Abstract
Background: Certain immunotoxic peptides from gluten are resistant to gastrointestinal digestion and can interact with celiac-patient factors to trigger an immunologic response. A gluten-free diet (GFD) is the only effective treatment for celiac disease (CD), and its compliance should be monitored to avoid cumulative damage. However, practical methods to monitor diet compliance and to detect the origin of an outbreak of celiac clinical symptoms are not available.
Objective: We assessed the capacity to determine the gluten ingestion and monitor GFD compliance in celiac patients by the detection of gluten and gliadin 33-mer equivalent peptidic epitopes (33EPs) in human feces.
Design: Fecal samples were obtained from healthy subjects, celiac patients, and subjects with other intestinal pathologies with different diet conditions. Gluten and 33EPs were analyzed by using immunochromatography and competitive ELISA with a highly sensitive antigliadin 33-mer monoclonal antibody.
Results: The resistance of a significant part of 33EPs to gastrointestinal digestion was shown in vitro and in vivo. We were able to detect gluten peptides in feces of healthy individuals after consumption of a normal gluten-containing diet, after consumption of a GFD combined with controlled ingestion of a fixed amount of gluten, and after ingestion of <100 mg gluten/d. These methods also allowed us to detect GFD infringement in CD patients.
Conclusions: Gluten-derived peptides could be sensitively detected in human feces in positive correlation with the amount of gluten intake. These techniques may serve to show GFD compliance or infringement and be used in clinical research in strategies to eliminate gluten immunotoxic peptides during digestion. This trial was registered at clinicaltrials.gov as NCT01478867.
See corresponding editorial on page 537.
INTRODUCTION
Gluten is the storage protein of wheat, rye, barley, and oats and is not well tolerated in genetically predisposed individuals who suffer from CD4. Although most dietary proteins are digested into simple amino acids, dipeptides, and tripeptides by gastrointestinal proteases, gluten proteins are not completely digested and remain in the gastrointestinal tract (1, 2). The α-gliadin 33-mer is one of the digestion-resistant gluten peptides that is highly reactive to isolated celiac T cells and is the main immunodominant toxic peptide in celiac patients (3–5).
A lifelong GFD is currently the only available treatment for CD patients. Clinical manifestations associated with untreated CD, such as osteoporosis, anemia, depression, and infertility, can ameliorate with a GFD. Therefore, strict adherence to a GFD is essential to reduce symptoms, avoid nutritional deficiencies, and improve quality of life. However, according to several reports, dietary transgression is relatively frequent (32.6–55.4%) in celiac patients (6). In addition, a part of the celiac population (5–10%) does not respond to a GFD and has persistent villous atrophy with continued malabsorption. Although it is possible for patients to relapse despite a strict GFD, involuntary infringement or hypersensitivity to small amounts of gluten can also trigger the symptoms of the disease. Thus, an accurate marker that allows short-term monitoring of GFD compliance is needed. Approximately 1–2% of patients, mainly adults, can develop RCD, which is characterized by symptomatic malabsorption and persistent villous atrophy despite a strict GFD (7, 8). A demonstration of a lack of gluten in the diet of RCD patients would help in the differential diagnosis (9). Direct measures of dietary transgressions, such as mucosal inflammation and antitissue transglutaminase or antigliadin antibody concentrations in serum could be used to confirm GFD compliance. However, because a decrease in antibody titers may take years to achieve even when an individual is supposedly compliant with a GFD, the effectiveness of these markers to monitor gluten ingestion is unclear (10–12). Furthermore, there is no effective method to rule out the possibility that the symptoms of RCD are not due to a hypersensitive intolerance to gluten traces or are associated with involuntary gluten exposure (13).
The G12 and A1 moAbs against the main immunogenic epitope of the α-gliadin 33-mer have proved to be very useful for the detection of toxic peptides in food samples and for the enzymatic detoxification of gluten in clinical research (14, 15). In this study, we evaluated the feasibility of monitoring gluten ingestion by detection of 33EPs in feces. Detection of whole and digested gluten in feces would be useful in clinical research studies and GFD monitoring, as well as for the precise diagnosis of RCD. Our results suggested that immunologic methods that use the G12 moAb could be a specific and reliable novel tool for the detection of prolamine immunotoxic peptides in feces and, thus, useful in research and clinical monitoring of CD.
SUBJECTS AND METHODS
Subjects
Subjects were recruited at 2 different referral centers after approval by the local ethics committee. Twenty-six healthy subjects (16 female and 10 male subjects; age range: 3–42 y) were enrolled in the study. Exclusion criteria included the presence of known medical disease, digestive disease symptoms, family history of CD, use of prescription medications, and use of antibiotics or probiotics in the previous 2 mo to the inclusion in the study. All subjects had normal hemoglobin levels, serum biochemical analysis, and renal and hepatic function.
A total of 53 celiac patients (35 female and 18 male subjects; age range: 1–12 y), 7 subjects of whom had active disease and 46 subjects of whom consumed a GFD for >2 y, were included in the study. The diagnosis of CD was based on the detection of IgA antiendomysial and IgA antitissue transglutaminase antibodies in serum and confirmed by a small intestinal biopsy. Biopsy specimens were obtained by using gastrointestinal endoscopy and evaluated according to the Marsh criteria for diagnosis of CD (16). All patients included in the study showed Marsh 3 grade villous atrophy at the time of the diagnosis.
Six control subjects (5 women and 1 man; age range: 33–65 y) with functional dyspepsia according to the Rome III criteria were also included in the study. All subjects showed normal duodenal mucosa (Marsh 0) and negative serology for CD.
The Local Ethics Committee of the Hospital Universitario de León (Leon, Spain) and the “Hospital de Valme” (Seville, Spain) approved the study, and informed consent was obtained from subjects.
Fecal sampling
Fresh stools from healthy subjects were obtained during different diet conditions as follows: 1) before initiation of the GFD while on a normal gluten-containing diet, 2) on each day of the GFD, 3) on the fourth day after administration of 9 g gluten/d, 4) on the fourth day after administration of 30 g gluten/d, and 5) on each day of administration of a gluten microdose (50 mg to 1 g/d).
In celiac patients, fecal samples were collected at different stages of the disease as follows: before the start of a GFD in active patients, while following a GFD in patients in remission, and during a gluten challenge in patients in remission. In nonceliac controls with other intestinal pathologies, fecal samples were collected while subjects followed a normal gluten-containing diet. All samples were homogenized and aliquoted within 3 h of defecation and stored at −80°C.
Experimental design
Analysis of the presence of gluten and 33EPs in feces of subjects was carried out blinded, and dietary compliance was assessed retrospectively by a patient or parent interview.
Extraction of peptides/prolamines from feces
Prolamines from feces were extracted by mixing 1 g feces with 10 mL of 60% (vol:vol) ethanol in a rotary shaker for 1 h at room temperature. Then, the suspension was centrifuged at 2500 × g for 10 min, and the supernatant fluid was obtained.
G12 moAb and 33-mer peptide
The G12 moAb, its derived horseradish peroxidase–conjugated moAb, and the 33-mer peptide used in the study were from Biomedal SL.
Simulated gastrointestinal digestion
To reproduce gastric digestion in vitro, PWG gliadin was incubated at 37°C for 60 min in HCl solution (pH 2.0) that contained 0.60 mg pepsin/mL (Sigma). Gastric digests were adjusted to pH 6.0 with sodium phosphate buffer and subjected to a simulated duodenal digestion by sequential addition of the pancreatic enzymes trypsin (0.375 mg/mL) and chymotrypsin (0.375 mg/mL) at 37°C for 30 min (both from Sigma) (17).
G12 moAb immunochromatographic test for detection of gluten
The immunochromatographic assay was modified from the guidelines of the manufacturer (GlutenTox Stick; Biomedal SL). After prolamine extraction with ethanol 60%, samples were serially diluted (1:10–1:20,000) in the buffer solution provided by the manufacturer for concentrations that corresponded to 6, 25, 50, 100, 250, and 500 ppm. The immunochromatographic strips were dipped into the sample solutions (300 μL each) for 10 min and allowed to air dry afterward.
SDS-PAGE fractionation and Western blotting
SDS-PAGE was prepared with 15–18% acrylamide, and tricine–SDS-PAGE was performed as described by Sousa et al (18). Proteins and peptides in the gel were silver stained or transferred to a polyvinylidene fluoride membrane. Membranes were incubated with G12 moAb after washing and incubating with antimouse IgG-phosphatase antibody (Sigma) (17).
Competitive ELISA
Relative amounts of samples that contained immunotoxic epitopes were quantified by a G12 competitive ELISA (15, 17). Maxisorp microtitre plates (Nunc) were coated with PWG gliadin solution and incubated overnight at 4°C. The plates were washed with PBS-Tween 20 buffer and blocked with blocking solution (PBS and 5% nonfat milk) for 1 h at room temperature. Serial dilutions were made of the 33-mer peptide and the samples in PBS-bovine serum albumin 3%, to each of which was added horseradish peroxidase–conjugated G12 moAb solution. The samples were preincubated at room temperature and added to the wells. After 30 min of incubation at room temperature, the plates were washed, and substrate solution (TMB; Sigma) was added. The reaction was stopped with 1 M sulfuric acid, and the absorbance at 450 nm was measured (microplate reader UVM340; Asys Hitech GmbH).
Statistical analysis
All experiments were carried out in triplicate, and all statistical analyses were performed with SPSS 18.0 for Windows (SPSS Inc). PWG gliadin and 33-mer assays were expressed as means ± SDs. Differences between groups were examined by 1-factor ANOVA, and individual means were compared by using a Bonferroni-corrected Student's t test. The fecal 33-mer peptide assays were expressed as medians and ranges and 25th and 75th percentiles. Differences between groups were examined by using the Friedman's and Wilcoxon's tests. A statistical probability of P < 0.05 was considered significant for all analyses.
RESULTS
Quantification of PWG gliadin toxic peptides by the G12 moAb after simulated gastrointestinal digestion
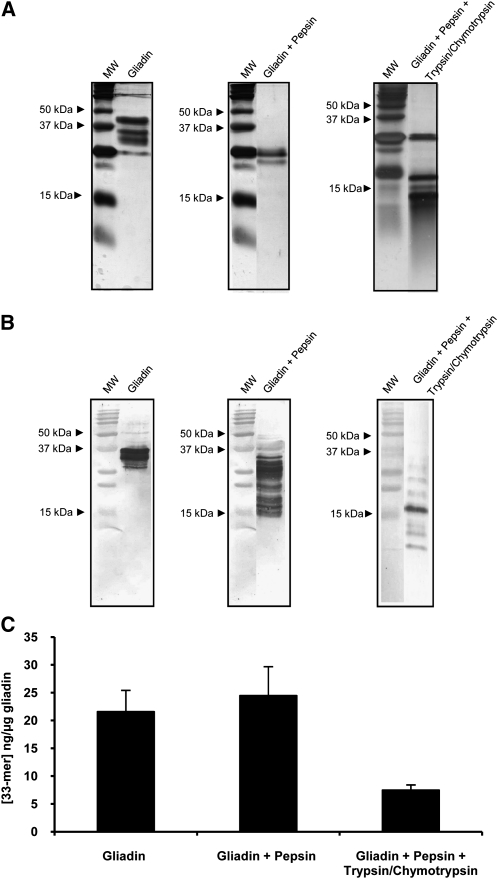
The gliadin 33-mer peptide contains 3 copies of the QPQLPY epitope. The G12 moAb specifically recognizes the QPQLPY epitope as well as other immunoreactive peptides in toxic prolamines (15). To evaluate the ability of the G12 moAb to detect the reactive peptides formed after gastrointestinal digestion of gliadin, we first subjected PWG gliadin to simulated gastric digestion by using pepsin (the major protease present in the stomach) followed by simulated intestinal digestion by using the pancreatic enzymes trypsin and chymotrypsin. The prolamine protein profile obtained after simulated digestion and the extent of the digestion were confirmed by SDS-PAGE. Electrophoretic separation of intact gliadin revealed intense bands of α, β, and γ gliadins (molecular weight: 33–45 kDa) and weak bands of omega gliadin (molecular weight: 50–67 kDa). Digestion of gliadin with pepsin gave rise to smaller peptidic fragments <25 kDa each, and additional digestion with trypsin and chymotrypsin generated even smaller peptides as a result of the enzymatic hydrolysis (Figure 1A). Additional Western blot analysis confirmed the recognition of the different reactive peptides by the anti–33-mer G12 moAb, even after treatment with gastric and pancreatic enzymes (Figure 1B).
FIGURE 1.
The mean (±SD) relative affinity of G12 moAb to immunotoxic peptides derived from PWG gliadin after simulated gastrointestinal digestion. A and B: SDS-PAGE and Western blot of PWG gliadin, PWG gliadin + pepsin, and PWG gliadin + pepsin + trypsin/chymotrypsin. Samples were stained with silver (A) or after transfer to a polyvinylidene fluoride membrane and incubated with G12 moAbs (B). C: Competition assay that measured the affinity of the horseradish peroxidase–conjugated G12 moAb to gliadin peptides present in gastrointestinal digests. Three assays were performed with samples run in triplicate. moAb, monoclonal antibody; MW, molecular weight marker; PWG, Prolamin Working Group.
To test the capacity of the G12 moAb to quantify the gliadin toxic peptides obtained after gastrointestinal digestion, a G12 competitive ELISA to measure the concentration of G12-moAb–related epitopes was performed on simulated gastrointestinal digests of PWG gliadin. This technique was chosen over sandwich ELISA because of its capacity to detect both intact and small protein fragments. The latter detection could be underestimated by sandwich ELISAs because they require ≥2 different epitopes per molecule. Gastric digestion of gliadin led to a slight increase in the concentration of reactive 33-mer peptides with respect to that of intact PWG gliadin (intact PWG gliadin: 21.6 ng 33-mer/μg; gastric-digested PWG gliadin: 24.5 ng 33-mer/μg). This increase could have been due to hidden epitopes of gliadin that became more accessible and allowed the interaction with the anti–33-mer moAb after digestion with gastric enzymes. Although with lower intensity (7.5 ng 33-mer/μg), the G12 moAb continued to recognize the gliadin peptides formed after intestinal digestion, which possibly indicated that two-thirds of the G12-recognized epitopes were destroyed by intestinal enzymes (Figure 1C).
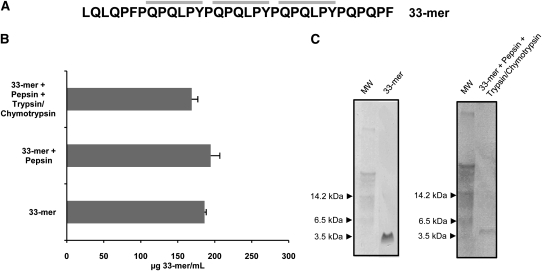
To verify that the 33-mer peptide remained intact after gastric and intestinal proteolysis, the 33-mer peptide was subjected to simulated gastrointestinal digestion, and its concentration was determined after each digestive step by using a G12 competitive ELISA. The 33-mer peptide concentration after gastric digestion (194 μg/mL) or intestinal digestion (169 μg/mL) did not vary significantly from that of nondigested 33-mer peptide (186 μg/mL) (Figure 2, A and B). These results were confirmed by Western blot analysis in which a single band of ~3.5 kDa in size was seen in the undigested and the sequentially digested 33-mer samples (33-mer theoretical molecular weight: 3.9 kDa) (Figure 2C).
FIGURE 2.
Mean (±SD) resistance of 33-mer peptides to cleavage by gastrointestinal enzymes. A: Amino acid sequences of the 33-mer peptide. The G12 moAb recognition sequence in the 33-mer peptide is in bold-face type. B: Competition assay for the detection of 33-mer peptide by G12 moAb after treatment with pepsin, trypsin, and chymotrypsin. Three assays were performed with samples run in triplicate. C: Western blot analysis of 33-mer peptide by the G12 moAb after treatment with gastrointestinal enzymes. moAb, monoclonal antibody; MW, molecular weight marker.
The ability of the G12 moAb to detect hydrolyzed gliadin was also evaluated by using G12-moAb–based immunochromatographic strips. The detection limit for gliadin and hydrolyzed gliadin was 30 ng/mL (6 ppm of gluten) and 50 ng/mL (10 ppm of hydrolyzed gluten), respectively; whereas for the 33-mer peptide and the 33-mer peptide subjected to digestion, the detection limit was 0.5 ng/mL.
These sets of experiments show that a substantial portion of G12-moAb detectable prolamine epitopes from ingested food were resistant to gastrointestinal digestion and, thus, could be detected at any level of the gastrointestinal tract (at least in the absence of gut microflora).
In vivo monitoring of gluten peptides and proteins in feces of healthy individuals after consumption of a controlled gluten-containing diet
To test the feasibility and sensitivity to detect gluten in feces of individuals after consumption of a normal gluten-containing diet, we collected fecal samples from healthy subjects (n = 15) who received a nonstandardized diet in which gluten was consumed daily (at least one piece of white bread at breakfast and normal ingestion of, eg, pasta and cookies), and we analyzed the presence of gluten polypeptides by using G12 immunochromatographic strips and G12 competitive ELISA. The concentration of gluten peptides in feces by using G12 immunochromatographic strips increased ≥100-fold above the detection limit in all individuals after consumption of a normal gluten-containing diet. The G12 competitive ELISA confirmed the presence of 33EPs in the fecal samples of all subjects after consumption of a normal gluten-containing diet and showed a gluten peptide excretion that ranged between 226 and 87,092 ng 33EPs/g feces, which suggested that the degree of gluten hydrolysis was strongly affected by the diet, type of gluten-containing food, and/or individual characteristics such as the gut microflora.
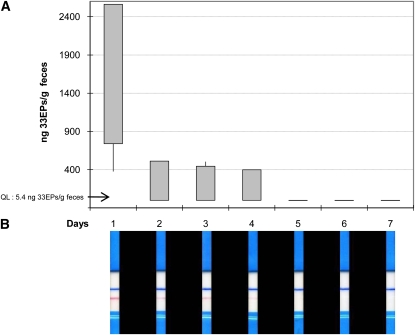
After proving the ability to detect gluten in feces by immunologic methods, we sought to determine the time needed for the reactive gluten peptide to be excreted. For this assessment, a group of healthy volunteers, who consumed a normal gluten-containing diet, received a GFD for 1 wk, and fecal samples were collected every day for the 1-wk-long gluten-free period. The time necessary for the gluten reactive peptide to become undetectable ranged from 2 to 4 d as determined by G12 immunochromatographic strips and G12 competitive ELISA (Figure 3).
FIGURE 3.
Determination of the time to elimination of ingested gluten in healthy individuals subjected to a gluten-free diet for 7 d. A: Competitive assay to determine the concentration of 33EPs (ng/g) in feces (n = 4). Each sample was analyzed in triplicate, and maximum, minimum, and 25th and 75th percentiles are shown. B: One representative immunochromatographic-strip example of the trial performed with the samples collected during the study period of one subject. Blue stripes represent an internal positive control that indicates that the stick worked properly; pink stripes indicate the presence of gluten. QL, quantification limit; 33EPs, 33-mer equivalent peptidic epitopes.
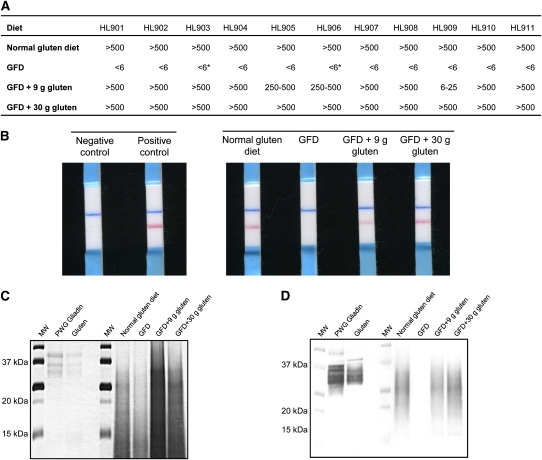
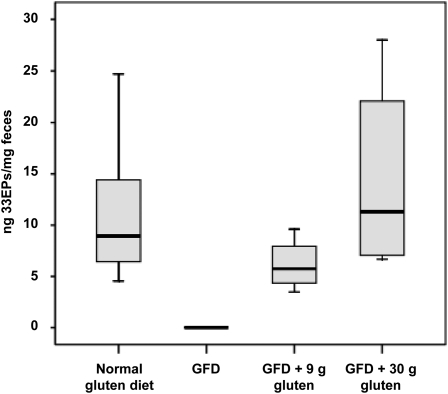
Given the variability shown in the concentration and time to excretion of gluten-reactive peptides across subjects after consumption of a normal gluten-containing diet, the next experiments to monitor the presence of 33EPs in feces were performed with a fixed amount of gluten. Thus, to examine whether the amount of gluten consumption correlated with the amount of gluten excretion, 11 healthy volunteers were subjected to a strict GFD for 1 wk, after which a fixed amount of 9 g of gluten, which was distributed across the main meals, was administered every day for 4 consecutive days followed by 30 g of gluten for 4 additional days. One single type of gluten (without previous heat treatment) was used in all cases to avoid variability because of the administration of gluten from different sources. For quantification by using G12 immunochromatographic strips, the stool samples were serially diluted after collection to cover a range between 6 and 500 ppm. After consumption of a normal gluten-containing diet, all subjects showed gluten excretion in feces, with values that exceeded 500 ppm (Figure 4), whereas all fecal samples collected during the period in which subjects consumed a GFD had gluten amounts below the detection limit of the method. When subjects consumed 9 g gluten/d, the amount of gluten detected in feces was >250 ppm in all but one case, which contained between 6 and 25 ppm, and when subjects consumed 30 g gluten/d, excreted amounts were >500 ppm (Figure 4, A and B), which represented a 100-fold increase over the detection limit. This result showed an approximate correlation between the amount of gluten consumed and the amount of gluten peptides with G12 epitopes excreted in feces. In addition, Western blot analysis showed G12 moAb reactivity for all samples (positive controls, GFD + 9 g gluten, and GFD + 30 g gluten), except for those obtained during the GFD period (Figure 4, C and D). G12 competitive ELISA, which was used to determine the concentration of toxic peptides in these samples, showed the presence of 33EPs in all stool samples collected during the normal gluten containing–diet period, whereas the amounts of toxic peptides were below the quantification limit of the method (5.4 pg 33EPs/mg sample) when the subjects were maintained on a GFD. The range of 33EPs in subjects who consumed 30 g gluten/d was 6.7–28.0 ng 33EPs/mg feces (1240–5185-fold above the detection limit), which was significantly higher (P = 0.018) than in subjects who consumed 9 g gluten/d, in whom the range was 3.5–9.6 ng 33EPs/mg feces (648–1778-fold above detection limit) (Figure 5). These results were in agreement with those obtained with immunochromatographic strips.
FIGURE 4.
Detection of gluten in feces of healthy individuals subjected to a controlled gluten-containing diet. A: Semiquantitative analysis of gluten peptides and proteins in feces of healthy individuals by using G12 immunochromatographic strips HL901–HL911 (studied subjects: n = 11). An asterisk indicates that gluten traces were detected. B: One representative immunochromatographic-strip example of the trial performed with the samples collected during the study period for one subject. Blue stripes represent an internal positive control that indicates that the stick worked properly; pink stripes indicate the presence of gluten. C and D: SDS-PAGE and Western blot of gluten peptides and proteins extracted from feces. GFD, gluten-free diet; MW, molecular weight marker; PWG, Prolamin Working Group.
FIGURE 5.
Competitive ELISAs that used horseradish peroxidase–conjugated G12 monoclonal antibodies to test the relation between the gluten proteins ingested and excreted and the 33-mer–related peptide content in feces. The concentration of 33EPs (ng/mg) in feces after a controlled gluten-containing diet is shown. The concentration of 33-mer peptide was determined by comparison with a synthetic 33-mer standard curve (n = 11). Median, maximum, minimum, and 25th and 75th percentiles are shown. Three assays were performed with samples run in triplicate. GFD, gluten-free diet; 33EPs, 33-mer equivalent peptidic epitopes.
In a significant proportion of celiac patients, an abnormal small bowel morphology persists despite a GFD, probably because of the persistent ingestion of trace amounts of gluten. Catassi et al (19) showed that treatment of CD required the ingestion of <50 mg gluten/d. To test whether the G12-moAb–based methods were capable of detecting these small amounts of gluten in fecal samples, increasing doses of gluten (from 50 mg to a maximum of 1g gluten/d) were administered after an initial 50-mg microdose. Subjects were given a standard piece of white bread that contained 25 mg gluten per piece of bread, and the gluten content in the stools was measured by using G12 immunochromatographic strips and G12 competitive ELISA. Gluten amounts in feces became detectable beyond the 50-mg dose of gluten, and the individual variations seen were probably due to individual differences and variations in daily food consumption.
Quantification of gluten peptides and proteins in feces of CD patients after consumption of a gluten-containing diet and GFD
To determine whether the G12-moAb–based methods were suitable for monitoring gluten ingestion in celiac patients, a study was conducted in CD patients to measure gluten peptides and proteins in feces. The study included 43 patients with CD in remission who had been consuming a long-term GFD (>2 y), 7 patients with active disease who were consuming normal gluten-containing diet up to the moment of diagnosis, and 3 CD patients who were consuming a GFD and were subjected to a gluten challenge. Non-CD subjects with other intestinal pathologies and healthy volunteers were used as control subjects. In 42 of 43 patients with CD in remission the amounts of toxic gluten peptides in feces were below the detection limit of the method. The remaining patient had gluten peptide amounts >6 ppm. When interviewed at the end of the study, this supposedly noncompliant patient confirmed consumption of cake (which contained gluten) during the study. In patients with active CD, the gluten shown in feces collected during the normal gluten-containing diet period was >6 ppm (range: 118–135 ng 33EPs/g feces).
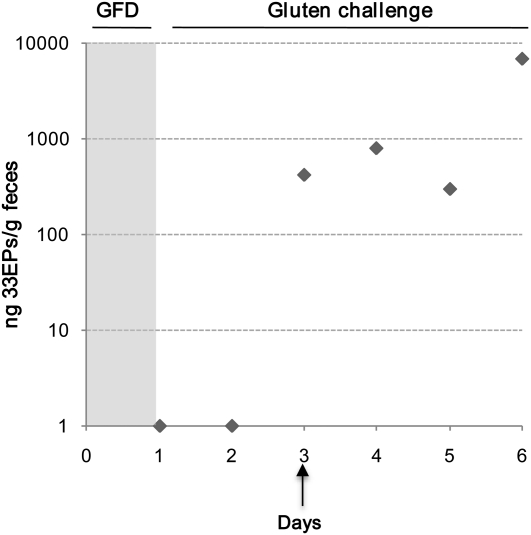
To assess the presence of gluten in feces of CD patients in remission, patients in remission after consumption of a GFD were given a gluten-containing diet for 6 d, and their stools were collected each day of the challenge period. The gluten challenge was carried out to confirm the diagnosis of CD and at the request of parents who were uncertain about the diagnosis. Out of the 3 patients who received the gluten challenge, 2 patients developed symptoms of vomiting, abdominal pain, and diarrhea within the first days of the administration of gluten and were excluded from the study. However, in the patient who tolerated the gluten challenge, fecal samples were collected for each of the 6 d that the challenge lasted, and the amount of gluten excreted was analyzed by using G12 antibody-based immunological methods to determine whether there was gluten excretion and the amounts of toxic peptides excreted during the period of gluten challenge. According to G12 competitive-ELISA results (Figure 6), excretion of gluten peptides started on the third day of the gluten challenge. Between days 3 and 6 of the challenge, fecal amounts of gluten reactive peptides ranged between 417 and 7037 ng 33EPs/g feces. These results were corroborated by using G12 immunochromatographic strips. In non-CD control subjects with other intestinal pathologies, the gluten content, although the subjects consumed a normal gluten-containing diet, ranged between 201 and 29,076 ng 33EPs/g feces. The intraindividual and interindividual differences seen in excretion could have been related to the diet, the type of gluten-containing food, and the variability in fermentative degradation.
FIGURE 6.
Analysis of mean (±SD) gluten amounts excreted in the feces of a celiac patient subjected to a gluten challenge for 6 d. Amounts of toxic peptide (ng/g feces) were determined by using a G12 competitive ELISA. Data were obtained from 3 independent experiments with samples run in triplicate. GFD, gluten-free diet; 33EPs, 33-mer equivalent peptidic epitopes.
These results confirmed that the G12 immunologic methods used for the detection of 33-mer gluten peptides were reliable tools for the detection of GFD transgressions in patients with active CD and with CD in remission after consumption of a GFD. These methods could, therefore, be used to verify GFD compliance to improve the diagnosis of cases refractory to the diet or with acute symptoms and for prolamine detoxification therapies in clinical research.
DISCUSSION
In this article, we describe a novel method to detect and monitor the presence of immunodominant gluten peptides in human feces on the basis of the use of the antigliadin 33-mer G12 antibody. To our knowledge, this was the first study to detect gluten-derived peptides in feces of patients with CD or other intestinal pathologies. The resistance of gluten peptides to gastrointestinal digestion, in particular that of the immunotoxic 33-mer peptide, ensures that a significant part of the ingested gluten peptides are excreted in feces, and thus recovery of measurable amounts of the immunotoxic fraction in feces would indicate that gluten has passed through the digestive tract and, therefore, that gluten has been consumed. In this study, the anti–33-mer G12-based immunoassays showed that >30% of the gliadin-reactive peptides remained intact after hydrolysis during in vitro simulated gastrointestinal digestion. However, the absorption of intact gliadin-reactive peptides along the gastrointestinal tract may vary between individuals. Diversity in the gut microflora (microbiome) and in the accompanying diet could have a dramatic impact on the resultant peptide concentration in feces (20). Although a low digestion rate of gluten peptides would reduce their absorption, most of the undigested gluten peptides would be excreted, and either rapid immunochromatographic strips or ELISAs could be used to detect gluten ingestion in feces.
A main application of these assays to measure gluten in feces could be the monitoring of GFD compliance in CD patients. In the current study, we observed that, despite a large variation between individuals, there was a rough correlation between the amount of gluten consumed and the amount of gluten excreted. Differences in the amount of ingested gluten could be estimated by using different methods, such as immunochromatographic strips, competitive ELISA, and Western blot. Immunochromatographic strips might be attractive as clinical standard assays in a broad range of laboratories as well as in point-of-care settings. However, the use of a G12 competitive ELISA could be more suitable in situations that require a more quantitative analysis of the sample such as when monitoring a GFD, identifying the source of dietary infringement in CD patients, or assessing the efficacy of novel therapies in the destruction of gluten toxic peptides (4, 21).
Some authors (19) have suggested that prolonged ingestion of 50 mg gluten/d causes significant damage to the architecture of the small intestine in patients who have begun treatment of CD. Furthermore, high consumption of gluten (1–5 g/d), although lower than that of the nonceliac population, causes a relapse of the disease at clinical, serologic, and histologic amounts. As a result, it is important to know the capacity of the analytic method used to detect ingestion of small amounts of gluten. Our results indicated that the ingestion of gluten amounts as low as 50 mg in processed bread could be detected in feces.
Currently, there are no validated methods to monitor adherence to a GFD. Antitissue transglutaminase antibodies have been proposed as markers of GFD compliance; however their effectiveness is still not clear (10). IgA-class antibodies can take several months to decrease, and IgG-class antibodies can take ≥1 y to decrease, after the antigen source has been eliminated from the diet. Therefore, serology does not predict recovery (8) and is not useful for follow-up. The use of serial endoscopies is not useful either because it is not considered an ethical practice. Other tests suggested as suitable diet-monitoring markers were the permeability test and fecal calprotectine (12, 13). We can measure the consequences of dietary transgressions, such as mucosal inflammation, but there is no way to show gluten intake and avoid the harmful aftermaths. Dietary interviews, although shown to be helpful in the determination of diet compliance, are often not possible to standardize, are subjective or rely on a truthful response from the patient, and cannot identify involuntary infringements. Our results showed that a broad range of gluten amounts (from 50 mg to 30 g) could be detected in feces of pediatric patients who were recently diagnosed with CD, were in remission, or were subjected to a gluten challenge; however, future studies that involved adult CD patients should be performed. These studies confirmed the use of G12 immunoassays to more accurately diagnose and monitor the disease and, thus, their value to study nonresponsive patients and to monitor adherence to a GFD.
In this study, we estimated the time of gluten toxic-peptide excretion to be between 2 and 4 d. These results should be taken into account if these methods are used in the diagnosis of gluten intoxication or if acute CD symptoms appear in an individual already treated with GFD.
Patients with RCD do not experience remission of symptoms while consuming a strict GFD. The diagnosis of RCD requires the exclusion of other diseases that can cause diarrhea and villous atrophy and the confirmation of adherence to a strict GFD (7, 8). A previous study showed that 82–90% of RCD patients referred to 2 large tertiary care centers were consuming gluten and had been improperly diagnosed of RCD (22). Moreover, there is no effective method to rule out if RCD symptoms are due to hypersensitive to trace amounts of gluten or to involuntary gluten exposure. The results obtained in the current study with patients with active CD patients, in remission, or after a gluten challenge demonstrated that these methods could be used to determine uncontrolled gluten ingestion and might be a useful for studying RCD (7, 8).
In a previous study, we showed that the reactivity of antigliadin 33-mer moAbs correlated with the potential immunotoxicity of gluten proteins. T cell–reactivity analysis and enzymatic detoxification of gluten by glutenases showed that the signal of the antigliadin 33-mer moAbs correlated with the sample's potential toxicity for celiac patients (4, 15, 21), which suggested that glutenases were able to cleave the epitopes detected by these antibodies. Therefore, the determination of the content of 33EPs in feces could be used to monitor gluten detoxification of celiac patients treated with novel enzymatic therapies (3, 21, 23), proving the utility of these methods in experimental therapies for CD.
In this article, we have shown how to monitor the consumption of gluten by simple immunologic assays in feces and thereby overcome some unresolved scientific and clinical problems in celiac-patient monitoring, including 1) the monitoring of short -term GFD compliance, 2) assessment of the efficacy of enzymatic therapies, and 3) how to diagnose gluten intoxication that is due to involuntary food contamination. Researchers may want to consider the use of these methods when designing clinical trials in CD and to correctly interpret acute or refractory symptoms in celiac patients. In conclusion, the resistance of 33EPs to gastrointestinal digestion and the use of G12 anti–33-mer-based tests may be useful for monitoring dietary compliance in CD patients due to its noninvasiveness, sensitivity, and significant correlation with consumed gluten.
Acknowledgments
We are grateful to Carolina Argüelles for her valuable suggestions in the writing of the manuscript and thank the generous volunteer subjects who enrolled in the study. We also thank Manuel Romero and Catherine N Torgler for comments on the manuscript and Verónica Segura for assistance with the ELISAs.
The authors’ responsibilities were as follows—IC, SV, AR-H, JC, ÁC, and CS: designed the research; IC, AR, MÁS, AC, and EN: conducted the research; SV, JC, ÁC, and CS: provided essential reagents or materials; IC, AR-H, ÁC, and CS: analyzed data or performed statistical analyses; IC, SV, AR-H, ÁC, and CS: wrote the manuscript; IC and CS: had primary responsibility for the final content of the manuscript; and all authors: read and approved the final manuscript. CS, IC, SV, AR, and ÁC are inventors of the patent “Monitoring the adherence to gluten-free diet in feces” (no. P201001633). MÁS, AC, EN, JC, ARH had no conflicts of interest.
Footnotes
Abbreviations used: CD, celiac disease; GFD, gluten-free diet; moAb, monoclonal antibody; PBS, phosphate-buffered saline; PWG, Prolamin Working Group; RCD, refractory celiac disease; 33EP, 33-mer equivalent peptidic epitope.
REFERENCES
- 1.Fasano A. Surprises from celiac disease. Sci Am 2009;301:54–61 [DOI] [PubMed] [Google Scholar]
- 2.Ganapathy V, Gupta N, Martindale RG. Physiology of the gastrointestinal tract. 4th ed. New York, NY: Johnson LR, 2006 [Google Scholar]
- 3.Shan L, Molberg Ø, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Structural basis for gluten intolerance in celiac sprue. Science 2002;297:2275–9 [DOI] [PubMed] [Google Scholar]
- 4.Bethune MT, Crespo-Bosque M, Bergseng E, Mazumdar K, Doyle L, Sestak K, Sollid LM, Khosla C. Noninflammatory gluten peptide analogs as biomarkers for celiac sprue. Chem Biol 2009;16:868–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tye-Din JA, Stewart JA, Dromey JA, Beissbarth T, van Heel DA, Tatham A, Henderson K, Mannering SI, Gianfrani C, Jewell DP, et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci Transl Med 2010;2:41ra51 [DOI] [PubMed] [Google Scholar]
- 6.Silvester JA, Rashid M. Long-term follow-up of individuals with celiac disease: an evaluation of current practice guidelines. Can J Gastroenterol 2007;21:557–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadithi M, Peña AS. Current methods to diagnose the unresponsive and complicated forms of coeliac disease. Eur J Intern Med 2010;21:247–53 [DOI] [PubMed] [Google Scholar]
- 8.Walker MM, Murray JA. An update in the diagnosis of disease. Histopathology 2011;59:166–79 [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Calle P, Codoceo R, Polanco I, Gómez-Cerezo J, Orsi M, Tenias JM. Is an intestinal permeability test a valid marker for slight dietary transgressions in adolescents with coeliac disease? Gut 1993;34:774–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tack GJ, Verbeek WH, Schreurs MW, Mulder CJ. The spectrum of celiac disease: epidemiology, clinical aspects and treatment. Nat Rev Gastroenterol Hepatol 2010;7:204–13 [DOI] [PubMed] [Google Scholar]
- 11.Selimoğlu MA, Karabiber H. Celiac disease: prevention and treatment. J Clin Gastroenterol 2010;44:4–8 [DOI] [PubMed] [Google Scholar]
- 12.Duerksen DR, Wilhelm-Boyles C, Parry DM. Intestinal permeability in long-term follow-up of patients with celiac disease on a gluten-free diet. Dig Dis Sci 2005;50:785–90 [DOI] [PubMed] [Google Scholar]
- 13.Ertekin V, Selimoğlu MD, Turgut A, Bakan N. Fecal calprotectin concentration in celiac disease. J Clin Gastroenterol 2010;44:544–6 [DOI] [PubMed] [Google Scholar]
- 14.Morón B, Cebolla A, Manyani H, Alvarez-Maqueda M, Megías M, Thomas M C, López MC, Sousa C. Sensitive detection of cereal fractions that are toxic to celiac disease patients by using monoclonal antibodies to a main immunogenic wheat peptide. Am J Clin Nutr 2008;87:405–14 [DOI] [PubMed] [Google Scholar]
- 15.Morón B, Bethune M, Comino I, Manyani H, Ferragud M, López MC, Cebolla A, Khosla C, Sousa C. Toward the assessment of food toxicity for celiac patients: Characterization of monoclonal antibodies to a main immunogenic gluten peptide. PLoS ONE 2008;3:e2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh MN, Crowe PT. Morphology of the mucosal lesion in gluten sensitivity. Baillieres Clin Gastroenterol 1995;9:273–93 [DOI] [PubMed] [Google Scholar]
- 17.Comino I, Real A, de Lorenzo L, Cornell H, López-Casado MÁ, Barro F, Lorite P, Torres MI, Cebolla A, Sousa C. Diversity in oat potential immunogenicity: basis for the selection of oat varieties with no toxicity in celiac disease. Gut 2011;60:915–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sousa C, Johansson C, Charon C, Manyani H, Sautter C, Kondorosi A, Crespi M. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol Cell Biol 2001;21:354–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catassi C, Fabiani E, Iacono G, D'Agate C, Francavilla R, Biagi F, Volta U, Accomando S, Picarelli A, De Vitis I, et al. A prospective, double-blind, placebo-controlled trial to establish a safe gluten threshold for patients with celiac disease. Am J Clin Nutr 2007;85:160–6 [DOI] [PubMed] [Google Scholar]
- 20.Sonnenburg JL, Fischbach MA. Community health care: therapeutic opportunities in the human microbiome. Sci Transl Med 2011;3:78ps12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehren J, Morón B, Martin E, Bethune MT, Gray GM, Khosla C. A food-grade enzyme preparation with modest gluten detoxification properties. PLoS ONE 2009;4:e6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leffler DA, Dennis M, Hyett B, Kelly E, Schuppan D, Kelly CP. Etiologies and predictors of diagnosis in nonresponsive celiac disease. Clin Gastroenterol Hepatol 2007;5:445–50 [DOI] [PubMed] [Google Scholar]
- 23.Shan L, Qiao SW, Arentz-Hansen H, Molberg Ø, Gray GM, Sollid LM, Khosla C. Identification and analysis of multivalent proteolytically resistant peptides from gluten: implications for celiac sprue. J Proteome Res 2005;4:1732–41 [DOI] [PMC free article] [PubMed] [Google Scholar]