Abstract
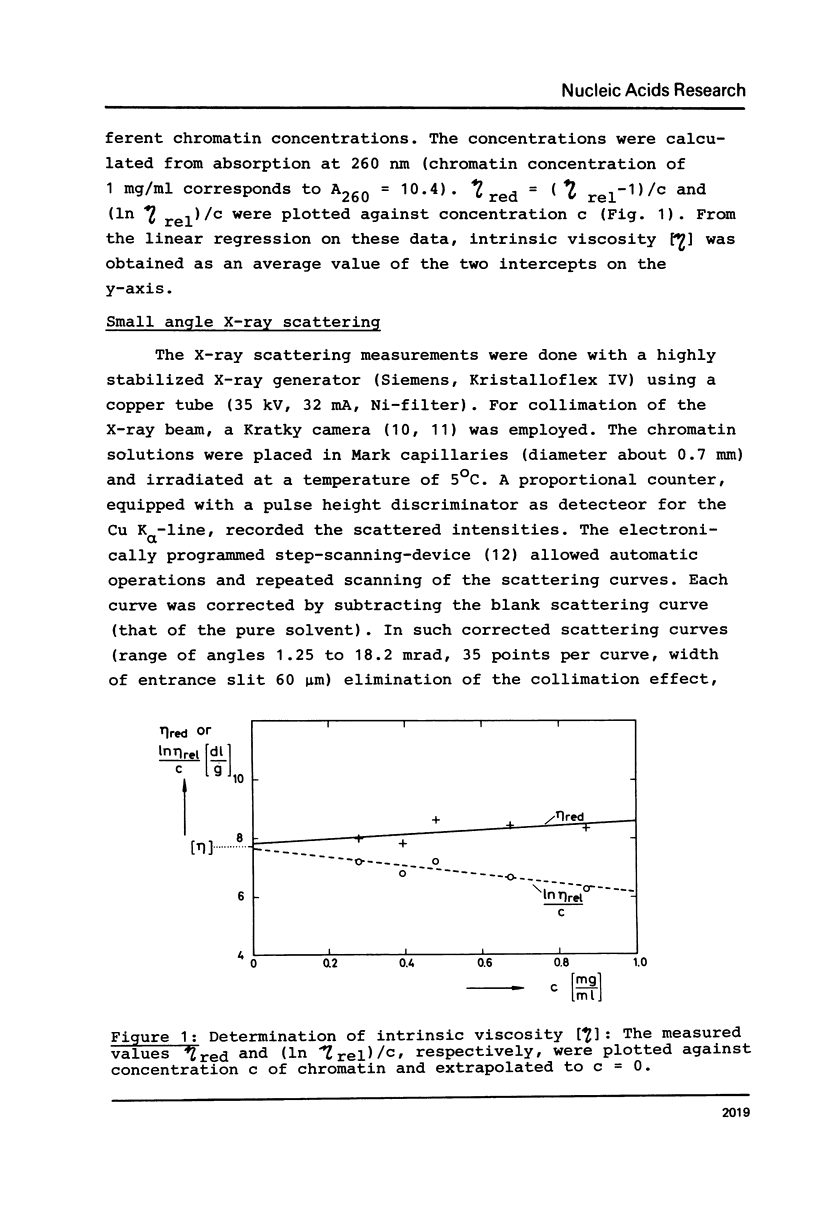
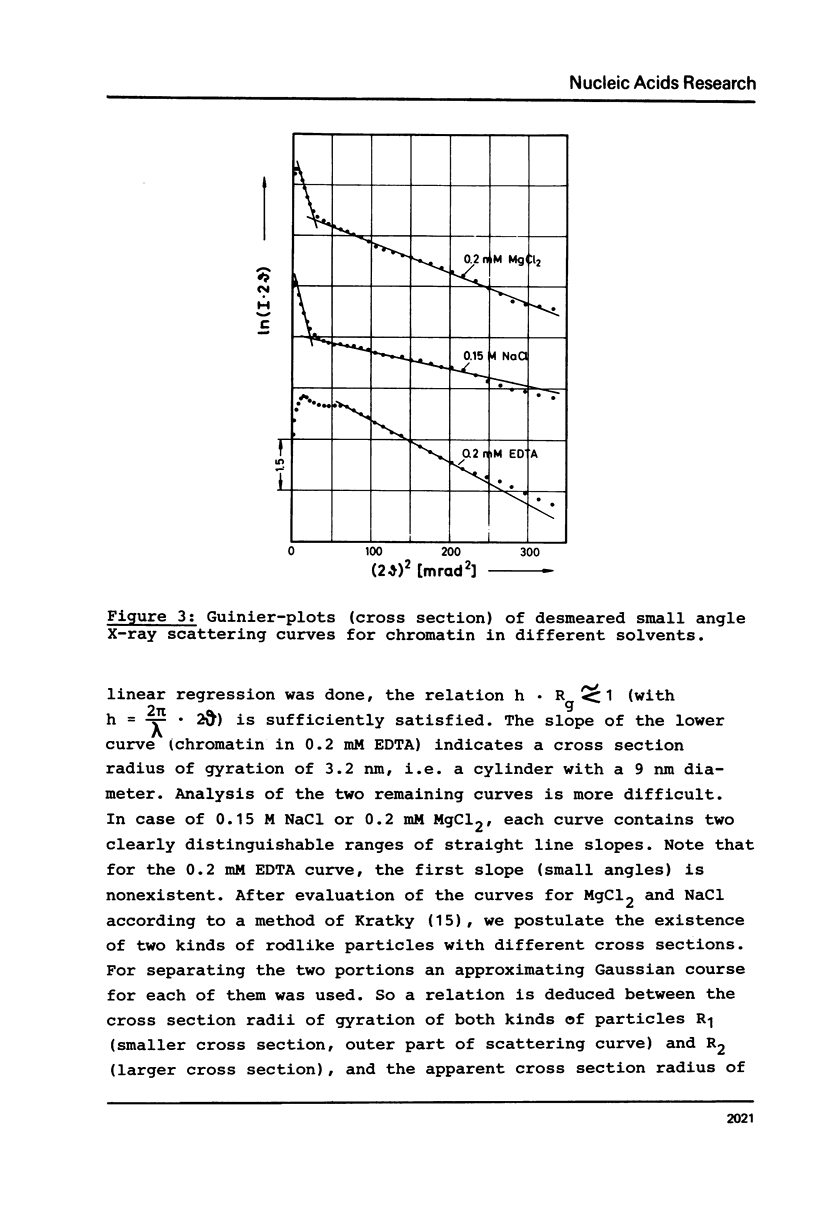
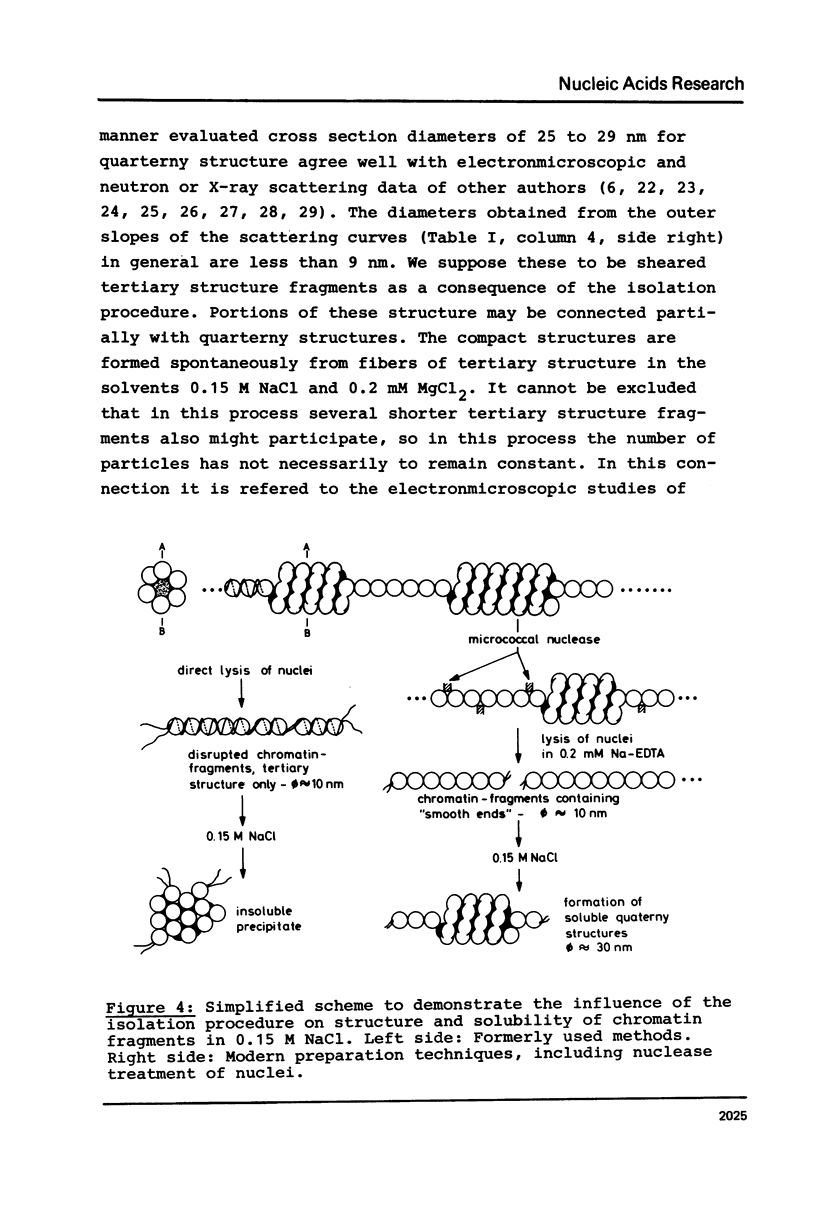
Properties of calf thymus chromatin, prepared by mild procedures, have been studied in various solvents. In 0.2 mM EDTA s-values ranged from 20 to 30 S and intrinsic viscosities from 5 to 24 dl/g. Dialysis against 0.15 M NaCl or 0.2 mM MgCl2 changed these values to 80 to 100 S and 0.2 to 5 dl/g, respectively, indicating an essentially more compact structure. In 0.2 mM EDTA X-ray scattering yielded a cross section diameter of 9 nm, which is associated with the tertiary structure of chromatin fiber (M/L = 21200 Dalton/nm). By dialysis against 0.15 M NaCl or 0.2 mM MgCl2 part of the material spontaneously formed quarterny structures (cross section diameters 25-29 nm). The rest of the material with cross section diameters less than 9 nm is supposed to be more strongly sheared tertiary structure which seems to be unable to form quarterny structure due to artificial conformational changes.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bak A. L., Zeuthen J., Crick F. H. Higher-order structure of human mitotic chromosomes. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1595–1599. doi: 10.1073/pnas.74.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudy P., Bram S. Chromatin fiber dimensions and nucleosome orientation: a neutron scattering investigation. Nucleic Acids Res. 1978 Oct;5(10):3697–3714. doi: 10.1093/nar/5.10.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram S., Baudy P., Lepault J., Hermann D. Chromatin very small angle neutron scattering: further evidence for a 30 nm diameter super coil in dilute solutions. Nucleic Acids Res. 1977 Jul;4(7):2275–2282. doi: 10.1093/nar/4.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram S., Ris H. On the structure of nucleohistone. J Mol Biol. 1971 Feb 14;55(3):325–336. doi: 10.1016/0022-2836(71)90321-4. [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Cotter R. I., Pardon J. F. Light scattering measurements supporting helical structures for chromatin in solution. Nucleic Acids Res. 1978 May;5(5):1571–1580. doi: 10.1093/nar/5.5.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson R. D., Olins D. E. Chromatin model calculations: Arrays of spherical nu bodies. Nucleic Acids Res. 1976 Jan;3(1):89–100. doi: 10.1093/nar/3.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B. G., Baldwin J. P., Bradbury E. M., Ibel K. Organisation of subunits in chromatin. Nucleic Acids Res. 1976 Jul;3(7):1739–1746. doi: 10.1093/nar/3.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H. G., Haynes M. E. Electron-microscope observations on cell nuclei in various tissues of a teleost fish: the nucleolus-associated monolayer of chromatin structural units. J Cell Sci. 1976 Jul;21(2):315–327. doi: 10.1242/jcs.21.2.315. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRATKY O. X-RAY SMALL ANGLE SCATTERING WITH SUBSTANCES OF BIOLOGICAL INTEREST IN DILUTED SOLUTIONS. Prog Biophys Mol Biol. 1963;13:105–173. doi: 10.1016/s0079-6107(63)80015-2. [DOI] [PubMed] [Google Scholar]
- Lewis E. A., DeBuysere M. S., Rees A. W. Configuration of unsheared nucleohistone. Effects of ionic strength and of histone F1 removal. Biochemistry. 1976 Jan 13;15(1):186–192. doi: 10.1021/bi00646a029. [DOI] [PubMed] [Google Scholar]
- Li H. J., Hu A. W., Maciewicz R. A., Cohen P., Santella R. M., Chang C. Structural transition in chromatin induced by ions in solution. Nucleic Acids Res. 1977 Nov;4(11):3839–3854. doi: 10.1093/nar/4.11.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees A. W., Debuysere M. S., Lewis E. A. Soluble nucleohistone of compact configuration. Biochim Biophys Acta. 1974 Aug 15;361(1):97–108. doi: 10.1016/0005-2787(74)90212-3. [DOI] [PubMed] [Google Scholar]
- Sperling L., Klug A. X-ray studies on "native" chromatin. J Mol Biol. 1977 May 15;112(2):253–263. doi: 10.1016/s0022-2836(77)80142-3. [DOI] [PubMed] [Google Scholar]
- Sperling L. The mass per unit length of chromatin by low-angle x-ray scattering. FEBS Lett. 1976 Apr 15;64(1):89–91. doi: 10.1016/0014-5793(76)80256-6. [DOI] [PubMed] [Google Scholar]
- Vengerov Y. Y., Popenko V. I. Changes in chromatin structure induced by EDTA treatment and partial removal of histone H1. Nucleic Acids Res. 1977 Sep;4(9):3017–3027. doi: 10.1093/nar/4.9.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Benyajati C. Higher order coiling of DNA in chromatin. Cell. 1977 Sep;12(1):83–100. doi: 10.1016/0092-8674(77)90187-8. [DOI] [PubMed] [Google Scholar]
- Zimm B. H., Crothers D. M. SIMPLIFIED ROTATING CYLINDER VISCOMETER FOR DNA. Proc Natl Acad Sci U S A. 1962 Jun;48(6):905–911. doi: 10.1073/pnas.48.6.905. [DOI] [PMC free article] [PubMed] [Google Scholar]


