Abstract
Whereas moderate drinking may have health benefits, excessive alcohol consumption causes many important acute and chronic diseases and is the third leading contributor to preventable death in the United States. Twin studies suggest that alcohol-consumption patterns are heritable (50%); however, multiple genetic variants of modest effect size are likely to contribute to this heritable variation. Genome-wide association studies provide a tool for discovering genetic loci that contribute to variations in alcohol consumption. Opportunities exist to identify susceptibility loci with modest effect by meta-analyzing together multiple studies. However, existing studies assessed many different aspects of alcohol use, such as typical compared with heavy drinking, and these different assessments can be difficult to reconcile. In addition, many studies lack the ability to distinguish between lifetime and recent abstention or to assess the pattern of drinking during the week, and a variety of such concerns surround the appropriateness of developing a common summary measure of alcohol intake. Combining such measures of alcohol intake can cause heterogeneity and exposure misclassification, cause a reduction in power, and affect the magnitude of genetic association signals. In this review, we discuss the challenges associated with harmonizing alcohol-consumption data from studies with widely different assessment instruments, with a particular focus on large-scale genetic studies.
INTRODUCTION
Excessive alcohol consumption is among the leading contributors to morbidity and mortality worldwide (1). According to the Centers for Disease Control and Prevention, an average of 79,000 deaths/y in the United States between 2001 and 2005 were attributable to the acute and chronic effects of alcohol. Whereas moderate alcohol consumption (≤2 drinks/d for men and ≤1 drink/d for women) may have some health benefits (2, 3) and disadvantages (4), heavy drinkers are at increased risk of alcohol-related health conditions (5, 6) and alcoholism—a serious and fairly common psychiatric disorder.
Alcohol consumption, including excessive drinking, is influenced by heritable factors (7–10). Approximately 50% of the variance in alcohol consumption is attributable to additive genetic factors (11, 12). A recent genetic study of an Asian population produced highly significant results (P < 10−58) for SNPs6 neighboring the ALDH gene, ALDH2 (13), which is involved in alcohol metabolism. Despite this success, the initial GWAS of alcohol consumption (14) and alcohol dependence (15–17) in European-American samples identified few variants approaching genome-wide significance (P < 5 × 10−8). It is likely that multiple genetic variants of modest effect contribute to the amount of alcohol consumed, particularly in non-Asian samples, and we anticipate that extremely large sample sizes will be needed to have the statistical power to detect them. Such large sample sizes must be obtained by pooling together data from multiple studies via meta-analysis—an approach that has yielded considerable success for identifying genetic variants that contribute to heaviness of cigarette smoking (18–22) and many other phenotypes.
Why are GWAS of alcohol consumption so challenging? One of the major challenges is phenotype heterogeneity due to differences in the ways that studies assess alcohol intake. Individual studies vary markedly in their objectives for collecting alcohol-related information and therefore differ in their means of measuring consumption and in the period of interest. For instance, a study of alcoholism may collect data on a participant's lifetime history of heavy drinking, whereas a study of breast cancer may query respondents on light to moderate levels of alcohol intake to model its effects on breast cancer (23, 24). This measurement heterogeneity has significant implications for genetic studies. In this review, we discuss the genetic underpinnings of alcohol consumption, potential sources of measurement heterogeneity, and the extent to which differences between studies may affect genetic analyses.
IS ALCOHOL CONSUMPTION HERITABLE?
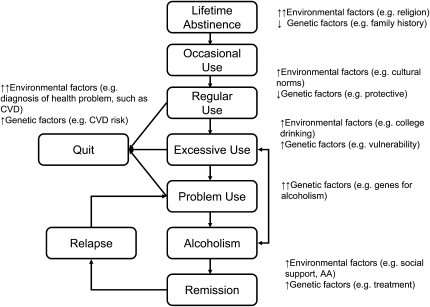
Alcohol use can be conceptualized as a series of different stages starting from initiation of use, transitioning to regular use, and potentially leading to excessive use and alcoholism (25), as shown in Figure 1. Whereas most aspects of alcohol use are heritable (ie, influenced, to some degree, by genetic factors) (26), drinking initiation (or conversely lifetime abstention) tends to be the least influenced by genetic factors (27–29) and rather appears to be determined primarily by familial environment (30, 31). For example, religion and culture have strong influences on whether and when a person starts to drink. However, once alcohol use is initiated, many domains of alcohol use—including current use, pattern of use, and heaviest use—are influenced by underlying genetic variation and are heritable (h2 ≈ 50%) (8, 32–34). There is substantial correlation between each of these domains and, similarly, there is estimated to be overlap of genetic factors, up to 90%, across them (8, 9, 33, 34). Furthermore, these heritable characteristics of alcohol intake overlap with genetic vulnerability to alcoholism (genetic correlations ≥0.9) (8, 32). Environment, of course, continues to be an important contributor to all these domains, and the context of these environmental influences vary across the stages (Figure 1).
FIGURE 1.
Stages of alcohol involvement, the role of genetic and environmental influences, and their importance in genetic studies. The arrows indicate the relative hypothesized magnitude of effect of genetic and environmental factors (up arrow: increasing effect, with number of arrows indicating strength of effect; down arrow: decreasing effect). AA, Alcoholics Anonymous; CVD, cardiovascular disease.
Understanding the different stages of alcohol use and abuse and the biases that affect their measurement can aid us in improving genetic studies. For instance, given the stages of alcohol use and the proposed common and specific genetic and environmental influences that contribute to each stage, we anticipate that the likelihood of identifying genetic influences that contribute to alcohol consumption will decrease if lifetime alcohol abstainers are combined with light drinkers or former drinkers when measures of alcohol consumption are constructed (eg, a never drinker was coded as having drunk 0 drinks/wk) (35), because nondrinkers at a particular time point include both those who have never drank, those who drank a lot in the past but have quit, and those who drink alcohol occasionally but only drink a small amount.
WHAT IS KNOWN ABOUT GENETIC VARIATION IN ALCOHOL CONSUMPTION?
Genes in the ADH and ALDH complex that regulate alcohol metabolism (36) are among the most established genetic contributors to all aspects of drinking. The metabolism of alcohol involves conversion of alcohol to acetaldehyde, by ADH and then acetaldehyde to acetate, by ALDH. Variants in the genes encoding ADH and ALDH, some of which are more common in Asians (37), are known to regulate alcohol metabolism to varying degrees. Defects in this pathway can lead to high concentrations of the intermediate metabolite acetaldehyde, which causes flushing (facial reddening) accompanied by nausea, dizziness, and other unpleasant physiologic symptoms); thus, individuals who experience this reaction are considerably less likely to continue to drink (38). Although there are several intriguing polymorphisms in the ADH and ALDH gene families, 2 coding SNPs are of note. In the ADH1B gene, one allele of rs1229984 leads to accelerated oxidation of alcohol to acetaldehyde; in ALDH2, one allele of rs671 leads to nearly no activity of the mitochondrial ALDH (36). These polymorphisms are robustly correlated with alcohol consumption and dependence (39–41). However, the allele frequencies of the most protective variants are very low in European populations, which leaves most of the heritability in these populations to be explained by other genes. Despite this, a recent study found strong support for the protective effect of rs1229984 on alcohol dependence in Europeans and African Americans (42).
Many other genes that may be associated with the personality characteristics of drinkers (eg, impulsivity and disinhibition) and loss of control and other neurobiological consequences of drinking (eg, alcohol-induced blackouts and tolerance) are likely to also be involved (12, 26). Because these genetic variants are likely to individually exert only modest effects on alcohol consumption, it is a considerable challenge to identify them. Accumulating very large samples that draw phenotypic and genomic data from multiple sources provides one such exciting way to address this challenge.
THE ADVANTAGE OF GWAS META-ANALYSIS
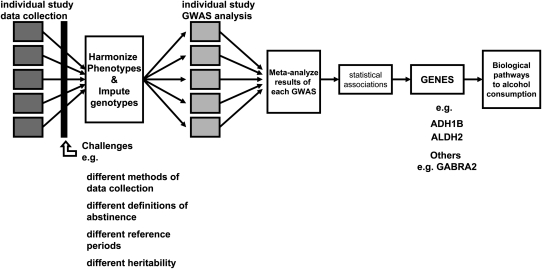
ow a meta-analysis of GWAS proceeds is outlined in Figure 2. In addition to harmonizing phenotypes, as is typically performed in all meta-analyses, genotypes from various platforms also need to be harmonized, via imputation, to a common scale (eg, HapMap, 1000 Genomes). Three large-scale GWAS meta-analyses of cigarette smoking have now definitively validated the role of a missense mutation (rs16969968 or its proxy rs1051730) (43) in the cluster of CHRNA5-CHRNA3-CHRNB4 genes on chromosome 15 (19, 21, 22). However, to achieve levels of appropriate statistical significance that overcome potential type I error from multiple comparisons, samples exceeding 40,000 (with the largest sample exceeding 140,000) were necessary. The success with GWAS of smoking behavior offers the hope that alcohol consumption and alcohol dependence can be similarly studied in large population samples. Recently, such a meta-analysis of alcohol consumption identified an SNP in the AUTS2 (autism susceptibility) gene to be associated with alcohol consumption (daily ethanol intake in g/kg body mass; P = 4.1 × 10−9) (44). They also reported significant expression differences in this gene across high compared with low alcohol-preferring mice. More than 42,000 subjects were included in this meta-analysis, and the overall effect size was modest. Although this is an exciting development for genetic studies of alcohol consumption, unlike rs16969968 for smoking, the role of AUTS2 in alcohol consumption remains to be validated. Of course, additional loci are most likely involved in the genetics of both alcohol and smoking.
FIGURE 2.
Stages of meta-analysis of genetic data—an illustration. ADH, alcohol dehydrogenase; ALDH, aldehyde dehydrogenase; GABRA2, γ-aminobutyric acid receptor subunit A2; GWAS, genome-wide association studies.
Meta-analyses, whether epidemiologic or genetic, implicitly require a minimum level of comparability across studies (Figure 2). Even though a meta-analysis aggregates across results from within-study analyses, across-study heterogeneity influences the comparability of effect sizes (eg, regression coefficients and ORs) when there is heterogeneity in the measured endpoint. The effect of measurement heterogeneity on gene finding is affected by the extent to which common loci underlie the genetic contribution to the distinct aspects of alcohol use (eg, drinking frequency, quantity, and alcohol dependence) being meta-analyzed. In the following sections, we discuss key contributors to heterogeneity in the measurement of alcohol consumption and describe their potential effect on the GWAS meta-analyses of these phenotypes.
MEASURING ALCOHOL CONSUMPTION
Measures of alcohol consumption typically include drinking quantity (eg, how many drinks in a given reference period, such as drinks/wk or drinks/d) and frequency (eg, how often one drinks in a given reference period, such as drinking days/wk). These measures can be assessed during various reference periods: in the past 30 d, in the past 12 mo, over a lifetime, during a period of “typical” or “usual” alcohol consumption, or during a period of heaviest use. Although assessed by fewer studies, differences between drinking a certain amount of alcohol over the week or at one sitting are also important. For example, the maximum number of drinks consumed within a single 24-h period is also heritable and is highly correlated with a person's vulnerability to excessive alcohol consumption (8, 32, 45).
WHY MEASUREMENT HETEROGENEITY OCCURS
Alcohol consumption is an aspect of normative diet, a risk factor for a variety of health-related outcomes, and an addictive substance. Thus, depending on the goals of a study, measures of alcohol consumption can be collected by using food-frequency questionnaires, from medical charts, via self-report in case-control studies of health-related outcomes, and from interviews in studies for addiction and other psychiatric disorders. To illustrate this remarkable variation in the assessment of alcohol consumption, representative alcohol-consumption measures from individual studies of the GENEVA Consortium are shown in Table 1 (46, 47).
TABLE 1.
Illustration of across-study variation in data collected on alcohol consumption in the Gene, Environment Association (GENEVA) Consortium1
| Study | Primary outcome | Alcohol consumption item | Alcoholism | Source |
| Study of Addiction: Genes and Environment | Alcohol dependence | What is the typical number of drinks consumed in a week when you were drinking? | Diagnostic | Psychiatric Interview (SSAGA) |
| Health Professionals Follow-Up Study, Nurses’ Health Study | Type 2 diabetes, open-angle glaucoma | For each X listed, fill in the circle indicating how often on average you have used the amount specified during the past year: never, <1 time/mo, 1–3 times/mo, 1 time/wk, 2–4 times/wk, 5–6 times/wk, 1 time/d, 2–3 times/d, 4–5 times/d, or ≥6 times/d (Note: “X” represents light beer, beer, red wine, white wine, or hard liquor; categories were converted to drinks/wk) | Self-report (ever been diagnosed with alcoholism by a physician?) | Food-frequency questionnaire (for alcohol consumption assessment) and lifestyle/medical history questionnaire (for alcoholism assessment) |
| Atherosclerosis Risk In Communities | Atherosclerosis, cardiovascular disease | Current drinkers were asked “How many glasses of wine do you usually have per week (4-ounce glasses)?” and “How many bottles or cans of beer do you usually have per week (12-ounce bottles or cans)?”; and “How many drinks of hard liquor do you usually have per week (1.5-ounce shots)?” | — | Interview self-report (single item) |
| Genetics of Early Onset Stroke | Early-onset ischemic stroke | Cases: during the year before your stroke, did you drink X? How often did you usually drink X? (possible answers: drinks/d, drinks/wk, drinks/mo, or drinks/y) (Note: “X” represents light beer, beer, wine, or hard liquor) | — | Interview self-report (single item) |
| Controls: during the year before the reference date, did you drink X? How often did you usually drink X? (possible answers: drinks/d, drinks/wk, drinks/mo, or drinks/y) (Note: X represents light beer, beer, wine, or hard liquor) | ||||
| Dental Caries | Oral health | 1) During the time in your life when you were drinking most often, how many days each week were you drinking? During the time in your life when you were drinking most often, how many drinks did you have on a typical day? | — | Interview self-report (single item) |
| 2) Which of these statements comes closest to describing how often you drank any alcoholic beverages in the past 30 d? Every day, 5–6 d/wk, 3–4 d/wk, 1–2 d/wk, 2–3 times, once, or not at all. During the past 30 d, how many drinks did you usually have on days when you drank alcoholic beverages? |
1 ounce = ∼29.6 mL. SSAGA, Semi-Structured Assessment for the Genetics of Alcoholism.
Within each of these realms of research, significant effort has been made to harmonize measures of alcohol consumption. For example, disparate measures of alcohol can be converted into exposure categories (48), and categories of safe compared with unsafe consumption (49) have been developed for studies of all-cause mortality. Similar efforts have been directed at developing harmonized alcohol-consumption measures for studies of cancer (50). Alcohol intake data collected by using various food-frequency questionnaires tend to correlate highly (51, 52). However, reducing measurement heterogeneity within a set of studies with a common theme (eg, alcohol and cancer) does not ensure that these alcohol measures can themselves be combined with alcohol use assessed in studies focused on other endpoints, ie, if one is interested in identifying the genetic underpinnings of alcohol consumption, variations in the alcohol measures presented in Table 1 may still pose a challenge.
SOURCES OF MEASUREMENT HETEROGENEITY
The challenges associated with measurement heterogeneity for alcohol consumption, particularly in the context of genomic studies, are listed in Table 2. Some of these challenges are not unique to alcohol consumption (eg, recall bias), and others are fairly straightforward (eg, conversion of ethanol grams in a standard drink across different countries). However, some key concerns that are unique to alcohol consumption (eg, episodic nature of drinking and recent abstention) can produce spurious results because of the misclassification of subjects. Whereas we emphasize those issues that bear major concerns for genetic analyses, readers interested in issues related to the general measurement of alcohol consumption are invited to read a review of this topic by Dawson (53).
TABLE 2.
Challenges associated with assessment of alcohol consumption from various sources
| Challenge | Example |
| Distinguishing between lifetime and recent abstention | A heavy drinker recently (past 30 d) stops drinking because of a health condition for which the study is ascertained. |
| Episodic nature of drinking | An individual drinks 10 drinks/d on weekends but only 2 drinks/d on weekdays except during the holidays. Individual may also report a period of risky drinking in college but not anymore. |
| Reference periods for drinking can vary | Because of the episodic nature of drinking, an individual reporting consuming 3 drinks/d, on average, cannot be assumed to be drinking 21 drinks/wk (ie, assumption that drinking is constant every day of the week). |
| Cultural norms | Drinking regularly may be normative in some cultures but unacceptable in others. |
| Assessing different beverage forms | An individual reports greater alcohol intakes when responding to questions on beer, wine, and spirits compared with a single question on “alcoholic drinks.” |
| Conversion to grams of ethanol requires participant understanding of a “standard” drink | An individual drinks red wine at home, and unless prompted with visual cue, it is unclear whether their pour of wine reflects a standard pour. |
| Recall bias and bias attributable to other respondent characteristics (eg, underreporting by overweight/obese individuals) | An overweight individual reports fewer drinks because of weight concerns. |
Interpretation of abstention
Lifetime abstainers are uncommon in several populations. They are worth excluding from genetic studies of alcohol use and alcohol dependence, because alcohol initiation is more strongly affected by religious and cultural norms. Individuals with a family history of alcoholism may also abstain from alcohol to avoid developing the disorder, because these individuals are at higher genetic risk. Lifetime drinkers may also report recent abstention due to a host of environmental factors (eg, life events and pregnancy) (54), due to health factors (eg, gastrointestinal disease) (55), or due to enrollment in a treatment program for alcohol dependence (eg, Alcoholics Anonymous) (56). Thus, an individual with alcohol dependence may have susceptibility loci for alcohol use, yet be inadvertently misclassified as a light or nondrinker because of limitations in the design of the study in which they participated. Studies that do not assess lifetime use are particularly affected by this issue, because they cannot distinguish between lifetime and recent abstainers.
The episodic nature of alcohol consumption
Drinking, unlike smoking, is often episodic and can vary considerably across periods of recent (past 12 mo, past 30 d, or a shorter reference period) and lifetime use (average across the lifetime, during periods of typical or heaviest use). Even within a single week, an individual may drink more heavily on weekends but less frequently during the week, which produces marked variation in their report of drinks per week. It is likely that the magnitude and nature of genetic influences on these various periods of drinking only partially overlap. Whereas there is overlap across these genetic factors, this overlap diminishes when current drinking (eg, past 30 d) is examined along with lifetime heavy drinking (34, 57).
Quantity and frequency across reference periods
Uniform measures of alcohol consumption have not been universally adopted. Attempts to harmonize the varying reference periods (drinks/d, drinks/wk, or drinks/mo) used in different studies without a measure of drinking frequency (eg, How many days a week do you drink?) can lead to biases, particularly because of the episodic nature of alcohol consumption. For instance, assume that a study assesses drinks/d, and the respondent reports drinking 2 drinks/d. We are interested in a measure of drinks/wk, but we do not know whether the individual drinks only on weekends. Do we compute 14 drinks/wk (2 drinks/d × 7 d), which is a significant inflation over the accurate 4–6 drinks/wk, or do we simply use drinks/d as the measure from that study?
Differences in cultural norms across studies
Alcohol intake varies substantially by population and is affected by sex, geographic region, and cultural practices. For example, intake among US Seventh-Day Adventists (58) varies substantially from that among indigenous Canadian and Alaskan native populations, such as Inuit (59–61). The substantial range in alcohol intake across populations is another challenge when pooling data. For example, although drinking 1 or 2 glasses of wine per day with dinner may be very common in some populations, it would be considered very extreme in other populations. Thus, susceptibility loci for alcohol intake are affected by the broader environmental context. These complicated interactions are challenging to account for within the meta-analytic framework.
Beverage forms
Considerable sociocultural and individual-specific variation in the choice of alcoholic beverage consumed also exist, and it is well recognized that reported alcohol consumption is higher when participants are asked about individual beverage types (beer, wine, and liquor) separately rather than about “alcoholic beverages” as a group (62, 63). On the other hand, for many alcohol-associated outcomes, such as cancer, it is thought that ethanol is the important factor, regardless of its source. Should measures drawn from specific alcoholic beverages be combined with data drawn from single item assessments of “drinks”? One possible mechanism for harmonizing these varying assessments is via conversion to ethanol grams, which as discussed below, requires many assumptions.
Standard drinks and conversion to ethanol grams
Although cigarettes can vary in their nicotine content, most smokers consume cigarettes in a pack of standard size, thus establishing a “metric” for their report. Alcohol, in contrast, is consumed as a variety of beverages with varying ethanol contents, forms (eg, mixed drinks and coolers), and quantities (eg, can of beer and beer on tap) that render an estimate of the number of standard drinks (ie, amount of ethanol) (64) difficult to quantify. Drink size and ethanol content may also vary by country. The ability to reconcile these differences by conversion relies heavily on the participant's understanding of the definition of a “standard drink.” For instance, a drinker may report drinking a glass of wine every day but drinks the wine in a large “balloon” glass, which can contain the equivalent of 2 to 3 standard drinks; hence, alcohol consumption and ethanol grams are underestimated. Picture cards or verbal/visual references of standard drinks by beverage type (eg, beer compared with malted beer; table wine compared with fortified wine) might be used to orient respondents to the meaning of a standard drink. However, this may not always be feasible within the context of a food-frequency questionnaire.
Recall and other respondent biases
Finally, although not unique to alcohol, much like most self-reported measures, general elements of recall bias may also contribute to measurement inaccuracy. Recall can be biased by characteristics correlated with drinking. For instance, underweight individuals have been found to overestimate and overweight/obese individuals tend to underreport certain aspects of the diet (eg, fats and simple carbohydrates) (65, 66); this may apply to alcohol consumption as well. Although statistical approaches have been developed to address these issues, these efforts, even if successful, can have an effect on study power.
THE FUTURE OF GWAS META-ANALYSES OF ALCOHOL CONSUMPTION
Understanding the etiology of a behavior that is, at once, both a feature of normal diet and a serious and common psychiatric disorder, and a substantial cause of chronic disease, is a critical public health issue. In this regard, it is reassuring that GWAS meta-analyses have proven to be successful at addressing for other complex phenotypes, including behavioral measures (67, 68). Furthermore, simulation studies suggest that, even with exposure misclassification, very large studies can succeed at identifying susceptibility loci (69). Importantly, if even a subset of genetic influences is common across each alcohol metric, then a meta-analysis should be able to identify genes contributing to this shared genetic propensity, given a large enough data set. In contrast, those genetic influences that are specific to particular aspects of alcohol consumption will require careful harmonization of intake measures.
For future studies that plan on including alcohol-consumption measures, several resources are available to guide investigators with data collection, and they are listed in Table 3. For instance, the PhenX Toolkit (70) offers questions on a variety of alcohol-consumption measures, including definitions of standard drinks as well as visual references of sizes of standard beverage servings. PhenX also offers items to assess lifetime use, age at first use, and symptoms of alcohol dependence for studies interested in a more detailed assessment of maladaptive drinking. The National Institute on Alcohol Abuse and Alcoholism offers several assessments (eg, AUDIT) that can be used to screen for and measure quantity, frequency, and liability to abuse and dependence.
TABLE 3.
List of resources for assessing alcohol consumption in US populations1
| Resource | Utility/features |
| https://www.phenxtoolkit.org | Items to assess 30-d quantity and frequency; includes visual aids for standard drink sizes |
| http://pubs.niaaa.nih.gov/publications/aa65/aa65.htm | Assessments that can be used to screen for alcohol-related problems |
| http://rethinkingdrinking.niaaa.nih.gov/IsYourDrinkingPatternRisky/WhatsAtRiskOrHeavyDrinking.asp | Items to assess risky drinking |
| http://www.cdc.gov/alcohol/fact-sheets/alcohol-use.htm | Information on standard drink sizes and some sample questions |
| http://pubs.niaaa.nih.gov/publications/arh27-1/18-29.htm | Discussion of measurement-related issues for alcohol (53) |
For non-US populations, standard drink size and grams of ethanol may vary.
We recognize that investigators collect measures of alcohol consumption that maximally inform their outcome of interest and that efforts to minimize respondent burden may limit the extent to which common assessments are used across studies. It is hoped that the concerns raised in this review will encourage investigators to consider the inclusion of a well-validated measure that indexes heritable variation in the tendency to drink alcohol in their study. We recommend the inclusion of maximum drinks consumed in a single 24-h period. This single low-burden item was heritable (h2 = 50%) (8, 32), associated with ADH variants (40), was found to correlate strongly with liability to alcohol consumption and alcoholism, and may also indicate alcohol tolerance. Even in cultures in which maximum drinks may reflect a “rite of passage,” such as drinking on one's 21st birthday in the United States, symptoms of alcohol dependence have been found to correlate strongly even with this isolated instance of heavy drinking (71). Undoubtedly, cultural, societal, and other environmental factors influence this measure and its correspondence with lifetime alcoholism risk. Not all individuals with a lifetime high number of maximum drinks consumed in a single 24-h period may subsequently develop alcoholism. In detailed assessments, indexes of quantity and frequency of drinking augment information from maximum drinks.
Existing and ongoing studies should recognize the context of genomic findings that arise from the combination of such heterogeneous sources and model random effects or calculate other metrics of heterogeneity (see reference 72 for a review of these metrics) when conducting genetic analyses. Even though there has been discussion surrounding the lower power in smaller studies to detect heterogeneity (73), this is necessary for alcohol intake. In fact, evidence for between-study heterogeneity can ultimately lead to new discoveries (eg, genes relevant only in individuals exposed to certain environments) (74). There is often a trade-off between heterogeneity and the sample size necessary to detect the effects of genes (69); thus, augmenting the sample size can help. From a technical perspective, investigators may wish to consider statistical methods developed for the analysis of covariates as outcomes in genomic studies, as is typically the case for alcohol consumption (75). Biases in such studies are most pronounced when the primary and secondary traits are correlated with one another or if the marker jointly influences both traits. In these instances, methods such as inverse-probability-of-sampling weighted regression may be more appropriate. Many such meta-analytic efforts are currently ongoing and are accounting for these technical caveats and amassing tens of thousands of subjects, heralding a brand new era of discovery for the genetics of alcohol consumption. We invite investigators with relevant alcohol-related information and GWAS data to join and participate in these ongoing efforts.
Acknowledgments
We thank the staff and participants of the ARIC and all other GENEVA studies for their important contributions.
The authors’ responsibilities were as follows—AA and LJB: wrote the first draft of the manuscript; NDF: provided extensive feedback and conceptual support concerning all versions of the manuscript; Y-CC, PL, JRS, QS, KT, BY, RSD, GH, and JHK; provided summaries of alcohol data from the GENEVA studies; JO, SB, EB, JWC, MCC, AF, NC, RC, SK, LQ, and RMvD: provided feedback on preparation of variables and assisted with study-specific measurement concerns; KKB, DMD, HJE, AG, VH, JK, JIN, MS, JPR, and LJB: assisted with preparation of the measurement heterogeneity table and text and advised on genetic implications of measurement heterogeneity; E Boerwinkle, FH, SL, MM, BDM, LRP, and LJB: collected data for the GENEVA studies; SB and NC: managed the phenotype harmonization working group with AA, LJB, NDF, Y-CC, JRS, QS, KT, BY, JWC, MCC, RSD, AF, GH, JHK, JO, E Bookman, KKB, DMD, VH, SK, RMvD, E Boerwinkle, FH, SL, MM, BDM, and LRP as members. All authors reviewed and edited all versions of the manuscript. All authors represent the alcohol working group of the GENEVA Consortium. LJB, JPR, and AG are listed as inventors on the patent “Markers for Addiction” (US 20070258898): covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. LJB acted as a consultant for Pfizer Inc in 2008. None of the other authors reported competing interests. The funding sources were not involved in any aspect of manuscript preparation.
Footnotes
Abbreviations used: ADH, alcohol dehydrogenase; ALDH, aldehyde dehydrogenase; GENEVA, Gene, Environment Association; GWAS, genome-wide association studies; SNP, single nucleotide polymorphism.
REFERENCES
- 1.Centers for Disease Control Alcohol-attributable deaths and years of potential life lost—United States, 2001. MMWR Morb Mortal Wkly Rep 2004;53:866–70 [PubMed] [Google Scholar]
- 2.Marmot MG, Rose G, Shipley MJ, Thomas BJ. Alcohol and mortality: a U-shaped curve. Lancet 1981;1:580–3 [DOI] [PubMed] [Google Scholar]
- 3. US Department of Agriculture, US Department of Health and Human Services. Nutrition and your health: dietary guidelines for Americans. Washington, DC: US Government Printing Office, 1990.
- 4.Holman CD, English DR. Ought low alcohol intake to be promoted for health reasons? J R Soc Med 1996;89:123–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marmot M, Brunner E. Alcohol and cardiovascular disease: the status of the U shaped curve. BMJ 1991;303:565–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klatsky AL. Alcohol and cardiovascular mortality: common sense and scientific truth. J Am Coll Cardiol 2010;55:1336–8 [DOI] [PubMed] [Google Scholar]
- 7.Heath AC, Meyer J, Jardine R, Martin NG. The inheritance of alcohol consumption patterns in a general population twin sample: II. Determinants of consumption frequency and quantity consumed. J Stud Alcohol 1991;52:425–33 [DOI] [PubMed] [Google Scholar]
- 8.Kendler KS, Myers J, Dick D, Prescott CA. The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcohol Clin Exp Res 2010;34:1058–65. [DOI] [PMC free article] [PubMed]
- 9.Agrawal A, Grant JD, Littlefield A, Waldron M, Pergadia ML, Lynskey MT, Madden PA, Todorov A, Trull T, Bucholz KK, et al. Developing a quantitative measure of alcohol consumption for genomic studies on prospective cohorts. J Stud Alcohol Drugs 2009;70:157–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaprio J, Rose RJ, Romanov K, Koskenvuo M. Genetic and environmental determinants of use and abuse of alcohol: the Finnish Twin Cohort studies. Alcohol Alcohol Suppl 1991;1:131–6. [PubMed]
- 11.Bierut LJ, Saccone NL, Rice JP, Goate A, Foroud T, Edenberg H, Almasy L, Conneally PM, Crowe R, Hesselbrock V, et al. Defining alcohol-related phenotypes in humans. The Collaborative Study on the Genetics of Alcoholism. Alcohol Res Health 2002;26:208–13 [PMC free article] [PubMed] [Google Scholar]
- 12.Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol 2006;11:386–96 [DOI] [PubMed] [Google Scholar]
- 13.Baik I, Cho NH, Kim SH, Han BG, Shin C. doi: 10.3945/ajcn.110.001776. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr 2011;93:809–16. [DOI] [PubMed] [Google Scholar]
- 14.Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, et al. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry 2011;70:513–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA 2010;107:5082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 2009;66:773–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res 2010;34:840–52. [DOI] [PMC free article] [PubMed]
- 18.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 2010;42:448–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 2010;42:441–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet 2010;6:e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE 2009;4:e4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet 2010;42:436–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan SF, Cantwell MM, Cardwell CR, Velentzis LS, Woodside JV. Dietary patterns and breast cancer risk: a systematic review and meta-analysis. Am J Clin Nutr 2010;91:1294–302 [DOI] [PubMed] [Google Scholar]
- 24.Mukamal KJ, Rimm EB. Alcohol consumption: risks and benefits. Curr Atheroscler Rep 2008;10:536–43 [DOI] [PubMed] [Google Scholar]
- 25.Bierut LJ. Genetic vulnerability and susceptibility to substance dependence. Neuron 2011;69:618–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dick DM, Bierut LJ. The genetics of alcohol dependence. Curr Psychiatry Rep 2006;8:151–7 [DOI] [PubMed] [Google Scholar]
- 27.Dick DM, Pagan JL, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. Changing environmental influences on substance use across development. Twin Res Hum Genet 2007;10:315–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose RJ. A developmental behavior-genetic perspective on alcoholism risk. Alcohol Health Res World 1998;22:131–43 [PMC free article] [PubMed] [Google Scholar]
- 29.McGue M. The behavioral genetics of alcoholism. Curr Dir Psychol Sci 1999;8:109–15 [Google Scholar]
- 30.Pagan JL, Rose RJ, Viken RJ, Pulkkinen L, Kaprio J, Dick DM. Genetic and environmental influences on stages of alcohol use across adolescence and into young adulthood. Behav Genet 2006;36:483–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heath AC, Martin NG. Genetic influences on alcohol consumption patterns and problem drinking: results from the Australian NH&MRC twin panel follow-up survey. Ann N Y Acad Sci 1994;708:72–85. [DOI] [PubMed]
- 32.Grant JD, Agrawal A, Bucholz KK, Madden PA, Pergadia ML, Nelson EC, Lynskey MT, Todd RD, Todorov AA, Hansell NK, et al. Alcohol consumption indices of genetic risk for alcohol dependence. Biol Psychiatry 2009;66:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dick DM, Prescott C, McGue M. The genetics of substance use and substance use disorders. In: Handbook of behavior genetics. 1st ed. New York, NY: Springer, 2010:433–54 [Google Scholar]
- 34.Dick DM, Meyers JL, Rose RJ, Kaprio J, Kendler KS. doi: 10.1111/j.1530-0277.2011.01564.x. Measures of current alcohol consumption and problems: two independent twin studies suggest a complex genetic architecture. Alcohol Clin Exp Res 2011;35:2152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heath AC, Martin NG, Lynskey MT, Todorov AA, Madden PA. Estimating two-stage models for genetic influences on alcohol, tobacco or drug use initiation and dependence vulnerability in twin and family data. Twin Res 2002;5:113–24 [DOI] [PubMed] [Google Scholar]
- 36.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health 2007;30:5–13 [PMC free article] [PubMed] [Google Scholar]
- 37.Han Y, Gu S, Oota H, Osier MV, Pakstis AJ, Speed WC, Kidd JR, Kidd KK. Evidence of positive selection on a class I ADH locus. Am J Hum Genet 2007;80:441–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomasson HR, Crabb DW, Edenberg HJ, Li TK. Alcohol and aldehyde dehydrogenase polymorphisms and alcoholism. Behav Genet 1993;23:131–6 [DOI] [PubMed] [Google Scholar]
- 39.Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet 2009;18:580–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet 2006;15:1539–49 [DOI] [PubMed] [Google Scholar]
- 41.Li D, Zhao H, Gelernter J. doi: 10.1016/j.biopsych.2011.02.024. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biol Psychiatry 2011;34:1058–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 2011;Oct 4 (Epub ahead of print; DOI: 10.1038/mp.2011.124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 2007;16:36–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivières S, Aliev FA, Khan AA, Amin N, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA 2011;108:7119–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, et al. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet 2000;96:632–7 [DOI] [PubMed] [Google Scholar]
- 46.Cornelis MC, Agrawal A, Cole JW, Hansel NN, Barnes KC, Beaty TH, Bennett SN, Bierut LJ, Boerwinkle E, Doheny KF, et al. The Gene, Environment Association Studies Consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol 2010;34:364–72. [DOI] [PMC free article] [PubMed]
- 47.Bennett SN, Caporaso N, Fitzpatrick AL, Agrawal A, Barnes K, Boyd HA, Cornelis MC, Hansel NN, Heiss G, Heit JA, et al. Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience. Genet Epidemiol 2011;35:159–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holman CD, English DR, Milne E, Winter MG. Meta-analysis of alcohol and all-cause mortality: a validation of NHMRC recommendations. Med J Aust 1996;164:141–5 [DOI] [PubMed] [Google Scholar]
- 49.Holman CD, English DR. An improved aetiologic fraction for alcohol-caused mortality. Aust J Public Health 1995;19:138–41 [DOI] [PubMed] [Google Scholar]
- 50.Smith-Warner SA, Spiegelman D, Ritz J, et al. Methods for pooling results of epidemiologic studies: the Pooling Project of Prospective Studies of Diet and Cancer. Am J Epidemiol 2006;163:1053–64 [DOI] [PubMed] [Google Scholar]
- 51.Larsson SC, Giovannucci E, Wolk A. Alcoholic beverage consumption and gastric cancer risk: a prospective population-based study in women. Int J Cancer 2007;120:373–7 [DOI] [PubMed] [Google Scholar]
- 52.Brunner EJ, Mosdol A, Witte DR, Martikainen P, Stafford M, Shipley MJ, Marmot MJ. Dietary patterns and 15-y risks of major coronary events, diabetes, and mortality. Am J Clin Nutr 2008;87:1414–21 [DOI] [PubMed] [Google Scholar]
- 53.Dawson DA. Methodological issues in measuring alcohol use. National Institute on Alcohol Abuse and Alcoholism. 2003. Available from: http://pubs.niaaa.nih.gov/publications/arh27-1/18-29.htm (cited 6 February 2011) [PMC free article] [PubMed]
- 54.O'Leary CM, Heuzenroeder L, Elliott EJ, Bower C. A review of policies on alcohol use during pregnancy in Australia and other English-speaking countries, 2006. Med J Aust 2007;186:466–71 [DOI] [PubMed] [Google Scholar]
- 55.Chan AT, Giovannucci EL. Primary prevention of colorectal cancer. Gastroenterology 2010;138:2029–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferri M, Amato L, Davoli M. Alcoholics Anonymous and other 12-step programmes for alcohol dependence. Cochrane Database Syst Rev 2006;3:CD005032. [DOI] [PubMed] [Google Scholar]
- 57.Agrawal A, Lynskey M, Heath A, Chassin L. Developing a genetically informative measure of alcohol consumption using past 12 months indices. J Stud Alcohol Drugs 2011;72:444–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butler TL, Fraser GE, Beeson WL, Knutsen SF, Herring RP, Chan J, Sabate J, Montgomery S, Haddad E, Preston-Martin S, et al. Cohort profile: the Adventist Health Study-2 (AHS-2). Int J Epidemiol 2008;37:260–5 [DOI] [PubMed] [Google Scholar]
- 59.Hesselbrock VM, Hesselbrock MN, Segal B. Alcohol dependence among Alaskan natives and their health care utilization. Alcohol Clin Exp Res 2003;27:1353–5 [DOI] [PubMed] [Google Scholar]
- 60.Segal B. Drinking and drinking-related problems among Alaska natives. Alcohol Health Res World 1998;22:276–80 [PMC free article] [PubMed] [Google Scholar]
- 61.Tjepkema M, Wilkins R, Senecal S, Guimond E, Penney C. Mortality of urban Aboriginal adults in Canada, 1991-2001. Chronic Dis Can 2010;31:4–21 [PubMed] [Google Scholar]
- 62.Russell M, Welte JW, Barnes GM. Quantity-frequency measures of alcohol consumption: beverage-specific vs global questions. Br J Addict 1991;86:409–17 [DOI] [PubMed] [Google Scholar]
- 63.Dawson DA. Volume of ethanol consumption: effects of different approaches to measurement. J Stud Alcohol 1998;59:191–7 [DOI] [PubMed] [Google Scholar]
- 64. International Center for Alcohol Policies. What is a ldquostandard drinkrdquo? Washington, DC: ICAP, 1–7. 1998.
- 65.Tooze JA, Subar AF, Thompson FE, Troiano R, Schatzkin A, Kipnis V. Psychosocial predictors of energy underreporting in a large doubly labeled water study. Am J Clin Nutr 2004;79:795–804 [DOI] [PubMed] [Google Scholar]
- 66.Mendez MA, Popkin BM, Buckland G, et al. Alternative methods of accounting for underreporting and overreporting when measuring dietary intake-obesity relations. Am J Epidemiol 2011;173:448–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med 2010;363:166–76 [DOI] [PubMed] [Google Scholar]
- 68.Hirschhorn JN. Genomewide association studies—illuminating biologic pathways. N Engl J Med 2009;360:1699–701 [DOI] [PubMed] [Google Scholar]
- 69.Bennett SN, Caporaso N, Fitzpatrick A, et al. Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience. Genet Epidemiol 2011;35:159–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stover PJ, Harlan WR, Hammond JA, Hendershot T, Hamilton CM. PhenX: a toolkit for interdisciplinary genetics research. Curr Opin Lipidol 2010;21:136–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rutledge PC, Park A, Sher KJ. 21st birthday drinking: extremely extreme. J Consult Clin Psychol 2008;76:511–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet 2008;123:1–14 [DOI] [PubMed] [Google Scholar]
- 73.Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Stat Sci 2009;24:561–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS ONE 2007;2:e841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Monsees GM, Tamimi RM, Kraft P. Genome-wide association scans for secondary traits using case-control samples. Genet Epidemiol 2009;33:717–28 [DOI] [PMC free article] [PubMed] [Google Scholar]