Abstract
Background
Increasing evidence suggests a role for inflammation in the development of type 2 diabetes. Elevated levels of inflammatory cytokines, including interleukin-6, have been associated with insulin resistance, and dietary lipids can increase cytokine production. The objective of this study was to determine whether a single nucleotide polymorphism near the IL6 gene (rs7801406) modifies the relationship between dietary fat and markers of insulin sensitivity.
Methods
Subjects were healthy men and women aged 20–29 years from the Toronto Nutrigenomics and Health Study. Dietary intake was estimated using a one-month semiquantitative food frequency questionnaire. Fasting blood samples were taken for genotyping and biomarker measurement.
Results
The single nucleotide polymorphism was not associated with any of the measures of insulin sensitivity. However, it modified the relationship between total dietary fat and the homeostasis model assessment of insulin resistance (P = 0.053 for interaction). Total fat intake was positively related to HOMA-IR in individuals homozygous for the G allele (β = 0.005 ± 0.002, P = 0.03), but not among heterozygotes. There was an inverse relationship between total fat intake and HOMA-IR in individuals who were homozygous for the A allele (β = −0.012 ± 0.006, P = 0.047).
Conclusion
These findings suggest that dietary fat influences insulin sensitivity differently depending on genotype.
Keywords: interleukin-6, insulin sensitivity, nutrigenomics, dietary fat
Introduction
Type 2 diabetes mellitus (T2DM) is a growing global health problem, representing one of the leading preventable causes of mortality worldwide.1 T2DM is preceded by insulin resistance, whereby the biological effects of insulin are less than expected for glucose disposal and suppression of endogenous glucose production.2 This decrease in insulin action is compensated for by an increase in insulin secretion3 and, over time, insulin resistance leads to beta-cell dysfunction and eventual destruction.3 Several factors contribute to the progression of T2DM, including both genetics and the environment.
One such environmental factor that contributes to insulin resistance is high availability of fatty acids.4 With prolonged exposure to excess dietary fat, the storage capacity of adipose tissue is exceeded and fatty acids are released into the circulation to be stored in nonadipose tissues, including skeletal muscle.5,6 Within the muscle, fatty acid metabolites can contribute to insulin resistance directly and also activate inflammatory signalling.4
It has been proposed that T2DM is an acute-phase disease in which increased concentrations of cytokines are secreted under the influence of stimuli such as overnutrition in predisposed individuals.7 The acute-phase response is an inflammatory outcome with a specific cytokine profile that is activated in response to exogenous threats, such as pathogens or dietary factors, in particular fatty acid consumption. The cytokines released are primarily derived from activated macrophages and can directly enhance insulin resistance in adipocytes, and muscle and liver cells.8,9 Interleukin-6 is a proinflammatory cytokine that is increased in T2DM, independent of obesity. Chronically elevated levels of interleukin- 6 have been related to a number of metabolic abnormalities associated with insulin resistance.10–13 Prolonged exposure to interleukin-6 may cause insulin resistance in muscles14 and increases the subsequent risk of developing T2DM.10–13 Levels of interleukin-6 and other inflammatory markers can be modified by diet, specifically dietary fat, and contribute to the risk and manifestation of T2DM.
Consumption of a high saturated fat diet is associated with higher concentrations of proinflammatory markers,15,16 and has also been shown to increase the expression of proinflammatory genes.17 Conversely, in a cross-sectional study, increased n-3 polyunsaturated fatty acid consumption was associated with decreased plasma concentrations of interleukin-6.18 However, these results are not consistent, suggesting that other factors may play a role in the relationship between fat consumption and cytokine synthesis.
Genetic polymorphisms in IL6 can affect circulating levels of interleukin-619 and have been associated with a modified risk of T2DM.20 We recently showed that the rs7801406 single nucleotide polymorphism near the IL6 gene is inversely related to insulin levels in a population of monozygotic and dizygotic twins.21 This single nucleotide polymorphism is in linkage disequilibrium with two common intronic polymorphisms in IL6, the rs1474347 (linkage disequilibrium = 0.85) and rs1474348 (linkage disequilibrium = 0.84), that exhibit linkage disequilibrium = 1.21 While lifestyle and genetics each play a role in the development of T2DM, study results have been inconsistent and interactions between these two types of risk factors may help explain some of the interindividual variation. The objective of the present study was to determine whether the rs7801406 single nucleotide polymorphism that was recently shown to modify the serum levels of insulin21 is associated with markers of T2DM and influences the association between dietary fat intake and insulin sensitivity in a population of young adults.
Methods
Study population
The Toronto Nutrigenomics and Health study is an investigation of the role of genetics and nutrition in health and biomarkers of chronic disease. This population consists of men and women aged 20–29 years recruited from the University of Toronto, Canada.22,23 In this population, 47% of subjects are Caucasian, 35% are East Asian, 11% are South Asian, and 7% are considered “other” (eg, mixed ethnicities or smaller sizes of ethnocultural groups). Individuals who may have under-reported (≤800 kcal/day) or over-reported (≥3500 kcal/day female, ≥4000 kcal/day male, n = 94) their energy intakes, those with diabetes or other conditions characterized by increased inflammation (n = 57; Crohn’s disease, ulcerative colitis, arthritis) and individuals with possible acute inflammation (n = 9), as determined by C-reactive protein ≥ 10 mg/L, were excluded from the analyses. Individuals who were missing relevant dietary and biomarker data were also excluded (n = 28). The final population consisted of 707 individuals. Written informed consent was obtained from all participants, and the study was approved by the Ethics Review Committee at the University of Toronto.
Dietary assessment
A 196-item Toronto-modified Willett food-frequency questionnaire was used to assess habitual dietary intake over the past month. Each subject was instructed on how to complete the food-frequency questionnaire using visual aids for portion sizes in order to improve the accuracy of self-reported food intake.
Anthropometrics and energy expenditure
Anthropometric measurements, including height, weight, and waist circumference, were measured and body mass index (kg/m2) was calculated. Modifiable activity was measured by questionnaire and metabolic equivalent hours per day was calculated. This measurement represents both leisure and occupational activity, not including sedentary hours of sleeping or sitting. One metabolic equivalent is equal to 1 kcal expended per kg body weight per hour sitting at rest.24
Laboratory measurements
Each subject had venous blood drawn after a 12-hour overnight fast to measure biomarkers of glucose and lipid metabolism as well as highly sensitive C-reactive protein using standard clinical procedures. Insulin and glucose measurements were used to calculate insulin sensitivity as homeostasis model assessment of insulin resistance (HOMA-IR), using the following equation:
Genotyping and allele frequency
The rs7801406 polymorphism was genotyped in the study subjects based upon its high frequency in the general population, its association with lower serum insulin levels as previously reported,21 and its strong linkage disequilibrium (>80%) with two common polymorphisms in the IL6 gene (rs1474347 and rs1474348). DNA was isolated from whole blood using the GenomicPrep Blood DNA isolation kit (Amersham Pharmacia Biotech, Piscataway, NJ). The rs7801406 polymorphism was detected using TaqMan® allelic discrimination assays (ABI C-29151539-10) from Applied Biosystems (Foster City, CA), with real-time polymerase chain reaction on an ABI 7000 sequence detection system. Polymerase chain reaction conditions were 95°C for 10 minutes and 40 cycles of 95°C for 15 seconds and 60°C for one minute.
When genotyped, 41% of Caucasians were G/G homozygotes for rs7801406, 45% were G/A heterozygotes, and 14% were A/A homozygotes. Of the South Asian subjects, 64% were G/G, 32% were G/A, and 4%, were A/A. Among the East Asian subjects, 98.9% were G/G, 0.9% were G/A, and 0.2% were A/A. Because of the low frequency of this polymorphism among East Asians, they were not included in any of the analyses.
Statistical analysis
All statistical analyses were performed using SAS version 9.1 (SAS Institute Inc, Cary, NC). The GLM procedure in SAS was used to perform a one-way analysis of variance to test for differences in the characteristics between genotypes. The CONTRAST statement was used for pairwise group comparisons and the χ2 test was used to analyze categorical variables. Non-normally distributed variables were loge-transformed for analysis and their antilogs are reported. High-sensitivity C-reactive protein was transformed following a gamma distribution using the GENMOD procedure, and the median and interquartile range values for this variable are given.
The GLM procedure was also used to test whether the association of dietary total, saturated fat, monounsaturated, or polyunsaturated fatty acids, as continuous or categorical variables, with insulin sensitivity varied across the three genotypes for each polymorphism. A diet-gene interaction was found for total dietary fat and the rs7801406 polymorphism on HOMA-IR. Covariates that were associated with the outcome and reduced the variance of the relationship between total dietary fat and HOMA-IR were included in the model. Potential covariates that were associated with HOMA-IR or differed between genotypes were added individually to the model to test whether the interaction changed. These variables were not included in the final model. No differences or interactions were found between the genotypes and any of these potential confounders. Total dietary fat intake was adjusted for total energy intake using the nutrient density method (percent of energy from dietary fatty acids).25 The slopes of the three lines were estimated using the GLM procedure on a fully adjusted model including the genotype and genotype × total fat interaction term. Departure of genotype distributions from Hardy-Weinberg equilibrium was assessed using a χ2 test with one degree of freedom. Significant P values are two-sided and ≤0.05.
Results and discussion
The minor allele frequency in the final study population was 20.8% and the single nucleotide polymorphism was in Hardy-Weinberg equilibrium (P = 0.94). Individuals who were homozygous for the G allele were younger than the heterozygotes; however, this absolute difference was only 0.5 years. Those who were homozygous for the A allele consumed more calories than those who were heterozygotes. Therefore, age and calorie consumption, as well as ethnicity, were all controlled for when analyzing the association of the examined single nucleotide polymorphism with metabolic characteristics. There were no significant differences between the genotypes for any of the measured metabolic characteristics, including HOMA indices, insulin and glucose, or family history of diabetes (see Table 1).
Table 1.
Clinical and metabolic characteristics and dietary intake by rs7801406 genotype*
| rs7801406 | ||||
|---|---|---|---|---|
|
|
||||
| G/G (n = 333) | G/A (n = 305) | A/A (n = 69) | P** | |
| Age (years) | 22.6 ± 0.1a | 23.1 ± 0.1b | 23.1 ± 0.3a,b | 0.02 |
| Gender (% female) | 61.3% | 72.5% | 71.0% | 0.008 |
| BMI (kg/m2) | 23.1 ± 0.2 | 23.0 ± 0.2 | 23.4 ± 0.4 | 0.67 |
| Smoking status (%) | ||||
| Never | 82.6% | 79.7% | 85.5% | |
| Past | 9.3% | 13.4% | 2.9% | |
| Present | 8.1% | 6.9% | 11.6% | 0.07 |
| Physical activity (MET · hour/week) | 8.2 ± 0.2 | 7.8 ± 0.2 | 8.6 ± 0.4 | 0.06 |
| Waist circumference (cm) | 76.6 ± 0.5 | 76.6 ± 0.6 | 77.7 ± 1.1 | 0.58 |
| Systolic blood pressure (mmHg) | 116.8 ± 0.6 | 117.3 ± 0.7 | 118.4 ± 1.2 | 0.39 |
| Diastolic blood pressure (mmHg) | 69.9 ± 0.5 | 70.2 ± 0.6 | 69.3 ± 1.1 | 0.69 |
| Total cholesterol (mmol/L) | 4.1 ± 0.05 | 4.3 ± 0.06 | 4.2 ± 0.10 | 0.17 |
| LDL cholesterol (mmol/L) | 2.3 ± 0.04 | 2.3 ± 0.05 | 2.3 ± 0.09 | 0.67 |
| HDL cholesterol (mmol/L) | 1.5 ± 0.02 | 1.5 ± 0.03 | 1.4 ± 0.05 | 0.40 |
| Triglycerides (mmol/L) | 0.94 ± 0.03 | 1.0 ± 0.03 | 1.0 ± 0.06 | 0.19 |
| Free fatty acids (μmol/L) | 453 ± 14.9 | 484 ± 17.3 | 513 ± 32.4 | 0.11 |
| hs-CRP (mg/L) | 0.5 ± 0.9 | 0.5 ± 1.0 | 0.6 ± 1.6 | 0.49 |
| Glucose (mmol/L) | 4.8 ± 0.02 | 4.8 ± 0.02 | 4.8 ± 0.05 | 0.47 |
| Insulin (pmol/L) | 52.7 ± 1.7 | 52.8 ± 2.0 | 53.0 ± 3.8 | 0.93 |
| HOMA-IR | 1.6 ± 0.05 | 1.6 ± 0.06 | 1.6 ± 0.1 | 0.94 |
| HOMA-β | 116.8 ± 4.5 | 116.0 ± 5.2 | 113.3 ± 9.8 | 0.74 |
| Family history of diabetes (%) | 14.1% | 11.5% | 7.3% | 0.24 |
| Energy intake (kcal/day) | 1989 ± 35.1a,b | 1967 ± 36.7a | 2187 ± 77.2b | 0.03 |
| Total fat (% energy) | 30.0 ± 0.4 | 30.0 ± 0.4 | 31.0 ± 0.8 | 0.52 |
| SFA (% energy) | 9.6 ± 0.1 | 9.9 ± 0.1 | 10.1 ± 0.3 | 0.19 |
| MUFA (% energy) | 12.0 ± 0.2 | 11.9 ± 0.2 | 12.5 ± 0.5 | 0.36 |
| PUFA (% energy) | 5.7 ± 0.1 | 5.6 ± 0.1 | 5.5 ± 0.2 | 0.46 |
| Cholesterol (mg/d) | 242.4 ± 7.3 | 235.1 ± 7.6 | 252.3 ± 15.9 | 0.58 |
| Carbohydrates (% energy) | 52.9 ± 0.5 | 53.0 ± 0.5 | 51.9 ± 1.0 | 0.60 |
| Fiber (g/100 g carbohydrates) | 9.2 ± 0.2 | 9.4 ± 0.2 | 9.3 ± 0.4 | 0.60 |
| Protein (% energy) | 16.9 ± 0.2 | 16.9 ± 0.2 | 16.9 ± 0.4 | 0.98 |
| Alcohol (g/day) | 6.9 ± 0.5 | 7.1 ± 0.6 | 7.9 ± 1.2 | 0.57 |
Notes: Metabolic characteristics are adjusted for subject characteristics and dietary variables that were different between each genotype (ethnicity, age, gender, energy intake). Metabolic characteristics include: waist circumference, systolic and diastolic blood pressures, total, LDL and HDL cholesterol, triglycerides, free fatty acids, hs-CRP, glucose, insulin, HOMA-IR and HOMA-β.
P values for differences between genotypes were obtained using one-way analysis of variance, and the Chi-square test was used to test for differences between genotypes in categorical variables. Median ± interquartile range given for hs-CRP.
Different letters represent statistically significant differences between groups (P < 0.05).
Abbreviations: hs-CRP, high-sensitivity C-reactive protein; HOMA-IR, homeostasis model assessment of insulin resistance; HOMA-β, homeostasis model assessment of beta cell function; LDL, low-density lipoprotein; HDL, high-density lipoprotein; SFA, saturated fatty acids; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids.
We evaluated whether the rs7801406 polymorphism modifies the association between dietary fat and biomarkers of insulin sensitivity (HOMA-IR and serum insulin levels). A significant interaction was found between the genotypes and total dietary fat on serum insulin (P = 0.02, adjusted for waist circumference, triglycerides, ethnicity, and serum glucose) and HOMA-IR (P = 0.053, adjusted for waist circumference, ethnicity, triglycerides, and total cholesterol:HDL-cholesterol). Dietary fat intake was inversely related to HOMA-IR in individuals homozygous for the A allele (β = −0.012 ± 0.006, P = 0.047); however, a positive relationship was found among G/G homozygotes (β = 0.005 ± 0.002, P = 0.03, Figure 1). The association between dietary fat and HOMA-IR in the G/G homozygotes was significantly different from the A/A homozygotes (P = 0.02), but not the heterozygotes (P = 0.78). However, the relationship in A/A homozygotes differed significantly from that in the heterozygotes (P = 0.03). Furthermore, the relationship between dietary total fat intake and HOMA-IR was not significant among heterozygotes (β = 0.005 ± 0.093, P = 0.51) and the slope of this relationship did not differ significantly from the other genotype groups. No interactions were observed with any of the other covariates in the model.
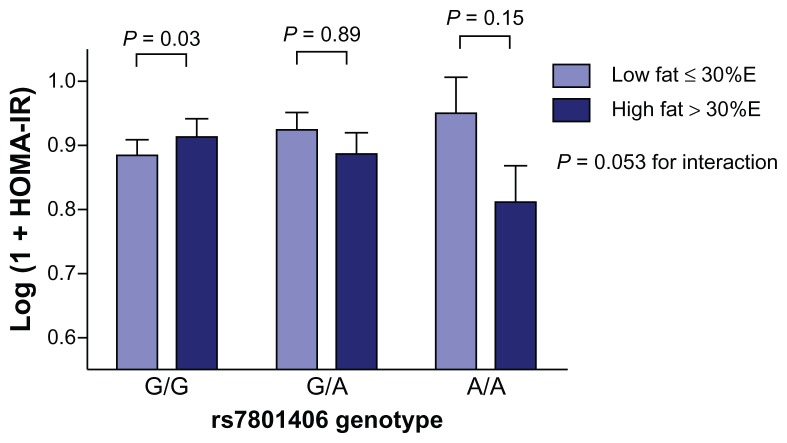
Figure 1.
Interaction between dietary fat, the rs7801406 polymorphism, and HOMAIR levels.
Notes: Values are means ± standard error of the mean adjusted for total: HDL cholesterol ratio, plasma triglycerides, waist circumference, and ethnocultural group. Dietary fat was inversely related to HOMA-IR in A/A carriers (β = −0.012 ± 0.006, P = 0.047) and positively among G/G homozygotes (β = 0.005 ± 0.002, P = 0.03). This relationship (in the G/G carriers) was significantly different from the A/A homozygotes (P = 0.02).
Abbreviations: HDL, high-density lipoprotein; HOMA-IR, homeostasis model assessment of insulin resistance.
As illustrated in Figure 1, the interaction between rs7801406 genotype and dietary fat intake on HOMA-IR continued to be significant when total fat intake was categorized according to recommended intake levels (≤30% of energy, P = 0.05). Comparing those who consumed total fat above the recommendation cutoff with individuals who met the recommendations in each genotype, mean HOMA-IR levels were significantly higher in subjects with the G/G genotype who consumed diets high in total fat (P = 0.03). No differences were observed between intake groups in the other genotypes (G/A, P = 0.89; A/A, P = 0.15). The inverse relationship observed in the A/A homozygotes was not significant, probably due to small sample size in this subgroup after stratification. We investigated whether ethnicity affected the associations between IL6 genotype and dietary fat; however, there was no interaction between the three factors. Moreover, stratification by ethnicity within the genotypes produced a sample size in the A/A group which did not permit a statistically meaningful conclusion.
The objective of this study was to determine whether the rs7801406 single nucleotide polymorphism is associated with factors related to predisposition to T2DM and if it can modify the relationship between dietary fat and disease risk factors. As mentioned above, our group recently found the rs7801406 polymorphism to be linked to lower serum insulin levels.21 This single nucleotide polymorphism is located near the IL6 gene and is in linkage disequilibrium with two common polymorphisms in IL6 (see above). A common single nucleotide polymorphism in the promoter region of IL6 (rs1800795, −174 G > C), was previously reported, in a large meta-analysis study, to reduce the risk of developing T2DM by nearly 10%.20 Although these findings suggest a role for IL6 polymorphisms in T2DM, additional studies confirming these results and investigating other polymorphisms, both in the IL6 gene or in a strong linkage disequilibrium with common IL6 single nucleotide polymorphisms, are necessary. In addition, studies examining gene-environment interactions can also provide insight into the mechanisms by which IL6 plays a role in the etiology of T2DM.
In the present study, rs7801406 was not associated with markers of T2DM; however, it modified the association between dietary fat and HOMA-IR. While this study does not confirm the association between rs7801406 and serum insulin levels as previously reported,21 the interaction between dietary fat and insulin could help explain this inconsistency. Overall, we found that rs7801406, although not associated with metabolic phenotypes related to T2DM risk, modified the relationship between dietary fat intake and insulin sensitivity in a population of young adults. Further studies are needed to confirm these results in other populations and to determine what effects this polymorphism has on interleukin-6 levels and function in the presence of a high-fat or low-fat diet.
Acknowledgments
BGB, MK, and AB received support from the Public Health Agency of Canada. The authors thank Andre Villages, Office of Biotechnology, Genomics and Population Health, Public Health Agency of Canada, for his help in identifying the proxy single nucleotide polymorphisms and the linkage disequilibrium analysis. AE-S holds a Canada Research Chair in Nutrigenomics.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wild S, Roglic G, Green A, et al. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Dinneen S, Gerich J, Rizza R. Carbohydrate metabolism in non-insulin-dependent diabetes mellitus. N Engl J Med. 1992;327:707–713. doi: 10.1056/NEJM199209033271007. [DOI] [PubMed] [Google Scholar]
- 3.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 4.Kewalramani G, Bilan PJ, Klip A. Muscle insulin resistance: assault by lipids, cytokines and local macrophages. Curr Opin Clin Nutr Metab Care. 2010;13:382–390. doi: 10.1097/MCO.0b013e32833aabd9. [DOI] [PubMed] [Google Scholar]
- 5.Storlien LH, Jenkins AB, Chisholm DJ, et al. Influence of dietary fat composition on development of insulin resistance in rats. Relationship to muscle triglyceride and omega-3 fatty acids in muscle phospholipid. Diabetes. 1991;40:280–289. doi: 10.2337/diab.40.2.280. [DOI] [PubMed] [Google Scholar]
- 6.Kraegen EW, Clark PW, Jenkins AB, Daley EA, Chisholm DJ, Storlien LH. Development of muscle insulin resistance after liver insulin resistance in high-fat-fed rats. Diabetes. 1991;40:1397–1403. doi: 10.2337/diab.40.11.1397. [DOI] [PubMed] [Google Scholar]
- 7.Pickup JC, Crook MA. Is type II diabetes mellitus a disease of the innate immune system. Diabetologia. 1998;41:1241–1248. doi: 10.1007/s001250051058. [DOI] [PubMed] [Google Scholar]
- 8.Bilan PJ, Samokhvalov V, Koshkina A, Schertzer JD, Samaan MC, Klip A. Direct and macrophage-mediated actions of fatty acids causing insulin resistance in muscle cells. Arch Physiol Biochem. 2009;115:176–190. doi: 10.1080/13813450903079314. [DOI] [PubMed] [Google Scholar]
- 9.Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest. 1995;95:2409–2415. doi: 10.1172/JCI117936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wannamethee SG, Lowe GD, Rumley A, Cherry L, Whincup PH, Sattar N. Adipokines and risk of type 2 diabetes in older men. Diabetes Care. 2007;30:1200–1205. doi: 10.2337/dc06-2416. [DOI] [PubMed] [Google Scholar]
- 11.Duncan BB, Schmidt MI, Pankow JS, et al. Low-grade systemic inflammation and the development of type 2 diabetes: the atherosclerosis risk in communities study. Diabetes. 2003;52:1799–1805. doi: 10.2337/diabetes.52.7.1799. [DOI] [PubMed] [Google Scholar]
- 12.Hu FB, Meigs JB, Li TY, Rifai N, Manson JE. Inflammatory markers and risk of developing type 2 diabetes in women. Diabetes. 2004;53:693–700. doi: 10.2337/diabetes.53.3.693. [DOI] [PubMed] [Google Scholar]
- 13.Spranger J, Kroke A, Mohlig M, et al. Inflammatory cytokines and the risk to develop type 2 diabetes: results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes. 2003;52:812–817. doi: 10.2337/diabetes.52.3.812. [DOI] [PubMed] [Google Scholar]
- 14.Jove M, Planavila A, Laguna JC, Vazquez-Carrera M. Palmitate-induced interleukin 6 production is mediated by protein kinase C and nuclear-factor kappaB activation and leads to glucose transporter 4 down-regulation in skeletal muscle cells. Endocrinology. 2005;146:3087–3095. doi: 10.1210/en.2004-1560. [DOI] [PubMed] [Google Scholar]
- 15.Basu A, Devaraj S, Jialal I. Dietary factors that promote or retard inflammation. Arterioscler Thromb Vasc Biol. 2006;26:995–1001. doi: 10.1161/01.ATV.0000214295.86079.d1. [DOI] [PubMed] [Google Scholar]
- 16.King DE, Egan BM, Geesey ME. Relation of dietary fat and fiber to elevation of C-reactive protein. Am J Cardiol. 2003;92:1335–1339. doi: 10.1016/j.amjcard.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 17.van Dijk SJ, Feskens EJ, Bos MB, et al. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am J Clin Nutr. 2009;90:1656–1664. doi: 10.3945/ajcn.2009.27792. [DOI] [PubMed] [Google Scholar]
- 18.He K, Liu K, Daviglus ML, et al. Associations of dietary long-chain n-3 polyunsaturated fatty acids and fish with biomarkers of inflammation and endothelial activation (from the Multi-Ethnic Study of Atherosclerosis [MESA]) Am J Cardiol. 2009;103:1238–1243. doi: 10.1016/j.amjcard.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stryjecki C, Mutch DM. Fatty acid-gene interactions, adipokines and obesity. Eur J Clin Nutr. 2011;65:285–297. doi: 10.1038/ejcn.2010.277. [DOI] [PubMed] [Google Scholar]
- 20.Huth C, Heid IM, Vollmert C, et al. IL6 gene promoter polymorphisms and type 2 diabetes: joint analysis of individual participants’ data from 21 studies. Diabetes. 2006;55:2915–2921. doi: 10.2337/db06-0600. [DOI] [PubMed] [Google Scholar]
- 21.Arora P, Garcia-Bailo B, Dastani Z, et al. Genetic polymorphisms of innate immunity-related inflammatory pathways and their association with factors related to type 2 diabetes. BMC Med Genet. 2011;12:95. doi: 10.1186/1471-2350-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontaine-Bisson B, El-Sohemy A. Genetic polymorphisms of tumor necrosis factor-alpha modify the association between dietary polyunsaturated fatty acids and plasma high-density lipoprotein-cholesterol concentrations in a population of young adults. J Nutrigenet Nutrigenomics. 2008;1:215–223. doi: 10.1159/000149825. [DOI] [PubMed] [Google Scholar]
- 23.Fontaine-Bisson B, Wolever TM, Chiasson JL, et al. Genetic polymorphisms of tumor necrosis factor-alpha modify the association between dietary polyunsaturated fatty acids and fasting HDL-cholesterol and apo A-I concentrations. Am J Clin Nutr. 2007;86:768–774. doi: 10.1093/ajcn/86.3.768. [DOI] [PubMed] [Google Scholar]
- 24.Ainsworth BE, Haskell WL, Leon AS, et al. Compendium of physical activities: classification of energy costs of human physical activities. Med Sci Sports Exerc. 1993;25:71–80. doi: 10.1249/00005768-199301000-00011. [DOI] [PubMed] [Google Scholar]
- 25.Willet W. Nutritional Epidemiology. 2nd ed. New York, NY: Oxford University Press; 1998. [Google Scholar]