the latter interpretation, the slow rate of exchange observed is consistent with the low Kd of these ligands. The ability of MRL20 and MRL24 to sample multiple binding modes would certainly impact the dynamics of the receptor, which is apparent in the relative receptor populations observed in our NMR studies. When considered with graded transactivation profiles and differences in efficacy for blocking Ser245 phosphorylation, the data suggest that the dynamics of the ligand and receptor would both affect function. One caveat to the docking results is that, because the bonds are rotatable during the in silico docking, programs such as AutoDock Vina and others can have difficultly finding a “correct” bound conformation, leading to artifactual results such as 180° flipped binding modes (Trott and Olson, 2010). However, our docking results (Figure 5b–e) are supported by our NMR results (Figures 3 and 4): (i) in terms of the ability to sample multiple binding modes; (ii) the relative populations of the binding modes (% ligand occupancy in the docking vs. ligand Pa and Pb in the NMR studies); and (iii) chemical shift differences of the functional groups of MRL24 that suggest two binding modes rotating about an axis of rotation that would result in a 180° flip. Our results perhaps challenge the notion that there could be one “correct” in silico docked conformation.
Despite the large differences in receptor stabilization induced by MRL24 binding, it is just as good as rosiglitazone, if not better, at blocking Cdk5-mediated phosphorylation of Ser245 in vitro and in vivo (Choi et al., 2010). Our NMR data reveal that MRL24 better stabilizes the β-sheet region and the Ser245 site compared to rosiglitazone, as inferred via relative NMR cross-peak intensities for residues in these regions. Also in agreement with our data showing that MRL20 is less efficacious than these ligands in blocking Ser245 phosphorylation in vitro, our NMR data reveal that MRL20 is less effective at stabilizing the β-sheet region and the Ser245 site. Furthermore, MRL20 and MRL24 both significantly and differentially impact the intermediate and slow time scale dynamics of PPARγ near the β-sheet region, H2, H2’ and H7. These are regions that are near or involved in a long-range contact between the PPARγ LBD and the RXRα DBD in the intact PPARγ/RXRα structure (Chandra et al., 2008). This observation, combined with our NMR observations revealing that these ligands regulate the dynamics of this surface, suggests a potential for long-range, ligand-mediated dynamic allostery between RXRα DBD and PPARγ LBD. However, a recent solution analysis using SAXS, SANS and FRET suggests this long-range interaction may only be stabilized by a minor transient conformation (Rochel et al., 2011).
There is an apparent correlation between the relative receptor NMR populations induced by ligand binding and the degree of ligand-induced transactivation. For example, rosiglitazone stabilizes PPARγ Lys230 (Figure 3A) into one chemical shift population (Pa≈1, Pb≈0), whereas two populations are observed for the graded agonists MRL20 (Pa=0.81, Pb=0.19) and MRL24 (Pa=0.76, Pb=0.24). Though the functional implications of the relative populations Pa and Pb are not completely apparent, generally the shift in population balance is associated with the degree of agonism. Furthermore, our observation that MRL20 and MRL24 sample multiple binding modes suggests that perhaps one binding mode may be more active in terms of classical transactivation. More apparent is the correlation between the degree of H11/H12 stabilization on the intermediate time scale and graded transactivation response. MRL20, which displays ~60% transactivation compared to rosiglitazone, only induces a partial stabilization of H11 and H12 compared to the full stabilization afforded by rosiglitazone. The degree of stabilization induced by MRL24 is even less, which is consistent with its ~25% transactivation response compared to rosiglitazone. As noted before, these observations are inconsistent with the single H12 conformation observed in the ligand-receptor co-crystal structures (Bruning et al., 2007). The combination of our NMR and HDX data support a model by which increased conformational mobility of H12 negatively affects coregulator interaction and transactivation (Bruning et al., 2007). Thus, the “active” conformation of H12 in the ligand-receptor co-crystal structures may indeed represent a conformational “snapshot” of the receptor-coregulator interaction, but is not fully representative of the receptor LBD conformation in solution.
There have been previous suggestions for a role of ligand dynamics in the mechanism of graded agonism. The regioisomeric PPARγ ligands studied here, MRL20 and MRL24, crystallized into two distinct conformations within the LBP of PPARγ (Figure 1F) with binding modes associated with their respective activity profiles (Bruning et al., 2007). However, we have shown here that the primary binding mode of one ligand (e.g. MRL20) is not the secondary binding mode for the other (e.g. MRL24). Our previous NMR study revealed that the PPARγ partial agonist nTZDpa induces a receptor conformation that is distinct from and less stable than the receptor conformation when bound to the full agonist rosiglitazone (Berger et al., 2003). An estrogen receptor (ER) partial agonist, WAY-166916, bound differently to the receptor when mutant proteins were used that stabilized the active and inactive conformations (Bruning et al., 2010). In this study, a series of WAY-166916-derived ligands were synthesized that had graded transcriptional response profiles and crystallized to the ER mutant proteins in a manner consistent with their activity profiles. Interestingly, the authors state that ligand-receptor co-crystal structures of ER partial agonists are hard to obtain, requiring protein mutations to stabilize receptor conformations. Indeed, this may arise due to the general ability of NR partial agonists to bind in multiple binding modes in a dynamic nature as opposed to a more static nature preferred for crystallization. This implies that ligand dynamics is likely involved in the mechanism of partial NR agonism. In terms of non-NR proteins, differences in binding modes for similar compounds have been noted (Teague, 2003) in crystal structures of transthyretin (Faig et al., 2001) and NAD(P)H:Quinone Oxidoreductase 1 (Faig et al., 2001), where similar ligands also display flipped binding modes. Technologically related to our work, a ligand-detected NMR study revealed that a single ligand can have multiple binding modes when bound to riboflavin synthase, whereas another ligand that binds the same protein has only one binding mode (Scheuring et al., 1996).
Our observations could have significant implications in the future development of PPARγ partial agonists, or ligands with no agonism, which are highly sought after modulators because they have potent insulin sensitizing activity but are anti-adipogenic with less propensity for edema, weight gain and other unwanted effects (Choi et al., 2011; Grether et al., 2010; Houtkooper and Auwerx, 2010; Kahn and McGraw, 2010). Our observations may also have implications for rational/structure-based drug design, where it is generally assumed that ligands have one binding mode, typically inferred via crystal structures (Guasch et al., 2011), and differences in binding modes between highly related ligands may be considered “surprising” (Teague, 2003). Future studies will determine if this is a general feature for graded/partial receptor agonism in other systems, as well as determine if selecting for ligands with a single binding mode vs. multiple binding modes can have impact on the desired pharmacological response of the ligand. Lastly, our work here shows the great utility in combining multidisciplinary approaches, including x-ray crystallography, HDX and NMR spectroscopy, to provide detailed insight into mechanism of action of small molecule ligands.
EXPERIMENTAL PROCEEDURES
Protein expression, purification and sample preparation
DNA encoding the PPARγ (NR1C3) LBD (residues 205–477; UniProt ID P37231, isoform 1) and RXRα (NR2B1) LBD (residues 223–462; UniProt ID P19793) was amplified by PCR and cloned into the expression vector pET-46 using the Ek/LIC system (EMD Chemicals/Novagen) as a tobacco etch virus (TEV) protease-cleavable N-terminal HisTag fusion protein. Expression and purification details are described in Supplementary Methods. The rosiglitazone-bound sample was 1.0 mM; the MRL20, MRL24 and apo samples were 0.25 mM; in 20 mM potassium phosphate (pH 7.4), 50 mM potassium chloride, 0.05% sodium azide (NMR buffer). A [2H,13C,15N]-PPARγ/unlabeled-RXRα LBD (RXRα complexed to 9-cis-retinoic acid) heterodimer sample was prepared from the NMR sample of [2H,13C,15N]-PPARγ LBD bound to MRL24. For the ligand-observe experiments, 1H experiments were performed on the aforementioned ligand-bound receptor samples; ligand only samples were at 50 µM in NMR buffer at 0.5% DMSO; and other unlabeled-receptor ligand experiments contained 200–500 µM protein. Ligands were added in 1.35 to 2 fold molar excess to approximately 10 µM protein and then buffer exchanged and/or concentrated.
NMR spectroscopy
NMR data were collected at 25 °C on a Bruker spectrometer at 700 MHz 1H frequency equipped with a TXI probe and Varian spectrometers at 600 and 800 MHz 1H frequency with a conventional (600 MHz) or cryogenically cooled (800 MHz) triple-resonance probe. The proton carrier frequency was set coincident with the water resonance for all experiments. Backbone assignments for the PPARγ LBD bound to rosiglitazone (1.0 mM protein), MRL24 (250 µM protein) and MRL20 (250 µM protein) were obtained using 3D TROSY-based assignment experiments available in Varian VNMRJ/BioPack or Bruker Topspin, including HNCO, HNCA, HN(CO)CA, HN(CA)CB and HN(COCA)CB, as well as 15N-NOESY-HSQC or 15N-NOESY-TROSY-HSQC (τmix= 120 ms), using [2H,13C,15N]-labeled protein. 2D [1H-15N]-TROSY-HSQC and 3D HNCO data were collected on apo/ligand-free [13C,15N]-Histag-PPARγ LBD. Data were processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRViewJ (Johnson, 2004) (One Moon Scientific, Inc.). Backbone chemical shift assignments were obtained for 245, 186 and 170 out of 264 non-proline PPARγ LBD residues in the presence of rosiglitazone (93%), MRL24 (70%) and MRL20 (64%), respectively. Chemical shift assignments for the PPARγ LBD complexed to rosiglitazone, MRL24 and MRL20 have been deposited in the BioMagResBank (BMRB; http://www.bmrb.wisc.edu) under accession codes 17975, 17976 and 17977, respectively. ZZ-exchange spectroscopy (EXSY) was used to attempt to quantify the rate of slow exchange (Farrow et al., 1994) using relaxation delays ranging from 5 to 900 ms. The heterodimer sample concentration was estimated to be ~75–100 µM and had relative decrease of ~75% in 1D 1H NMR signal compared to the [2H,13C,15N]-PPARγ sample bound to MRL24.
Ligand-detected NMR was performed using 1D 19F and 1D 1H NMR experiments, including a 19F NMR experiment without 1H decoupling at 659 MHz (free and receptor-bound ligand), a 1D 1H NMR experiment with water suppression (Hwang and Shaka, 1995) at 700 MHz (free ligand), and a [13C,15N]-isotope filtered experiment with water suppression (Ikura and Bax, 1992) at 700 MHz (ligand bound to [2H,13C,15N]-PPARγ LBD). Protein-ligand complex concentrations ranged from 200–500 µM. In the case of the receptor-bound ligand NMR data, which reveals the ligand is in slow exchange between multiple conformations, specific assignment of the receptor-bound ligand protons was complicated because NMR spin coupling patterns are not well resolved due to the slower ligand correlation time, comparable to that of the receptor, compared to free ligand. Receptor-bound chemical shift assignments were mapped using the relative peak intensity profiles and chemical shift values for groups 1, 3 and 4, leaving peaks for group 2, which does not strictly follow the same intensity profile as free ligand perhaps due to chemical shift line broadening. Data were processed and analyzed using dataChord Spectrum Analyst (One Moon Scientific, Inc.).
Ligands
Rosiglitazone was obtained commercially (Cayman Chemical). MRL20 and MRL24 were synthesized in-house using previously described methods (Acton et al., 2005).
HDX-MS
Solution phase amide HDX was performed on liganded and unliganded PPARγ LBD using a fully automated system and in-house software described previously (Chalmers et al., 2006; Chalmers et al., 2011; Pascal et al., 2009). Changes reported in HDX are from direct comparisons between unliganded and liganded PPARγ LBD. Details are described in Supplemental Methods.
Ligand docking
AutoDock Vina (Trott and Olson, 2010) was used for molecular docking studies of MRL20 and MRL24 to their respective crystal structures (Bruning et al., 2007) using standard parameters except for the degree of exhaustiveness (=30). The search space comprised the entire LBP of the PPARγ LBD. The ligand occupancy of the various states was calculated (1) assuming a canonical ensemble where the number of molecules, the volume and the temperature are fixed; and (2) all states were considered equally degenerate. The fraction of the total ligands occupying a docked state and energy was calculated using the canonical Boltzmann distribution:
where εo is the energy of the lowest energy level of the predicted ligand conformation, εi is the energy of the other energy levels, kB is the Boltzman constant, T is the temperature and the denominator is the sum of all the probabilities of the states analyzed for each ligand.
Cell-based reporter assay
293T cells were cotransfected in batch by adding 4.5 µg each of full-length Gal4-PPARγ and UAS-luciferase reporter using 27 µL X-treme Gene 9 transfection reagent (Roche) in Opti-mem reduced serum media (Gibco). Cells were plated at a density of 4×106 in 100×20mm polystyrene dishes (BD Falcon) with DMEM containing 10% FBS. Following 18 hour incubation at 37°C, 5% CO2, transfected cells were plated in triplicate in white 384-well plates (Perkin Elmer) at a density of 10,000 cells/well. Cells were treated with either 0.1% DMSO vehicle only or compound MRL-20, MRL-24 or Rosiglitazone in increasing doses from 333 pM-10 µM for 18 hours. The luciferase was quantitated with 20 µL Brite Lite Plus (Perkin Elmer) and read in 384-well Luminescence Perkin Elmer EnVision Multilabel plate reader. Graphs were plotted in GraphPad Prism Software as fold change of treated cells over DMSO control cells.
In vitro Cdk5/p35 kinase biochemical assay
In vitro Cdk5 kinase assay was performed as previously described (Choi et al., 2010). Briefly, active Cdk5/p35 kinase was purchased from Millipore. 0.5 mg of purified WT PPARγ was incubated with 0.25 mg active Cdk5 in kinase assay buffer (25 mM Tris-HCl pH 7.5, 5 mM β-glycerophosphate, 2 mM dithiothreitol (DTT), 0.1 mM Na3VO4, 10 mM MgCl2) for 2 hr at 30°C. 20 mM ATP was added to complex for 15 min at 30°C. PPARγ ligands were pre-incubated with receptor for 1 hr prior to addition of ATP and the assay was performed. Phosphorylation of substrates after SDS–PAGE was analyzed with an anti-phospho Ser245 (Ser273 in PPARγ2) polyclonal antibody. Densitometry analysis using ImageJ (NIH) was used to determine the relative efficacies for the ligands in blocking phosphorylation of Ser245.
Structural nomenclature and analysis
The sequence and secondary structure nomenclauture for the PPARγ LBD has been described (Uppenberg et al., 1998). Secondary structure nomenclature: helix is abbreviated as “H”; β-strand as “β”; and loop as “L” with numbers corresponding to the helix preceding and following the loop. PyMOL was used for structural visualization (DeLano Scientific, South San Francisco CA), and THESEUS (Theobald and Wuttke, 2006, 2008) was used for principal component analysis of PPARγ ligand-bound structures.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for support from B. Pascal and S. Willis for software analyzing the HDX data. This work was supported in part by James and Esther King Biomedical Research Program, Florida Department of Health (1KN-09 to D.J.K.), start-up funds from The Scripps Research Institute (to D.J.K.), the US National Institutes of Health (NIH) National Institute of General Medical Sciences (R01-GM084041 to P.R.G.; R01-GM063855 to M.R.) and the National Center for Research Resources (RR19077 and RR027755 to M.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acton JJ, 3rd, Black RM, Jones AB, Moller DE, Colwell L, Doebber TW, Macnaul KL, Berger J, Wood HB. Benzoyl 2-methyl indoles as selective PPARgamma modulators. Bioorganic & medicinal chemistry letters. 2005;15:357–362. doi: 10.1016/j.bmcl.2004.10.068. [DOI] [PubMed] [Google Scholar]
- Berger JP, Petro AE, Macnaul KL, Kelly LJ, Zhang BB, Richards K, Elbrecht A, Johnson BA, Zhou G, Doebber TW, et al. Distinct properties and advantages of a novel peroxisome proliferator-activated protein [gamma] selective modulator. Mol Endocrinol. 2003;17:662–676. doi: 10.1210/me.2002-0217. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc Natl Acad Sci U S A. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. X-ray crystallography and NMR reveal complementary views of structure and dynamics. Nat Struct Biol. 1997;4 Suppl:862–865. [PubMed] [Google Scholar]
- Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamenecka TM, He Y, Nettles KW, Griffin PR. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. doi: 10.1016/j.str.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Bruning JB, Parent AA, Gil G, Zhao M, Nowak J, Pace MC, Smith CL, Afonine PV, Adams PD, Katzenellenbogen JA, et al. Coupling of receptor conformation and ligand orientation determine graded activity. Nat Chem Biol. 2010;6:837–843. doi: 10.1038/nchembio.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- Chalmers MJ, Pascal BD, Willis S, Zhang J, Iturria SJ, Dodge JA, Griffin PR. Methods for the Analysis of High Precision Differential Hydrogen Deuterium Exchange Data. Int J Mass Spectrom. 2011;302:59, 68. doi: 10.1016/j.ijms.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR-alpha nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Banks A, Kamenecka T, Busby S, Chalmers M, Kumar N, Kuruvilla D, Shin Y, He Y, Bruning J, et al. Anti-diabetic actions of a non-agonist PPAR γ ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–481. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Banks AS, Estall JL, Kajimura S, Bostrom P, Laznik D, Ruas JL, Chalmers MJ, Kamenecka TM, Bluher M, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SY, Burris TP, Dodge JA, Montrose-Rafizadeh C, Wang Y, Pascal BD, Chalmers MJ, Griffin PR. Unique Ligand Binding Patterns Between Estrogen Receptor and Revealed by Hydrogen/Deuterium Exchange. Biochemistry. 2009;48:9668–9676. doi: 10.1021/bi901149t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SY, Chalmers MJ, Bruning J, Bramlett KS, Osborne HE, Montrose-Rafizadeh C, Barr RJ, Wang Y, Wang M, Burris TP, et al. Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method. Proc Natl Acad Sci U S A. 2008;105:7171–7176. doi: 10.1073/pnas.0710802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Faig M, Bianchet MA, Winski S, Hargreaves R, Moody CJ, Hudnott AR, Ross D, Amzel LM. Structure-based development of anticancer drugs: complexes of NAD(P)H:quinone oxidoreductase 1 with chemotherapeutic quinones. Structure. 2001;9:659–667. doi: 10.1016/s0969-2126(01)00636-0. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacological reviews. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- Grether U, Klaus W, Kuhn B, Maerki HP, Mohr P, Wright MB. New Insights on the mechanism of PPAR-targeted drugs. ChemMedChem. 2010;5:1973–1976. doi: 10.1002/cmdc.201000446. [DOI] [PubMed] [Google Scholar]
- Guasch L, Sala E, Valls C, Blay M, Mulero M, Arola L, Pujadas G, Garcia-Vallve S. Structural insights for the design of new PPARgamma partial agonists with high binding affinity and low transactivation activity. J Comput Aided Mol Des. 2011 doi: 10.1007/s10822-011-9446-9. [DOI] [PubMed] [Google Scholar]
- Hamuro Y, Coales SJ, Morrow JA, Molnar KS, Tuske SJ, Southern MR, Griffin PR. Hydrogen/deuterium-exchange (H/D-Ex) of PPARgamma LBD in the presence of various modulators. Protein Sci. 2006;15:1883–1892. doi: 10.1110/ps.062103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Ström A, Treuter E, Warner M, et al. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Auwerx J. Obesity: New life for antidiabetic drugs. Nature. 2010;466:443–444. doi: 10.1038/466443a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang T-L, Shaka AJ. Water Suppression That Works. Excitation Sculpting Using Arbitrary Waveforms and Pulse Field Gradients. J Mag Reson Ser A. 1995;112:275–279. [Google Scholar]
- Ikura M, Bax A. Isotope-filtered 2D NMR of a protein-peptide complex : study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J Am Chem Soc. 1992;114:2433–2440. [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Wilson EM, Li Y, Moller DE, Smith RG, Zhou G. Ligand-induced stabilization of PPARgamma monitored by NMR spectroscopy: implications for nuclear receptor activation. J Mol Biol. 2000;298:187–194. doi: 10.1006/jmbi.2000.3636. [DOI] [PubMed] [Google Scholar]
- Kahn BB, McGraw TE. Rosiglitazone, PPARgamma, and type 2 diabetes. The New England journal of medicine. 2010;363:2667–2669. doi: 10.1056/NEJMcibr1012075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochimica et biophysica acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Chen M, Stanley SE, Li E. Effect of heterodimer partner RXRalpha on PPARgamma activation function-2 helix in solution. Biochem Biophys Res Commun. 2008;365:42–46. doi: 10.1016/j.bbrc.2007.10.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauldin RV, Carroll MJ, Lee AL. Dynamic Dysfunction in Dihydrofolate Reductase Results from Antifolate Drug Binding: Modulation of Dynamics within a Structural State. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JT, Collins JL, Pearce KH. The nuclear receptor superfamily and drug discovery. ChemMedChem. 2006;1:504–523. doi: 10.1002/cmdc.200600006. [DOI] [PubMed] [Google Scholar]
- Pascal BD, Chalmers MJ, Busby SA, Griffin PR. HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom. 2009;20:601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JW. Communication breakdown: protein dynamics and drug design. Structure. 2009;17:319–320. doi: 10.1016/j.str.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mely Y, et al. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- Scheuring J, Fischer M, Cushman M, Lee J, Bacher A, Oschkinat H. NMR analysis of site-specific ligand binding in oligomeric proteins. Dynamic studies on the interaction of riboflavin synthase with trifluoromethyl-substituted intermediates. Biochemistry. 1996;35:9637–9646. doi: 10.1021/bi9600916. [DOI] [PubMed] [Google Scholar]
- Sheu SH, Kaya T, Waxman DJ, Vajda S. Exploring the binding site structure of the PPAR gamma ligand-binding domain by computational solvent mapping. Biochemistry. 2005;44:1193–1209. doi: 10.1021/bi048032c. [DOI] [PubMed] [Google Scholar]
- Teague SJ. Implications of protein flexibility for drug discovery. Nature reviews Drug discovery. 2003;2:527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. Accurate structural correlations from maximum likelihood superpositions. PLoS Comput Biol. 2008;4:e43. doi: 10.1371/journal.pcbi.0040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppenberg J, Svensson C, Jaki M, Bertilsson G, Jendeberg L, Berkenstam A. Crystal structure of the ligand binding domain of the human nuclear receptor PPARgamma. J Biol Chem. 1998;273:31108–31112. doi: 10.1074/jbc.273.47.31108. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Garcia-Ordonez RD, Pascal BD, Burris TP, Dodge JA, Griffin PR. Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within vitamin D receptor/retinoid X receptor heterodimer. Structure. 2010;18:1332–1341. doi: 10.1016/j.str.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.