Abstract
Background
Circulating levels of soluble intercellular adhesion molecule-1 (sICAM-1), soluble P-selectin (sP-selectin), and soluble E-selectin (sE-selectin) have been associated with variation at the ABO locus. To evaluate these associations and the effect sizes, we performed a meta-analysis with new and previous reported data for polymorphism rs579459.
Methods and Results
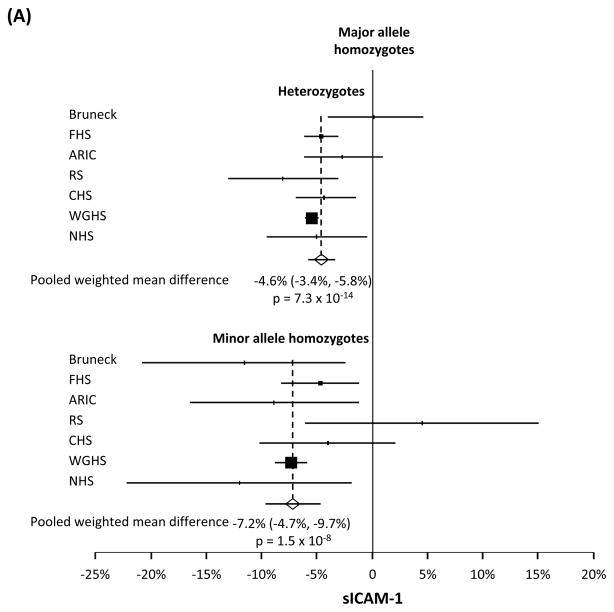
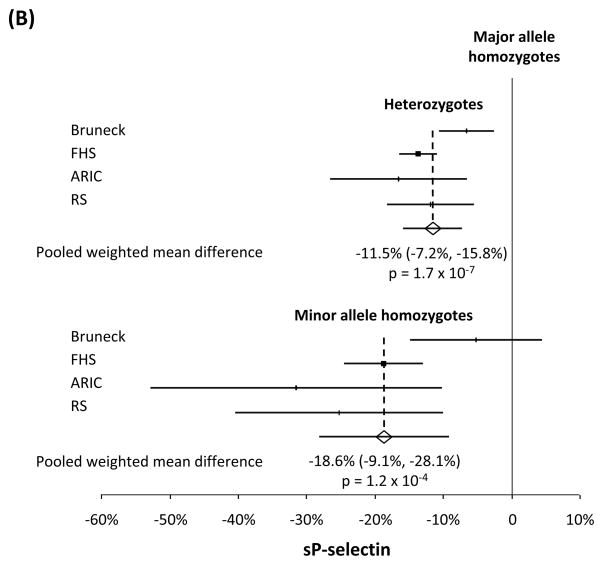
Compared with major allele homozygotes, heterozygotes and minor allele homozygotes had 4.6% (95%CI=3.4–5.8%, p=7.3×10−14) and 7.2% (95%CI=4.7–9.7%, p=1.5×10−8), respectively, lower sICAM-1 levels (n=33,671). An allele dose dependent association also was observed for sP-selectin (n=4,921), with heterozygotes and minor allele homozygotes having 11.5% (95%CI=7.2–15.8%, p=1.7×10−7) and 18.6% (95%CI=9.1–28.1%, p=1.2×10−4), respectively, lower levels than in major allele homozygotes. A larger effect size, again consistent with an additive genetic model, was seen for sE-selectin (n=2,860) whose level was 25.6% (95%CI=19.0–32.2%, p=2.1×10−14) lower in heterozygotes and 43.3% (95%CI=36.9–49.3%, p=4.3×10−42) lower in minor allele homozygotes, than in major allele homozygotes.
Conclusions
The data support the association of variation at the ABO locus with sICAM-1, sP-selectin and sE-selectin levels.
Keywords: Cell adhesion molecules, plasma, genetics, cardiovascular disease
Leukocyte recruitment plays an important role in inflammatory diseases.1 It typically begins with leukocyte rolling on the endothelium, followed by leukocyte attachment to endothelial cells and subsequently trans-endothelial migration. Rolling involves the interaction of leukocytes with P-selectin and E-selectin on endothelial cells, whilst leukocyte attachment to endothelial cells is mediated by intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1.1
Blood contains soluble forms of ICAM-1 (sICAM-1), P-selectin (sP-selectin) and E-selectin (sE-selectin), generated by shedding of ectodomains of the membrane-bound forms of these molecules or produced from transcript variants lacking the transmembrane domain.2 Increased circulating levels of sICAM-1, sP-selectin and/or sE-selectin have been associated with a number of diseases such as coronary heart disease and diabetes.3–7 The levels of sICAM-1, sP-selectin and sE-selectin are under genetic influences, with heritability estimates being 0.24–0.63, 0.45–0.70, and 0.50–0.64, respectively.8–10 Genome-wide association studies of sICAM-1, sP-selectin and sE-selectin levels have shown that they are associated with single nucleotide polymorphisms (SNPs) at the ABO locus.11–14 Interestingly, genome-wide association studies of coronary heart disease (CHD) have revealed an association between CHD and variation at the ABO locus.15;16
To more robustly evaluate the associations of sICAM-1, sP-selectin and sE-selectin with the ABO locus, and more reliably estimate the effect sizes, we performed a meta-analysis. We included new data from the Bruneck Study, data from several reported studies,11–14 and additional data from one of these reported studies.11
Methods
To identify association studies of SNPs at the ABO locus in relation to levels of sICAM-1, sP-selectin and/or sE-selectin, we performed systematic searches of PubMed, scanned the reference lists of original reports, and communicated with authors of the included studies. The electronic searches combined search terms related to polymorphisms at the ABO locus (e.g. ABO, polymorphism, SNP, variation, and variant) and ICAM-1, P-selectin, or E-selectin. The searches identified four publications. In two of these publications,12;13 SNP rs579459 showed the strongest association with sP-selectin or sE-selectin levels among all tested SNPs at the ABO locus. In another study (in which rs579459 was not directly typed),11 SNP rs507666 had the most significant association with sICAM-1 levels among all tested SNPs at this locus. In the fourth study (which also did not type rs579459 directly),14 SNP rs651007 was the top SNP at the ABO locus associated with sICAM-1 and sE-selectin levels. An analysis using the SNAP program (http://www.broadinstitute.org/mpg/snap/) with data from the 1000 Genomes Project showed that rs579459 was in perfect linkage disequilibrium (LD) with rs651007 and in near perfect LD (r2=0.96) with rs507666, in individuals of European ancestry.
We genotyped the Bruneck cohort17 for SNP rs579459 using the KASPar method. sICAM-1, sP-selectin, and sE-selectin levels in the Bruneck cohort had been measured by enzyme-linked immunosorbent assay as described previously.17;18 The Bruneck Study was approved by the local ethics committee and all participants gave their written informed consent.
We performed a meta-analysis with data from the Bruneck cohort and the four reported studies11–14 as well as additional data from one of these reported studies, the WGHS study11 for which ethic approval was granted by the institutional review board. For the meta-analysis, we only used summary statistic data from the cohorts and did not receive individual participant data. The meta-analysis included seven datasets for sICAM-1, four for sP-selectin, and four for sE-selectin. For the meta-analysis, data of unadjusted mean and standard deviation of sICAM-1, sP-selectin and sE-selectin levels according to genotypes were provided by authors of three of the previous studies11;12;14 where this information was not available in the papers, and were extracted from the report of the other study.13 With the use of the StatsDirect and Comprehensive Meta Analysis Version 2.0 software, we performed meta-analysis of weighted mean difference (wmd) in the percentage of and the unbiased standardized effect size (estimator d19) for each adhesion molecule, comparing minor allele homozygotes to heterozygotes and separately minor allele homozygotes to major allele homozygotes. The StatsDirect software provided the pooled mean effect size estimate (wmd+ or d+) with a 95% confidence interval, a chi-square statistic and probability of this pooled effect size being equal to zero.19 Consistency of findings across studies was assessed by the I2 statistic.20 Evidence of publication bias was assessed using funnel plots and the Egger test.21 Possible reasons for heterogeneity were investigated by meta-regression analysis.
Results and Discussion
The characteristics of study subjects are summarized in Table 1. A total of 33,671 subjects were available for the meta-analysis of sICAM-1, 4,921 for sP-selectin, and 2,860 for sE-selectin.
Table 1.
Summary of participating studies for the meta-analysis
| Participating studies | Subjects | Age (years)* | Female (%) | Assay | SNP | Number of subjects | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Method | Intra assay CV | Inter assay CV | A/A genotype | A/a genotype | a/a genotype | Total | ||||||
| sICAM-1 | Bruneck17 | Population-based | 63±11 | 51.1 | Plasma | ELISA | 5.1% | 6.9% | rs579459 | 440 | 268 | 33 | 741 |
| FHS12 | Community-based | 49±14 | 45.9 | Serum | ELISA | 3.9% | 3.9% | rs579459 | 4,176 | 2,340 | 329 | 6,845 | |
| ARIC12 | Community-based | 56±5 | 38.4 | Plasma | ELISA | 4.0% | 5.1% | rs579459 | 495 | 287 | 43 | 825 | |
| RS12 | Community-based | 70±9 | 53.3 | Plasma | ELISA | 6.9% | rs579459 | 351 | 214 | 35 | 600 | ||
| CHS12 | Population-based | 73±6 | 42.8 | Plasma | ELISA | 5.0% | rs579459 | 855 | 556 | 69 | 1,480 | ||
| WGHS11 | Population-based | 55±7 | 100 | Plasma | ELISA | 6.7% | rs507666† | 14,391 | 6,857 | 936 | 22,184 | ||
| NHS14 | Type 2 diabetes | 56±7 | 100 | Plasma | ELISA | 3.3–4.8% | rs651007‡ | 612 | 337 | 47 | 996 | ||
| sP-selectin | Bruneck17 | Population-based | 63±11 | 51.1 | Plasma | ELISA | 5.5% | 6.9% | rs579459 | 440 | 268 | 33 | 741 |
| FHS12 | Community-based | 61±10 | 45.6 | Plasma | ELISA | 3.2% | rs579459 | 1,872 | 1,000 | 164 | 3,036 | ||
| ARIC12 | Community-based | 57±5 | 35.7 | Plasma | ELISA | 3.9% | 5.8% | rs579459 | 432 | 265 | 41 | 738 | |
| RS12 | Community-based | 69±9 | 48.8 | Plasma | ELISA | <5% | <10% | rs579459 | 253 | 135 | 18 | 406 | |
| sE-selectin | Bruneck17 | Population-based | 63±11 | 51.1 | Plasma | ELISA | 4.8% | 7.4% | rs579459 | 440 | 268 | 33 | 741 |
| DCCT/EDIC13 | Type 1 diabetes | 39±7 | 46 | Serum | SLPA | <2% | 5% | rs579459 | 452 | 209 | 24 | 685 | |
| DCCT siblings13 | Non-diabetics | 45±9 | 57 | Serum | SLPA | <2% | 5% | rs579459 | 280 | 143 | 15 | 438 | |
| NHS14 | Type 2 diabetes | 56±7 | 100 | Plasma | ELISA | 4.5–6.2% | rs651007‡ | 612 | 337 | 47 | 996 | ||
All participants were of European ancestry. FHS, Framingham Heart Study; ARIC, Atherosclerosis Risk in Communities; RS, Rotterdam Study; CHS, Cardiovascular Health Study; WGHS, Women’s Genome Health Study; NHS, Nurses’ Health Study; DCCT, Diabetes Control and Complications Trial; EDIC, Epidemiology of Diabetes Intervention and Complications.
Genotype distributions in the various cohorts were all consistent with Hardy-Weinberg equilibrium except for WGHS (sICAM-1, p=0.001) and FHS (sP-selectin, p=0.046). SNP, single nucleotide polymorphism; A/A genotype, major allele homozygotes; A/a genotype, heterozygotes; a/a genotype, minor allele homozygotes; CV, coefficient of variation; ELISA, Enzyme-linked immunosorbent assay; SLPA; SearchLight™ Proteome Array.
mean ± standard deviation.
in nearly complete linkage disequilibrium with SNP rs579459 (r2 = 0.96) based on data from the 1000 Genomes Project;
in complete linkage disequilibrium with SNP rs579459 (r2 = 1) based on data from the 1000 Genomes Project
The meta-analysis showed that sICAM-1 levels were 4.6% (95% CI 3.4–5.8%) lower in heterozygotes and 7.2% (4.7–9.7%) lower in minor allele homozygotes, than in major allele homozygotes (p=7.3×10−14 and p=1.5×10−8, Figure 1A). Similarly, an allele dose dependent association was observed for sP-selectin, with heterozygotes and minor allele homozygotes having 11.5% (7.2–15.8%) and 18.6% (9.1–28.1%), respectively, lower levels than in major allele homozygotes (p=1.7×10−7 and p=1.2×10−4, Figure 1B). An allele dose dependent association also was seen for sE-selectin whose level was 25.6% (19.0–32.2%) lower in heterozygotes and 43.3% (36.9–49.3%) lower in minor allele homozygotes, than in major allele homozygotes (p=2.1×10−14 andp=4.3×10−42, Figure 1C). Standardized effect size was larger for sE-selectin than for sICAM-1 and sP-selectin (Supplemental Figures S1 to S3). We noted heterogeneity (Supplemental Table 1) which a meta-regression analysis indicated was not attributed to differences among individual studies in age, sex, type of subjects (population-based or diabetics), number of subjects (n>1000 or <1000), type of blood sample used (plasma or serum) or which SNP studied, although the meta-regression analysis had low power due to the relatively small numbers of individual studies. There was no evidence of publication bias. We observed correlations between sICAM-1, sP-selectin and sE-selectin levels (Supplemental Table 2).
Figure 1.
Weighted mean difference by genotype in soluble intercellular adhesion molecule-1 (sICAM-1), soluble P-selectin (sP-selectin), and soluble E-selectin (sE-selectin) levels. Data shown are weighted mean difference ± 95% confidence interval in circulating levels of sICAM-1 (panel A), sP-selectin (panel B) and sE-selectin (panel C), comparing heterozygotes or minor allele homozygotes, to major allele homozygotes, in a random-effects model.
SNP rs507666 is located within the ABO gene, and SNP rs579459 and rs651007 are in its proximity. The ABO gene encodes a glycosyltransferase that transfers sugar residues to the H antigen and determines the ABO blood group.22 Group A has two subtypes, i.e. A1 and A2, respectively. It has been shown that the A1 subtype has over 30-fold higher transferase activity than the A2 subtype.23 The A1 allele is perfectly tagged by the minor allele of SNP rs507666.11 SNP rs507666 is in near perfect LD (r2=0.96) with rs579459 and rs651007. Thus, the associations of these SNPs with sICAM-1, sP-selectin and sE-selectin levels may represent an effect of the ABO group A1 subtype. It has been suggested that the increased glycosyltransferase activity in individuals carrying the A1 allele might have an effect on the shedding, clearance or secretion of adhesion molecules, thereby influencing their levels in the circulation.11;12
Adhesion molecules are crucial to platelet leukocyte interaction and leukocyte migration into the vessel wall and thus important players in the atherosclerosis process underlying CHD.2;24 In a number of previous studies increased CHD risk has been associated with high sICAM-1, sP-selectin and sE-selectin levels.3;5;6 Unexpectedly, variants at the ABO locus conferring elevated CHD risk15;16;25, like the minor allele of SNP rs57945916, were associated with decreased levels of soluble adhesion molecules in our meta-analysis. One possible explanation for this seeming paradox may be that soluble adhesion molecules, although elevated in the case of endothelial dysfunction, actually compete with leukocyte adhesion to the endothelium (competition to cell surface adhesion molecules). Another possibility may be that the lower levels of soluble adhesion molecules might arise because of lower shedding of ectodomains, potentially leaving higher levels of intact cell surface adhesion molecules to recruit leukocytes to the blood vessel wall. To date, it is not known whether elevated levels of soluble adhesion molecules in vascular high-risk patients represent an epiphenomenon of vessel wall pathology, a true risk factor or a counter-regulatory per se protective mechanism as indicated by preliminary experimental data16. Experimental studies are required to further elaborate the pathophysiological role of soluble adhesion molecules and to clarify whether the prominent alterations in sICAM-1, sP-selectin and sE-selectin observed in this study are relevant to the recently discovered association between ABO SNPs and CHD risk.
Some limitations to our study warrant mentioning. First, the mechanism underlying the association of SNPs at the ABO locus with sICAM-1, sP-selectin and sE-selectin levels has remained unclear. Second, since SNP rs579459 is in strong LD with a number of other SNPs at this locus, it remains unknown which SNP is the causal variant. Third, since this study was conducted in individuals of European ancestry, the findings may not be generalizable to other races/ethnicity.
In conclusion, our study provides compelling evidence of an allele dose dependent association of variation at the ABO locus with circulating sICAM-1, sP-selectin and sE-selectin levels. These results contribute to the knowledge of genetic influences on these adhesion molecules which play important roles in many inflammatory diseases.
Supplementary Material
Acknowledgments
Funding Sources: We thank support from the British Heart Foundation (FS/07/021 and FS/11/28/28758). QX is the recipient of a British Heart Foundation Intermediate Basic Science Research Fellowship (FS/09/044/28007).
The work forms part of the research themes contributing to the translational research portfolio of Barts and the London Cardiovascular Biomedical Research Unit which is supported and funded by the National Institute of Health Research.
Framingham Heart Study (FHS) was supported by grants from the Boston University (N01-HC-25195) and NIH (RO1HL064753, R01HL076784, and R01AG028321). The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by a contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
The Atherosclerosis Risk in Communities Study (ARIC) is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are very grateful to the participants and staff from the Rotterdam Study, the participating general practitioners and the pharmacists. The genome-wide association study was funded by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Consortium for Healthy Aging (NCHA) project nr. 050-060-810. Abbas Dehghan is supported by Netherlands Organisation for Scientific Research (NOW) grant (vici, 918-76-619).
The Women’s Genome Health Study (WGHS) is supported by HL 043851 and HL69757 from the National Heart, Lung, and Blood Institute and CA 047988 from the National Cancer Institute, the Donald W. Reynolds Foundation and the Fondation Leducq, with collaborative scientific support and funding for genotyping provided by Amgen.
Footnotes
Conflict of Interest Disclosures:
Stefan Kiechl received research grants; TRP188-B12 FWF. Lu Qi received research grants from NIH. Russell P. Tracyreceived other research support from Celera. Paul M Ridker received research grants from AstraZeneca, Novartis, Merck, NHLBI, and NCI. He received other support from Amgen and Celera as well as honorarium from several universities. He has ownership/interest in a patents relate to inflammatory biomarkers. Also, he is a consultant or serves on the advisory board for ISIS, Merck, Vascular Biogenics, and Abbott. Emelia J. Benjamin received research grants; R01HL09257, RC1HL101056, RO1HL102214, RO1AG028321. She also consults or is on advisory committees for NIH and NHLBI.
References
- 1.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 2.Blankenberg S, Barbaux S, Tiret L. Adhesion molecules and atherosclerosis. Atherosclerosis. 2003;170:191–203. doi: 10.1016/s0021-9150(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 3.Ridker PM, Hennekens CH, Roitman-Johnson B, Stampfer MJ, Allen J. Plasma concentration of soluble intercellular adhesion molecule 1 and risks of future myocardial infarction in apparently healthy men. Lancet. 1998;351:88–92. doi: 10.1016/S0140-6736(97)09032-6. [DOI] [PubMed] [Google Scholar]
- 4.Blankenberg S, Rupprecht HJ, Bickel C, Peetz D, Hafner G, Tiret L, Meyer J. Circulating cell adhesion molecules and death in patients with coronary artery disease. Circulation. 2001;104:1336–1342. doi: 10.1161/hc3701.095949. [DOI] [PubMed] [Google Scholar]
- 5.Ridker PM, Buring JE, Rifai N. Soluble P-selectin and the risk of future cardiovascular events. Circulation. 2001;103:491–495. doi: 10.1161/01.cir.103.4.491. [DOI] [PubMed] [Google Scholar]
- 6.Hwang SJ, Ballantyne CM, Sharrett AR, Smith LC, Davis CE, Gotto AM, Jr, Boerwinkle E. Circulating adhesion molecules VCAM-1, ICAM-1, and E-selectin in carotid atherosclerosis and incident coronary heart disease cases: the Atherosclerosis Risk In Communities (ARIC) study. Circulation. 1997;96:4219–4225. doi: 10.1161/01.cir.96.12.4219. [DOI] [PubMed] [Google Scholar]
- 7.Meigs JB, Hu FB, Rifai N, Manson JE. Biomarkers of endothelial dysfunction and risk of type 2 diabetes mellitus. JAMA. 2004;291:1978–1986. doi: 10.1001/jama.291.16.1978. [DOI] [PubMed] [Google Scholar]
- 8.Lee M, Czerwinski SA, Choh AC, Demerath EW, Sun SS, Chumlea WC, Towne B, Siervogel RM. Quantitative genetic analysis of cellular adhesion molecules: the Fels Longitudinal Study. Atherosclerosis. 2006;185:150–158. doi: 10.1016/j.atherosclerosis.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Vistoropsky Y, Trofimov S, Pantsulaia I, Livshits G. Genetic and environmental determinants of variation of soluble adhesion molecules. Ann Hum Genet. 2006;70:749–758. doi: 10.1111/j.1469-1809.2006.00275.x. [DOI] [PubMed] [Google Scholar]
- 10.Schnabel RB, Lunetta KL, Larson MG, Dupuis J, Lipinska I, Rong J, Chen MH, Zhao Z, Yamamoto JF, Meigs JB, Nicaud V, Perret C, Zeller T, Blankenberg S, Tiret L, Keaney JF, Jr, Vasan RS, Benjamin EJ. The relation of genetic and environmental factors to systemic inflammatory biomarker concentrations. Circ Cardiovasc Genet. 2009;2:229–237. doi: 10.1161/CIRCGENETICS.108.804245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pare G, Chasman DI, Kellogg M, Zee RY, Rifai N, Badola S, Miletich JP, Ridker PM. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbalic M, Dupuis J, Dehghan A, Bis JC, Hoogeveen RC, Schnabel RB, Nambi V, Bretler M, Smith NL, Peters A, Lu C, Tracy RP, Aleksic N, Heeriga J, Keaney JF, Jr, Rice K, Lip GY, Vasan RS, Glazer NL, Larson MG, Uitterlinden AG, Yamamoto J, Durda P, Haritunians T, Psaty BM, Boerwinkle E, Hofman A, Koenig W, Jenny NS, Witteman JC, Ballantyne C, Benjamin EJ. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum Mol Genet. 2010;19:1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paterson AD, Lopes-Virella MF, Waggott D, Boright AP, Hosseini SM, Carter RE, Shen E, Mirea L, Bharaj B, Sun L, Bull SB. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. Arterioscler Thromb Vasc Biol. 2009;29:1958–1967. doi: 10.1161/ATVBAHA.109.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi L, Cornelis MC, Kraft P, Jensen M, van Dam RM, Sun Q, Girman CJ, Laurie CC, Mirel DB, Hunter DJ, Rimm E, Hu FB. Genetic variants in ABO blood group region, plasma soluble E-selectin levels and risk of type 2 diabetes. Hum Mol Genet. 2010;19:1856–1862. doi: 10.1093/hmg/ddq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, Horne BD, Stewart AF, Assimes TL, Wild PS, Allayee H, Nitschke PL, Patel RS, Martinelli N, Girelli D, Quyyumi AA, Anderson JL, Erdmann J, Hall AS, Schunkert H, Quertermous T, Blankenberg S, Hazen SL, Roberts R, Kathiresan S, Samani NJ, Epstein SE, Rader DJ, Qasim AN, DerOhannessian SL, Qu L, Cappola TP, Chen Z, Matthai W, Hakonarson HH, Wilensky R, Kent KM, Lindsay JM, Pichard AD, Satler L, Waksman R. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de FU, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O’Donnell CJ, McPherson R, Erdmann J, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiechl S, Lorenz E, Reindl M, Wiedermann CJ, Oberhollenzer F, Bonora E, Willeit J, Schwartz DA. Toll-like receptor 4 polymorphisms and atherogenesis. N Engl J Med. 2002;347:185–192. doi: 10.1056/NEJMoa012673. [DOI] [PubMed] [Google Scholar]
- 18.Targher G, Bonadonna RC, Alberiche M, Zenere MB, Muggeo M, Bonora E. Relation between soluble adhesion molecules and insulin sensitivity in type 2 diabetic individuals: role of adipose tissue. Diabetes Care. 2001;24:1961–1966. doi: 10.2337/diacare.24.11.1961. [DOI] [PubMed] [Google Scholar]
- 19.Hedges Olkin. Statistical methods for meta-analysis. London: Academic Press; 1985. [Google Scholar]
- 20.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 21.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990;345:229–233. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto F, McNeill PD, Hakomori S. Human histo-blood group A2 transferase coded by A2 allele, one of the A subtypes, is characterized by a single base deletion in the coding sequence, which results in an additional domain at the carboxyl terminal. Biochem Biophys Res Commun. 1992;187:366–374. doi: 10.1016/s0006-291x(05)81502-5. [DOI] [PubMed] [Google Scholar]
- 24.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 25.Teupser D, Baber R, Ceglarek U, Scholz M, Illig T, Gieger C, Holdt LM, Leichtle A, Greiser KH, Huster D, Linsel-Nitschke P, Schafer A, Braund PS, Tiret L, Stark K, Raaz-Schrauder D, Fiedler GM, Wilfert W, Beutner F, Gielen S, Grosshennig A, Konig IR, Lichtner P, Heid IM, Kluttig A, El Mokhtari NE, Rubin D, Ekici AB, Reis A, Garlichs CD, Hall AS, Matthes G, Wittekind C, Hengstenberg C, Cambien F, Schreiber S, Werdan K, Meitinger T, Loeffler M, Samani NJ, Erdmann J, Wichmann HE, Schunkert H, Thiery J. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ Cardiovasc Genet. 2010;3:331–339. doi: 10.1161/CIRCGENETICS.109.907873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.