Abstract
Previous studies suggest some effects of selenium on risk of several chronic diseases, which may be mediated through a small number of selenoenzymes with antioxidant properties. In this cross-sectional analysis of 195 participants from the Seattle Barrett’s Esophagus Study who were free of esophageal cancer at the time of blood draw, we examined whether the number of the minor alleles in 26 tagging single nucleotide polymorphisms (SNP) of five selenoenzyme genes [i.e., glutathione peroxidase 1–4 (GPX1–4) and selenoprotein P (SEPP1)] was associated with activity of GPX1 in white blood cells and GPX3 in plasma, and concentrations of SEPP1 and markers of oxidative stress [malondialdehyde (MDA) and protein carbonyl content] in plasma. At the gene level, associations were observed between overall variation in GPX1 and GPX1 activity (P = 0.02) as well as between overall variation in GPX2 and SEPP1 concentrations (P = 0.03). By individual SNP, two variants in GPX1 (rs8179164 and rs1987628) showed a suggestive association with GPX1 activity (P = 0.10 and 0.08, respectively) and two GPX2 variants (rs4902346 and rs2071566) were associated with SEPP1 concentration (P = 0.004 and 0.002, respectively). Furthermore, two SNP in the SEPP1 gene (rs230813 and rs230819) were associated with MDA concentrations (P = 0.03 and 0.02, respectively). Overall, our study supports the hypothesis that common genetic variants in selenoenzymes affect their activity.
Introduction
Experimental and epidemiologic studies suggest a protective effect of selenium on risk of several diseases that are associated with increased oxidative stress, such as cancer (1–3) and cardiovascular diseases (4). Important biological functions of selenium, including antioxidant properties, are exerted through selenoenzymes (5). Hence, genetic variants in these selenoenzymes may affect the activity of these selenoenzymes and subsequently oxidative stress and disease risk.
Previous studies that examined the impact of genetic variation in selenoenzymes focused on a nonsynonymous SNP8 in the GPX1 gene, which changed an amino acid from proline to leucine (GPX1 Pro200Leu; rs1050450; this SNP was previously known as Pro198Leu) (6–10). Whereas three larger studies showed lower erythrocyte GPX1 activity for the Leu allele (7, 9, 10), two smaller studies found no association (6, 8). In addition, recent studies suggest that variants in GPX4 (rs713041) and SEPP1 (rs3877899 and rs7579) genes are associated with lymphocyte GPX1 and GPX4 activities, plasma GPX3 activity, and/or plasma SEPP1 concentration before or after short-term selenium supplementation (11, 12) and a cross-sectional study of healthy individuals (13). These studies are limited by the fact that they focused on a few candidate SNP and hence may have missed the impact of other variants in the genes. Therefore, we investigated the association of genetic variants within five selenoenzyme genes (i.e., GPX1–4 and SEPP1) selected to cover common variation with the activity of GPX1 and GPX3, SEPP1 concentration, and oxidative stress measured by MDA and PCC.
Materials and Methods
Study population.
This cross-sectional study was conducted within the Seattle Barrett’s Esophagus Study (14, 15). On an ongoing basis, the study collected blood samples, esophageal tissue samples, anthropometric measures, and data on diet, lifestyle, and health at baseline and each study visit, which occurred between 0.5 and 3 y (mean of 1.6 y) (14, 16). Our analysis included 195 participants without esophageal cancer at the time of blood draw who had a blood sample and at least two buffy coat samples available from the first or second follow-up visit (to avoid depletion of the biospecimen depository) and whose genotyping was successful (only three samples failed genotyping). Blood samples were collected between 1995 and 2005. A standard questionnaire was administered in person by trained staff at baseline and follow-up visits to assess sociodemographic characteristics, medical history, dietary habits, and lifestyle (15–17). Height and body weight were measured by trained staff during baseline and follow-up visits. Overnight fasting blood samples were collected in lavendar K2 vacutainer tubes with or without EDTA as anticoagulant during study visits prior to endoscopy, separated into serum, plasma, and buffy coat under the standardized protocol, and stored at −70°C (17–19). All study participants provided written informed consent.
Laboratory measures.
All samples were restored from the biospecimen depository and all assays were conducted at once in the same laboratories. White blood cells were isolated from buffy coats prior to the assay using the standard method. We measured the activity of GPX1 in white blood cells and GPX3 in plasma applying our standardized protocol using OXItek commercial kit (ZMC catalogue no. 0805002, ZeptoMetric) based on the Paglia and Valentine method (20) and using cumene hydroperoxide as the substrate. QC of known activity were run at the beginning of the assay each day to ensure the quality of assay internally. The mean CV of GPX1 and GPX3 activity from all samples run as duplicates were 2.1 and 3.2%, respectively. Both GPX1 and GPX3 assays were conducted at the Fred Hutchinson Cancer Research Center.
SEPP1 concentration was measured in plasma samples using the sandwich ELISA method as previously described (21). The CV (mean ± SD) of blinded QC from two plasma samples each measured seven times was 6.8% (4.83 ± 0.33) and 17.1% (5.14 ± 0.88). SEPP1 assay was conducted at the Vanderbilt University Medical Center.
As an oxidative stress marker for lipid peroxidation, MDA in EDTA-treated plasma, which was restored from plasma samples collected in a vacutainer tube that included EDTA as an anticoagulant (17), was assayed spectrophotometrically by a standardized lipid peroxidation microplate-based procedure according to kit instructions (kit no. NWK-MDA01 from Northwest Life Science) (22). This assay was conducted at the Fred Hutchinson Cancer Research Center. As a second oxidative stress marker, we analyzed PCC (aldehyde or ketone) in plasma, indicative of protein oxidation, using the noncompetitive ELISA method that was previously developed (23). QC with known PCC was included in each plate. The CV (mean ± SD) from internal QC in all six plates was 10.0% (0.40 ± 0.04) and those of blinded QC from two plasma samples measured each seven times were 16.1% (0.37 ± 5.96) and 12.8% (0.39 ± 5.01). This assay was conducted at Columbia University.
To adjust for selenium intake in our analysis, we measured serum selenium concentration using atomic absorption spectrometry (Perkin Elmer) according to the standard protocol (24). The mean CV from all samples run as duplicates was 7.8%. This assay was conducted at the Fred Hutchinson Cancer Research Center.
Among selenoenzymes that were reported to be associated with oxidative stress and expressed in the gastrointestinal tract, we genotyped a set of tagging SNP in each of five selenoenzyme genes (i.e., GPX1–4 and SEPP1) to efficiently capture the common genetic variation of the entire gene. We first selected all SNP in the selenocysteine insertion sequence, which facilitate selenoenzyme synthesis by a unique stem-loop structure (5) and all nonsynonymous SNP in exons identified via sequencing (25). Second, we selected additional tagging SNP based on the criteria of LD of r2 ≥ 0.8 and minor allele frequency of ≥5% (26) based on our sequencing data (25) on European American HapMap (27) samples. A total of 35 SNP were genotyped using matrix-assisted laser desorption/ionization time-of-flight on the Sequenom MassARRAY 7K platform (Sequenom) and conducted at the Translational Genomics Research Institute. Each plate included blinded duplicates from 5% of the study samples as QC. Based on Hardy-Weinberg equilibrium, the SNP call rate, and the concordance of QC across plates, the final analysis included 26 SNP. In detail, the call rate was <90% for three SNP (rs75404373, rs2277501, and rs4807542), the P value for Hardy-Weinberg equilibrium was <0.01 for four SNP (rs2293627, rs6888691, rs3763011, and rs757229), and the concordance of the blinded QC (10 pairs) was <90% for two SNP (rs2074452 and rs7579).
Statistical analysis.
We used multiple linear regression to assess the association between genotypes in selenoenzymes and the activity of GPX1 and GPX3, SEPP1 concentration, MDA concentration, and PCC. We adjusted for age at blood draw, serum selenium concentrations, gender, smoking status (i.e., never, former, or current), nonsteroid antiinflammatory drug use (i.e., never, former, or current), and ethnicity (European ancestry or others), because these six variables were each associated with at least one of the selenoenzyme activity or concentration or oxidative stress variables. BMI was not associated with any of these five selenoenzyme and oxidative stress variables and hence was not included. GPX1 activity was log-transformed to yield a normal distribution. Likewise, because the distribution of the MDA concentration was not normal, nine outliers, which were outside of the upper and lower three IQR, were excluded and the rest of values were log-transformed. We used a log-additive model to assess the effect of SNP on selenoenzyme activity or concentration and oxidative stress markers; genotypes were evaluated by assigning the number of minor alleles and testing for a linear trend. To adjust for multiple comparisons within each gene and to account for the correlation known as LD between SNP, we conducted the global gene test by comparing the log-likelihood ratio statistics between the model including covariates and the model including covariates and all SNP within a given gene (28). The analyses were repeated for European ancestry (n = 186), men (n = 161), and those who did not have high-grade dysplasia at the time of blood draw (n = 157) on the basis of the reported gender difference in selenoenzyme activity (29) and decreased antioxidant activity among patients with inflammatory conditions (30). Statistical analyses were conducted by SAS 9.1 and STATA 11.
Results
Our study population included predominately men (83%) and was of European ancestry (95%) (Table 1). Approximately 80% of the participants were either overweight or obese. The majority of our participants were not smoking at the time of blood collection. High-grade dysplasia was observed in ~20% of the participants.
TABLE 1.
Characteristics of the study population (n = 195)1
| Characteristics | All |
| Male gender, n (%) | 161 (82.6) |
| Age, y | 64.3 ± 11.7 |
| BMI, kg/m2 | 29.0 ± 4.2 |
| Ancestry, n (%) | |
| European | 185 (94.9) |
| Others | 10 (5.1) |
| Smoking status, n (%) | |
| Never | 61 (31.3) |
| Former | 113 (57.9) |
| Current | 21 (10.8) |
| NSAID use, n (%) | |
| Never | 65 (33.3) |
| Former | 54 (27.7) |
| Current | 76 (39.0) |
| Selenium supplement use, n (%) | 11 (5.6) |
| GPX1 activity, U/g protein | 43.1 ± 21.9 |
| GPX3 activity, U/L | 727 ± 120 |
| SEPP1 concentration, μg/L | 5.8 ± 1.1 |
| MDA, μmol/L | 1.10 ± 1.16 |
| PCC, nmol/mg protein | 0.36 ± 0.06 |
| Serum Se, μmol/L | 1.73 ± 0.31 |
| Histological diagnosis, n (%) | |
| High-grade dysplasia | 38 (19.5) |
| Low-grade dysplasia | 41 (21.0) |
| Gastroesophageal reflux disease | 6 (3.1) |
| Negative/indefinite | 110 (56.4) |
The mean ± SD or (%) is provided. MDA, malondialdehyde; NSAID, nonsteroid antiinflammatory drug; PCC, protein carbonyl content.
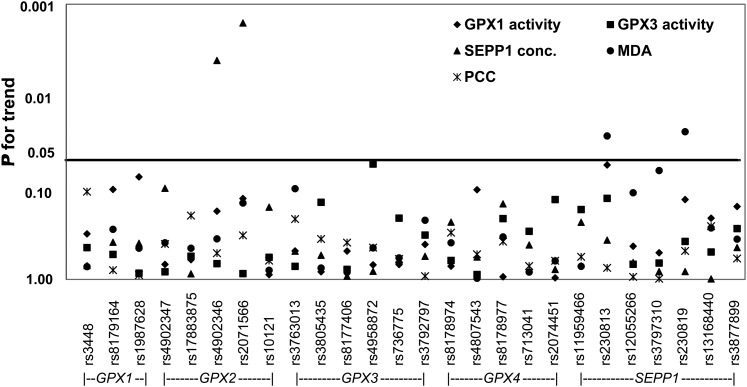
Overall variation in GPX1 was associated with GPX1 activity (global P = 0.02) (Table 2; Fig. 1). When investigating individual SNP in GPX1, there were suggestive associations of rs1987628 and rs8179164 with GPX1 activity (P-trend = 0.08 and 0.10, respectively). Carriers of the minor allele of rs1987628, CT and TT, had lower GPX1 activity than carriers of the common allele. Similarly, AT carriers of rs8179164 had higher GPX1 activity than AA carriers.
TABLE 2.
Associations between SNP in selenoenzymes and selenoenzyme activity or concentration and oxidative stress among all participants (n = 195)1
| Gene | SNP | Allele | n | GPX1 activity, U/g protein | GPX3 activity, U/L | SEPP1 concentration, μg/L | MDA, μmol/L | PCC, nmol/ mg protein |
| GPX1 | rs3448 | GG | 98 | 39.7 | 714 | 5.85 | 1.88 | 0.36 |
| AG | 86 | 39.5 | 726 | 5.70 | 1.86 | 0.34 | ||
| AA | 11 | 33.0 | 640 | 6.41 | 1.86 | 0.35 | ||
| P-trend | 0.34 | 0.45 | 0.72 | 0.67 | 0.12 | |||
| rs8179164 | AA | 187 | 39.0 | 716 | 5.83 | 1.86 | 0.35 | |
| AT | 8 | 48.1 | 689 | 5.50 | 1.99 | 0.35 | ||
| P-difference | 0.10 | 0.54 | 0.39 | 0.35 | 0.82 | |||
| rs1987628 | CC | 93 | 41.7 | 713 | 5.93 | 1.86 | 0.35 | |
| CT | 90 | 37.5 | 724 | 5.75 | 1.87 | 0.35 | ||
| TT | 11 | 38.6 | 696 | 5.91 | 1.96 | 0.36 | ||
| P-trend | 0.08 | 0.85 | 0.40 | 0.55 | 0.91 | |||
| Global P | 0.02 | 0.85 | 0.66 | 0.66 | 0.33 | |||
| GPX2 | rs4902347 | GG | 161 | 39.1 | 714 | 5.76 | 1.87 | 0.35 |
| GA | 33 | 40.3 | 723 | 6.13 | 1.88 | 0.36 | ||
| AA | 1 | 39.2 | 662 | 5.61 | 2.57 | 0.36 | ||
| P-trend | 0.66 | 0.82 | 0.10 | 0.49 | 0.39 | |||
| Rs17883875 | CC | 192 | 39.3 | 716 | 5.82 | 1.87 | 0.35 | |
| CG | 3 | 35.4 | 675 | 5.72 | 2.00 | 0.31 | ||
| P-difference | 0.62 | 0.56 | 0.86 | 0.50 | 0.21 | |||
| rs4902346 | TT | 142 | 38.7 | 715 | 5.69 | 1.86 | 0.35 | |
| CT | 47 | 40.5 | 713 | 6.16 | 1.87 | 0.35 | ||
| CC | 6 | 47.2 | 760 | 6.37 | 2.07 | 0.37 | ||
| P-trend | 0.17 | 0.67 | 0.004 | 0.43 | 0.50 | |||
| rs2071566 | GG | 125 | 38.4 | 716 | 5.67 | 1.85 | 0.35 | |
| GA | 59 | 40.1 | 709 | 6.03 | 1.90 | 0.35 | ||
| AA | 11 | 45.8 | 740 | 6.55 | 1.97 | 0.39 | ||
| P-trend | 0.14 | 0.87 | 0.002 | 0.12 | 0.34 | |||
| rs10121 | CC | 171 | 39.4 | 713 | 5.76 | 1.87 | 0.35 | |
| TC | 17 | 38.5 | 720 | 6.26 | 1.94 | 0.35 | ||
| TT | 1 | 53.6 | 812 | 5.08 | 1.48 | 0.43 | ||
| P-trend | 0.89 | 0.57 | 0.16 | 0.79 | 0.63 | |||
| Global P | 0.46 | 0.70 | 0.03 | 0.67 | 0.62 | |||
| GPX3 | rs3763013 | TT | 92 | 38.6 | 724 | 5.94 | 1.84 | 0.36 |
| TC | 77 | 39.6 | 699 | 5.63 | 1.88 | 0.35 | ||
| CC | 25 | 40.6 | 732 | 5.97 | 1.95 | 0.34 | ||
| P-trend | 0.47 | 0.72 | 0.48 | 0.12 | 0.23 | |||
| rs3805435 | CC | 164 | 39.3 | 721 | 5.82 | 1.87 | 0.35 | |
| CT | 30 | 38.8 | 691 | 5.79 | 1.88 | 0.36 | ||
| TT | 1 | 60.2 | 614 | 8.17 | 1.43 | 0.37 | ||
| P-trend | 0.86 | 0.14 | 0.54 | 0.90 | 0.38 | |||
| rs8177406 | TT | 138 | 38.8 | 714 | 5.83 | 1.88 | 0.35 | |
| CT | 53 | 40.5 | 722 | 5.73 | 1.84 | 0.35 | ||
| CC | 4 | 40.1 | 705 | 6.25 | 1.99 | 0.32 | ||
| P-trend | 0.52 | 0.78 | 0.94 | 0.98 | 0.38 | |||
| rs4958872 | TT | 106 | 39.9 | 734 | 5.81 | 1.86 | 0.35 | |
| TC | 80 | 38.4 | 691 | 5.81 | 1.88 | 0.35 | ||
| CC | 9 | 41.0 | 721 | 5.97 | 1.92 | 0.38 | ||
| P-trend | 0.71 | 0.06 | 0.82 | 0.48 | 0.44 | |||
| rs736775 | GG | 72 | 38.8 | 729 | 5.79 | 1.88 | 0.36 | |
| AG | 94 | 39.5 | 710 | 5.80 | 1.84 | 0.35 | ||
| AA | 29 | 39.9 | 700 | 5.93 | 1.95 | 0.35 | ||
| P-trend | 0.69 | 0.21 | 0.62 | 0.55 | 0.60 | |||
| rs3792797 | GG | 130 | 39.9 | 721 | 5.77 | 1.86 | 0.35 | |
| GT | 56 | 38.0 | 702 | 5.91 | 1.92 | 0.35 | ||
| TT | 3 | 38.3 | 701 | 5.58 | 1.87 | 0.41 | ||
| P-trend | 0.45 | 0.33 | 0.57 | 0.29 | 0.95 | |||
| Global P | 0.81 | 0.69 | 0.20 | 0.65 | 0.25 | |||
| GPX4 | rs8178974 | GGGGTG/GGGGTG | 127 | 39.1 | 712 | 5.77 | 0.63 | 0.35 |
| GGGGTG/- | 64 | 39.6 | 721 | 5.90 | 0.61 | 0.36 | ||
| −/− | 2 | 42.9 | 722 | 6.89 | 0.70 | 0.36 | ||
| P-trend | 0.67 | 0.62 | 0.25 | 0.30 | 0.28 | |||
| rs4807543 | GG | 174 | 39.8 | 715 | 5.83 | 0.63 | 0.35 | |
| GT | 21 | 34.9 | 719 | 5.69 | 0.63 | 0.34 | ||
| P-difference | 0.11 | 0.88 | 0.56 | 0.92 | 0.54 | |||
| rs8178977 | GG | 109 | 39.1 | 708 | 5.92 | 0.62 | 0.36 | |
| CG | 79 | 40.2 | 718 | 5.69 | 0.64 | 0.35 | ||
| CC | 7 | 34.6 | 781 | 5.69 | 0.62 | 0.33 | ||
| P-trend | 0.91 | 0.22 | 0.15 | 0.33 | 0.37 | |||
| rs713041 | CC | 53 | 40.2 | 711 | 5.90 | 0.62 | 0.35 | |
| CT | 94 | 38.5 | 735 | 5.83 | 0.63 | 0.35 | ||
| TT | 47 | 39.6 | 685 | 5.72 | 0.62 | 0.35 | ||
| P-trend | 0.81 | 0.30 | 0.42 | 0.75 | 0.73 | |||
| rs2074451 | GG | 53 | 39.7 | 725 | 5.89 | 0.64 | 0.36 | |
| GT | 91 | 38.7 | 726 | 5.76 | 0.62 | 0.35 | ||
| TT | 51 | 39.9 | 689 | 5.84 | 0.62 | 0.35 | ||
| P-trend | >0.99 | 0.13 | 0.79 | 0.82 | 0.60 | |||
| Global P | 0.63 | 0.82 | 0.34 | 0.43 | 0.44 | |||
| SEPP1 | rs11959466 | GG | 171 | 39.1 | 720 | 5.85 | 0.63 | 0.35 |
| AG | 22 | 40.5 | 688 | 5.59 | 0.60 | 0.35 | ||
| AA | 1 | 37.9 | 636 | 5.39 | 0.73 | 0.32 | ||
| P-trend | 0.71 | 0.17 | 0.24 | 0.73 | 0.57 | |||
| rs230813 | GG | 55 | 40.9 | 725 | 5.65 | 0.66 | 0.35 | |
| GC | 88 | 40.2 | 723 | 5.90 | 0.62 | 0.36 | ||
| CC | 52 | 35.8 | 689 | 5.83 | 0.60 | 0.34 | ||
| P-trend | 0.06 | 0.13 | 0.38 | 0.03 | 0.78 | |||
| rs12055266 | AA | 111 | 38.7 | 714 | 5.82 | 0.61 | 0.35 | |
| AG | 66 | 39.6 | 715 | 5.87 | 0.66 | 0.35 | ||
| GG | 18 | 41.5 | 730 | 5.59 | 0.63 | 0.35 | ||
| P-trend | 0.42 | 0.68 | 0.65 | 0.13 | 0.92 | |||
| rs3797310 | GG | 103 | 38.8 | 713 | 5.81 | 0.60 | 0.35 | |
| AG | 71 | 39.3 | 713 | 5.86 | 0.66 | 0.35 | ||
| AA | 19 | 41.4 | 730 | 5.69 | 0.63 | 0.35 | ||
| P-trend | 0.51 | 0.66 | 0.82 | 0.07 | 0.99 | |||
| rs230819 | GG | 65 | 36.8 | 709 | 5.75 | 0.60 | 0.34 | |
| GT | 81 | 40.2 | 712 | 5.92 | 0.63 | 0.36 | ||
| TT | 46 | 40.5 | 730 | 5.68 | 0.67 | 0.35 | ||
| P-trend | 0.14 | 0.39 | 0.83 | 0.02 | 0.50 | |||
| rs13168440 | AA | 139 | 38.7 | 711 | 5.84 | 0.62 | 0.35 | |
| GA | 50 | 40.2 | 727 | 5.73 | 0.64 | 0.36 | ||
| GG | 4 | 50.0 | 710 | 6.40 | 0.68 | 0.35 | ||
| P-trend | 0.21 | 0.50 | 0.99 | 0.28 | 0.26 | |||
| rs3877899 | GG | 118 | 38.3 | 707 | 5.86 | 0.62 | 0.35 | |
| GA | 61 | 41.0 | 722 | 5.70 | 0.63 | 0.36 | ||
| AA | 10 | 43.1 | 745 | 5.78 | 0.69 | 0.34 | ||
| P-trend | 0.16 | 0.28 | 0.44 | 0.37 | 0.58 | |||
| Global P | 0.71 | 0.34 | 0.56 | 0.19 | 0.85 |
Adjusted for age, serum selenium concentrations, nonsteroid antiinflammatory drug use, smoking status, gender, and ethnicity. MDA, malondialdehyde; PCC, protein carbonyl content.
FIGURE 1.
P-trend for individual SNP by selenoenzyme activity or concentrations or markers of oxidative stress in humans. Data were adjusted for age, serum selenium concentrations, nonsteroid antiinflammatory drug use, smoking status, gender, and ethnicity. MDA, malondialdehyde; PCC, protein carbonyl content.
The association between the overall genetic variation in GPX2, measured by five SNP, and SEPP1 concentration was significant (global P = 0.03). The minor allele of two variants in GPX2 (rs4902346 and rs2071566) were positively associated with SEPP1 concentration (P-trend = 0.004 and 0.002, respectively) (Table 2). The SEPP1 concentration was higher in carriers of CT and CC in rs4902346 than in carriers of TT. Likewise, carriers of GA and AA in rs2071566 had a higher SEPP1 concentration than carriers of GG.
Although the association between the overall variation in SEPP1 and MDA concentrations was not significant (global P = 0.19), the number of minor alleles in SEPP1 rs230813 was inversely (P-trend = 0.03) and in SEPP1 rs230819 was positively (P-trend = 0.02) associated with MDA concentrations (Fig. 1; Table 2). None of the other SNP was associated individually or overall at the gene level with any of the selenoenzyme activity or concentration or oxidative stress markers.
Results did not differ by subgroups of men (n = 161), European ancestry (n = 186), or those who did not have high-grade dysplasia at the time of blood draw (n = 157); the level of significance changed in some of the associations in selected subgroups, but overall, the direction of associations remained the same (data not shown). In particular, among men, the inverse association of one SNP in GPX1 (rs1987628) with GPX1 activity became significant (P-trend = 0.03). However, the number of women in our study was too small (n = 34) to investigate a potential gender difference. In addition, the overall results were similar with or without adjusting for serum selenium concentrations or selenium supplement use (data not shown).
Discussion
In this study, genetic variation in GPX1 was associated with GPX1 activity; specifically, two SNP (rs1987628 and rs8179164) were suggestively associated with GPX1 activity. In addition, the overall variation in GPX2 and two SNP in GPX2 (rs4902346 and rs2071566) were associated with SEPP1 concentrations. Two SEPP1 variants (rs230813 and rs230819) were individually associated with MDA concentrations.
The GPX1 candidate variant (rs1050450; Pro200Leu), which resides in the coding region and results in an amino acid substitution of proline with leucine, was associated with GPX1 activity in three larger studies (n = 231–1154) (7, 9, 10) but not in two smaller studies (n = 66 and 90) (6, 8). Further, it was associated with the risk of breast, bladder, or lung cancer (10, 29–31), although not all studies observed an association with various cancer sites (6, 9, 31–36). Our suggestive association for rs1987628-> T, which is in complete LD (r2 = 1.00) with rs1050450 (Pro200Leu) in European Americans (25) and thus both SNP are perfect proxies for each other, is consistent with the larger studies (7, 9, 10). This suggestive association became significant in men, which also agrees with the reported stronger association in men than in women (7); however, our study had the insufficient number of women to be investigated separately. Given the current and previous findings and the potential functional importance of an amino acid-changing variant and the reported interaction of a GPX1 variant, rs1050450, in the association between serum selenium and prostate cancer risk (37), future epidemiologic studies of selenium and selenoenzymes and disease risk should consider evaluating this variant and its interaction with selenium and GPX1 activity.
GPX2 is expressed in the gastrointestinal tract and is known to affect oxidative stress and inflammation in GPX2 knockout and GPX1/2 double knockout mice (38–40). Our study is the first to our knowledge to report an association between GPX2 variants, specifically rs4902346 and rs2071566, and SEPP1 concentrations. To our knowledge, no study examined this association of any GPX2 genetic variant with selenoenzyme activity or oxidative stress to date. Moreover, none of the four GPX2 SNP (rs4902346, rs2071566, rs2737844, and rs17881652) was associated with risk of gastrointestinal-related diseases (33, 34, 41, 42). Two GPX2 SNP (rs4902346 and rs2071566) were associated with the SEPP1 concentration in our study. The first SNP (rs4902346) is in complete LD (r2 = 1.00) with two GPX2 SNP (rs17880380 and rs2412065) and the second SNP (rs2071566) is in complete LD with rs2737844 in our own sequencing data (25) or in HapMap data (27) among European Americans. Thus, we further investigated the location and conservation scores not only of the two genotyped SNP but also the three tagged SNP, because any of them could be responsible for the observed effect due to LD. SNP rs17880380 is located in the 5′ region and could potentially affect GPX2 expression, whereas the other four variants (rs4902346, rs2071566, rs2412065, and rs2737844) are located in intron 1, the only intron of this gene. Because none of these four SNP is close to the exons, they are less likely to affect splicing. All five SNP are not highly conserved (conservation score <0.05) (43). All other GPX2 SNP in our sequencing data and HapMap data were in low LD (r2 < 0.70) with the two SNP associated with SEPP1 concentrations and hence we did not further explore them. Note that our primary hypotheses focused on associations between SNP in a gene and the activity or concentration of the same gene (e.g., SNP in GPX1 and GPX1 activity) or oxidative stress markers. As secondary hypotheses, based on previous findings (11–13), we tested associations between SNP in a gene and other selenoenzymes, such as the observed effect of GPX2 SNP on SEPP1 concentrations. Due to the limited esophageal tissue samples available, we were not able to measure GPX2 activity. Accordingly, this finding is likely due to chance and requires replication in future studies.
Although an antioxidant property of SEPP1 was previously suggested as well as selenium transport (44), to our knowledge, genetic variants in SEPP1 have not been investigated in relation to MDA concentrations. In our study, two SNP in SEPP1 (rs230813 and rs230819) were significantly associated with MDA concentration, which is indicative of lipid peroxidation, and hence this finding agrees with the postulated function of SEPP1 as phospholipid hydroperoxidase glutathione reductase (45).
Previously, the effects of selected SNP within GPX4 (rs713041) and SEPP1 (rs3877899 and rs7579) on selenoenzyme activity and concentrations were investigated in a supplementation trial of selenium (11, 12) and a cross-sectional study (13). Using baseline measures of the trial (n = 75), two SEPP1 SNP (rs3877899 and rs7579) in combination with sex and BMI influenced GPX1 activity, GPX3 activity, and/or SEPP1 concentrations (11). Moreover, the cross-sectional study (n = 261) found a significant difference in serum SEPP1 concentrations by one SEPP1 variant, rs3877899, but not the other variant (rs7579), and also the quadratic association between BMI and SEPP1 concentration (13). In contrast, our study did not find an association between SEPP1 SNP rs3877899 and selenoenzyme activity among all participants or men specifically. SNP rs7579 was excluded from our analysis due to its low concordance of blinded duplicates, suggesting a genotyping error. Furthermore, consistent with our finding, the trial did not find an association between the GPX4 variant rs713041 and activity of GPX1 and GPX3 (12) or SEPP1 concentrations (13). Although our study included a limited number of women, gender difference in the association between these SNP and selenoenzyme activity or concentrations, and potentially also interaction with BMI (11), warrants further investigation.
A strength of this study is the comprehensive approach for selecting SNP to capture the overall variation within the genes. Our study had a relatively large sample size (n = 195) compared to some of the previous studies (n = 66–90) (6, 8). Because our study included predominately individuals of European ancestry from the same region, the number of participants within the subgroups was limited. Although we were unable to rule out potential confounding by population substructure in our analyses, such confounding is expected to be small in well-designed studies like ours (46). The quality of our laboratory measurements and genotyping was well monitored and within the acceptable range, e.g., the mean call rate for SNP included in the final analysis was 98.9%.
This study also has limitations. First, the generalizability of our findings to a healthy population may be limited, because our study only included Barrett’s esophagus patients whose selenoenzyme activity or concentration might be affected by this condition. Nevertheless, our findings for all participants and those with high-grade dysplasia were comparable. Moreover, our selenoenzyme activity and concentrations and oxidative stress markers were similar to those reported in previous studies in healthy individuals (11, 47–51). Second, because we focused on three selenoenzymes, we did not measure other enzymes important for the overall antioxidant activity, including GPX2 and GPX4 and catalase and superoxide dismutase (52). However, GPX2 is expressed in the gut and we were not able to establish a reliable GPX4 assay, potentially due to reported difficulty of the assay (53). Third, the use of MDA to measure lipid peroxidation in blood samples may not be entirely adequate. Fourth, we conducted a large number of statistical tests, raising the possibility of false positive findings. Accordingly, we limited our primary hypotheses to test the effect of SNP in a selenoenzyme gene on the activity or concentration of the same gene and oxidative stress. The global gene test further served as multiple comparison adjustment within a gene while considering LD between SNP.
In conclusion, we observed an association between the genetic variation in GPX1 and GPX1 activity. Specifically, the inverse association of a GPX1 variant, rs1987628, with the GPX1 activity is consistent with previous studies. Two variants in GPX2, rs4902346 and rs2071566, were positively associated with SEPP1 concentrations, which to our knowledge have not been reported elsewhere and need to be replicated in future studies. Moreover, two SEPP1 variants (rs230813 and rs230819), although not the overall genetic variation, were associated with MDA concentrations. Overall, our findings support the hypothesis that common genetic variants in selenoenzymes may affect their activity.
Acknowledgments
U.P. and T.L.V. designed the study; R.F.B., K.E.H., I.B.K., R.M.S., and D.J.D. conducted laboratory measurements; T.L.V. provided study participants’ data; Y.T. conducted the statistical analysis and drafted the manuscript with the supervision of U.P., A.R.K., J.W.L., and T.L.V.; and U.P. had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Supported by the National Cancer Institute (grant nos. K22 CA118421, R01 CA120582, K05CA124911, and P01 CA091955) and by the NIH (grant no. DK58763).
Abbreviations used: LD, linkage disequilibrium; MDA, malondialdehyde; PCC, protein carbonyl content; QC, quality control; SNP, single nucleotide polymorphism
Literature Cited
- 1.Peters U, Takata Y. Selenium and the prevention of prostate and colorectal cancer. Mol Nutr Food Res. 2008;52:1261–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rayman MP. Selenium in cancer prevention: a review of the evidence and mechanism of action. Proc Nutr Soc. 2005;64:527–42 [DOI] [PubMed] [Google Scholar]
- 3.Zeng H, Combs GF., Jr Selenium as an anticancer nutrient: roles in cell proliferation and tumor cell invasion. J Nutr Biochem. 2008;19:1–7 [DOI] [PubMed] [Google Scholar]
- 4.Flores-Mateo G, Navas-Acien A, Pastor-Barriuso R, Guallar E. Selenium and coronary heart disease: a meta-analysis. Am J Clin Nutr. 2006;84:762–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moghadaszadeh B, Beggs AH. Selenoproteins and their impact on human health through diverse physiological pathways. Physiology (Bethesda). 2006;21:307–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arsova-Sarafinovska Z, Matevska N, Eken A, Petrovski D, Banev S, Dzikova S, Georgiev V, Sikole A, Erdem O, Sayal A, et al. Glutathione peroxidase 1 (GPX1) genetic polymorphism, erythrocyte GPX activity, and prostate cancer risk. Int Urol Nephrol. 2009;41:63–70 [DOI] [PubMed] [Google Scholar]
- 7.Bastaki M, Huen K, Manzanillo P, Chande N, Chen C, Balmes JR, Tager IB, Holland N. Genotype-activity relationship for Mn-superoxide dismutase, glutathione peroxidase 1 and catalase in humans. Pharmacogenet Genomics. 2006;16:279–86 [DOI] [PubMed] [Google Scholar]
- 8.Forsberg L. de Faire U, Marklund SL, Andersson PM, Stegmayr B, Morgenstern R. Phenotype determination of a common Pro-Leu polymorphism in human glutathione peroxidase 1. Blood Cells Mol Dis. 2000;26:423–6 [DOI] [PubMed] [Google Scholar]
- 9.Hansen RD, Krath BN, Frederiksen K, Tjonneland A, Overvad K, Roswall N, Loft S, Dragsted LO, Vogel U, Raaschou-Nielsen O. GPX1 Pro(198)Leu polymorphism, erythrocyte GPX activity, interaction with alcohol consumption and smoking, and risk of colorectal cancer. Mutat Res. 2009;664:13–9 [DOI] [PubMed] [Google Scholar]
- 10.Ravn-Haren G, Olsen A, Tjonneland A, Dragsted LO, Nexo BA, Wallin H, Overvad K, Raaschou-Nielsen O, Vogel U. Associations between GPX1 Pro198Leu polymorphism, erythrocyte GPX activity, alcohol consumption and breast cancer risk in a prospective cohort study. Carcinogenesis. 2006;27:820–5 [DOI] [PubMed] [Google Scholar]
- 11.Méplan C, Crosley LK, Nicol F, Beckett GJ, Howie AF, Hill KE, Horgan G, Mathers JC, Arthur JR, Hesketh JE. Genetic polymorphisms in the human selenoprotein P gene determine the response of selenoprotein markers to selenium supplementation in a gender-specific manner (the SELGEN study). FASEB J. 2007;21:3063–74 [DOI] [PubMed] [Google Scholar]
- 12.Méplan C, Crosley LK, Nicol F, Horgan GW, Mathers JC, Arthur JR, Hesketh JE. Functional effects of a common single-nucleotide polymorphism (GPX4c718t) in the glutathione peroxidase 4 gene: interaction with sex. Am J Clin Nutr. 2008;87:1019–27 [DOI] [PubMed] [Google Scholar]
- 13.Combs GF, Jr, Watts JC, Jackson MI, Johnson LK, Zeng H, Scheett AJ, Uthus EO, Schomburg L, Hoeg A, Hoefig CS, et al. Determinants of selenium status in healthy adults. Nutr J. 2011;10:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid BJ, Levine DS, Longton G, Blount PL, Rabinovitch PS. Predictors of progression to cancer in Barrett's esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudolph RE, Vaughan TL, Storer BE, Haggitt RC, Rabinovitch PS, Levine DS, Reid BJ. Effect of segment length on risk for neoplastic progression in patients with Barrett esophagus. Ann Intern Med. 2000;132:612–20 [DOI] [PubMed] [Google Scholar]
- 16.Vaughan TL, Dong LM, Blount PL, Ayub K, Odze RD, Sanchez CA, Rabinovitch PS, Reid BJ. Non-steroidal anti-inflammatory drugs and risk of neoplastic progression in Barrett's oesophagus: a prospective study. Lancet Oncol. 2005;6:945–52 [DOI] [PubMed] [Google Scholar]
- 17.Rudolph RE, Vaughan TL, Kristal AR, Blount PL, Levine DS, Galipeau PC, Prevo LJ, Sanchez CA, Rabinovitch PS, Reid BJ. Serum selenium levels in relation to markers of neoplastic progression among persons with Barrett's esophagus. J Natl Cancer Inst. 2003;95:750–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Risques RA, Vaughan TL, Li X, Odze RD, Blount PL, Ayub K, Gallaher JL, Reid BJ, Rabinovitch PS. Leukocyte telomere length predicts cancer risk in Barrett's esophagus. Cancer Epidemiol Biomarkers Prev. 2007;16:2649–55 [DOI] [PubMed] [Google Scholar]
- 19.Siahpush SH, Vaughan TL, Lampe JN, Freeman R, Lewis S, Odze RD, Blount PL, Ayub K, Rabinovitch PS, Reid BJ, et al. Longitudinal study of insulin-like growth factor, insulin-like growth factor binding protein-3, and their polymorphisms: risk of neoplastic progression in Barrett's esophagus. Cancer Epidemiol Biomarkers Prev. 2007;16:2387–95 [DOI] [PubMed] [Google Scholar]
- 20.Paglia DE, Valentine WN. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J Lab Clin Med. 1967;70:158–69 [PubMed] [Google Scholar]
- 21.Ericson SP, McHalsky ML, Rabinow BE, Kronholm KG, Arceo CS, Weltzer JA, Ayd SW. Sampling and analysis techniques for monitoring serum for trace elements. Clin Chem. 1986;32:1350–6 [PubMed] [Google Scholar]
- 22.Agarwal R, Chase SD. Rapid, fluorimetric-liquid chromatographic determination of malondialdehyde in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;775:121–6 [DOI] [PubMed] [Google Scholar]
- 23.Peng T, Li LQ, Peng MH, Liu ZM, Liu TW, Yan LN, Shen HM, Wang L, Wang Q, Wang KB, et al. Is correction for protein concentration appropriate for protein adduct dosimetry? Hypothesis and clues from an aflatoxin B1-exposed population. Cancer Sci. 2007;98:140–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satia JA, King IB, Morris JS, Stratton K, White E. Toenail and plasma levels as biomarkers of selenium exposure. Ann Epidemiol. 2006;16:53–8 [DOI] [PubMed] [Google Scholar]
- 25.Foster CB, Aswath K, Chanock SJ, McKay HF, Peters U. Polymorphism analysis of six selenoprotein genes: support for a selective sweep at the glutathione peroxidase 1 locus (3p21) in Asian populations. BMC Genet. 2006;7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31 [DOI] [PubMed] [Google Scholar]
- 29.Waters DJ, Chiang EC, Cooley DM, Morris JS. Making sense of sex and supplements: differences in the anticarcinogenic effects of selenium in men and women. Mutat Res. 2004;551:91–107 [DOI] [PubMed] [Google Scholar]
- 30.Mörk H, Scheurlen M, Al-Taie O, Zierer A, Kraus M, Schottker K, Jakob F, Kohrle J. Glutathione peroxidase isoforms as part of the local antioxidative defense system in normal and Barrett's esophagus. Int J Cancer. 2003;105:300–4 [DOI] [PubMed] [Google Scholar]
- 31.Choi JY, Neuhouser ML, Barnett M, Hudson M, Kristal AR, Thornquist M, King IB, Goodman GE, Ambrosone CB. Polymorphisms in oxidative stress-related genes are not associated with prostate cancer risk in heavy smokers. Cancer Epidemiol Biomarkers Prev. 2007;16:1115–20 [DOI] [PubMed] [Google Scholar]
- 32.Kote-Jarai Z, Durocher F, Edwards SM, Hamoudi R, Jackson RA, Ardern-Jones A, Murkin A, Dearnaley DP, Kirby R, et al. Association between the GCG polymorphism of the selenium dependent GPX1 gene and the risk of young onset prostate cancer. Prostate Cancer Prostatic Dis. 2002;5:189–92 [DOI] [PubMed] [Google Scholar]
- 33.Peters U, Chatterjee N, Hayes RB, Schoen RE, Wang Y, Chanock SJ, Foster CB. Variation in the selenoenzyme genes and risk of advanced distal colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2008;17:1144–54 [DOI] [PubMed] [Google Scholar]
- 34.Takata Y, Kristal AR, King IB, Song X, Diamond AM, Foster CB, Hutter CM, Hsu L, Duggan DJ, Langer RD, et al. Serum selenium, genetic variation in selenoenzymes, and risk of colorectal cancer: primary analysis from the Women's Health Initiative Observational Study and Meta-Analysis. Cancer Epidemiol Biomarkers Prev. 2011;20:1822–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox DG, Hankinson SE, Kraft P, Hunter DJ. No association between GPX1 Pro198Leu and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2004;13:1821–2 [PubMed] [Google Scholar]
- 36.Knight JA, Onay UV, Wells S, Li H, Shi EJ, Andrulis IL, Ozcelik H. Genetic variants of GPX1 and SOD2 and breast cancer risk at the Ontario site of the Breast Cancer Family Registry. Cancer Epidemiol Biomarkers Prev. 2004;13:146–9 [DOI] [PubMed] [Google Scholar]
- 37.Steinbrecher A, Meplan C, Hesketh J, Schomburg L, Endermann T, Jansen E, Akesson B, Rohrmann S, Linseisen J. Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol Biomarkers Prev. 2010;19:2958–68 [DOI] [PubMed] [Google Scholar]
- 38.Chu FF, Esworthy RS, Chu PG, Longmate JA, Huycke MM, Wilczynski S, Doroshow JH. Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res. 2004;64:962–8 [DOI] [PubMed] [Google Scholar]
- 39.Esworthy RS, Aranda R, Martin MG, Doroshow JH, Binder SW, Chu FF. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G848–55 [DOI] [PubMed] [Google Scholar]
- 40.Esworthy RS, Yang L, Frankel PH, Chu FF. Epithelium-specific glutathione peroxidase, Gpx2, is involved in the prevention of intestinal inflammation in selenium-deficient mice. J Nutr. 2005;135:740–5 [DOI] [PubMed] [Google Scholar]
- 41.Al-Taie OH, Uceyler N, Eubner U, Jakob F, Mork H, Scheurlen M, Brigelius-Flohe R, Schottker K, Abel J, Thalheimer A, et al. Expression profiling and genetic alterations of the selenoproteins GI-GPx and SePP in colorectal carcinogenesis. Nutr Cancer. 2004;48:6–14 [DOI] [PubMed] [Google Scholar]
- 42.Murphy SJ, Hughes AE, Patterson CC, Anderson LA, Watson RG, Johnston BT, Comber H, McGuigan J, Reynolds JV, Murray LJ. A population-based association study of SNPs of GSTP1, MnSOD, GPX2 and Barrett's esophagus and esophageal adenocarcinoma. Carcinogenesis. 2007;28:1323–8 [DOI] [PubMed] [Google Scholar]
- 43.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burk RF, Hill KE. Selenoprotein P-expression, functions, and roles in mammals. Biochim Biophys Acta. 2009;1790:1441–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein p. J Biol Chem. 1999;274:2866–71 [DOI] [PubMed] [Google Scholar]
- 46.Wacholder S, Rothman N, Caporaso N. Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J Natl Cancer Inst. 2000;92:1151–8 [DOI] [PubMed] [Google Scholar]
- 47.Brown KM, Pickard K, Nicol F, Beckett GJ, Duthie GG, Arthur JR. Effects of organic and inorganic selenium supplementation on selenoenzyme activity in blood lymphoctyes, granulocytes, platelets and erythrocytes. Clin Sci (Lond). 2000;98:593–9 [PubMed] [Google Scholar]
- 48.Dursun B, Dursun E, Suleymanlar G, Ozben B, Capraz I, Apaydin A, Ozben T. The effect of hemodialysis on accelerated atherosclerosis in diabetic patients: correlation of carotid artery intima-media thickness with oxidative stress. J Diabetes Complications. 2009;23:257–64 [DOI] [PubMed] [Google Scholar]
- 49.Hamer HM, Jonkers DM, Bast A, Vanhoutvin SA, Fischer MA, Kodde A, Troost FJ, Venema K, Brummer RJ. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clin Nutr. 2009;28:88–93 [DOI] [PubMed] [Google Scholar]
- 50.Hurst R, Armah CN, Dainty JR, Hart DJ, Teucher B, Goldson AJ, Broadley MR, Motley AK, Fairweather-Tait SJ. Establishing optimal selenium status: results of a randomized, double-blind, placebo-controlled trial. Am J Clin Nutr. 2010;91:923–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson CD, Robinson MF, Butler JA, Whanger PD. Long-term supplementation with selenate and selenomethionine: selenium and glutathione peroxidase (EC 1.11.1.9) in blood components of New Zealand women. Br J Nutr. 1993;69:577–88 [DOI] [PubMed] [Google Scholar]
- 52.Halliwell B, Cross CE. Oxygen-derived species: their relation to human disease and environmental stress. Environ Health Perspect. 1994;102 Suppl 10:5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kernstock RM, Girotti AW. New strategies for the isolation and activity determination of naturally occurring type-4 glutathione peroxidase. Protein Expr Purif. 2008;62:216–22 [DOI] [PMC free article] [PubMed] [Google Scholar]